Abstract

Hypercholesterolemia is a mediator for the etiology of cardiovascular diseases, which are characterized as the global leading cause of mortality. We aimed to investigate the inhibitory activity of Withania coagulans compounds against 3-hydroxy-3-methylglutaryl-coenzyme A reductase (Hmgcr) of Mus musculus using an extensive in silico approach. The 3D structure of the Hmgcr protein is not yet known, so we performed the homology modeling using MODELLER and SWISS-MODEL tools, followed with structural validation and assessment. The PROCHECK web server showed that the top-ranked homology model from SWISS-MODEL has 93.4% of residues in the most-favorable region, the quality factor was 98%, and the Verify3D score was 91.43%, compared to the other generated models. The druggable protein-binding cavities in a 3D model of Hmgcr were investigated with the aid of commonly prescribed statin compounds using the CB-dock approach. We compiled a 3D compound library of W. coagulans, followed by drug-likeness evaluation, and found 20 eligible compounds. The pattern of consensus residues obtained from the CB-dock procedure was then used for grid-box docking of W. coagulans compounds and statin drugs using AutoDock 4.2, respectively. The results showed that withanolide R (−10.77 kcal/mol), withanolide Q (−10.56 kcal/mol), withanolide J (−10.52 kcal/mol), atorvastatin (−8.99 kcal/mol), simvastatin (−8.66 kcal/mol), and rosuvastatin (−8.58 kcal/mol) were promising candidates that bind Hmgcr protein. The key residues involved in protein–ligand (withanolide R) interactions were Y516, C526, V529, I530, M533, I535, and V537, and the formation of a H-bond was at C526, M533, and I535 residues. M533 was the consensus residue having a tendency to form a H-bond with withanolide Q, too. Molecular dynamics simulations were used to validate the top-ranked docked complexes for the stability of the modeled protein. We also predicted the pharmacokinetic properties of binding affinity-based top-ranked compounds and concluded that they could be used as potential inhibitors of Hmgcr. However, further in vitro and in vivo studies are essential to completing the drug development process.
1. Introduction
Cholesterol is an organic sterol required by mammalian cells that serves as a structural component of the cell membrane, a substrate for cholecalciferol (vitamin D), bile acid, and a vital steroid for the biosynthesis of various essential hormones.1 The plasma insolubility of cholesterol necessitates its cellular transportation in spherical macromolecules known as lipoproteins.2 There are multiple reasons that trigger the abnormalities in cholesterol cellular homeostasis, which include genetic etiologies or increased food consumption that cause the disposal of this steroid in peripheral tissues.2,3 This steroid’s biochemical regulation is tightly regulated, and any change in cholesterol cellular homeostasis can lead to atherosclerosis and cardiovascular diseases (CVDs).2,3 Hypercholesterolemia remains one of the leading causes of death worldwide, so the principal mechanism of hypercholesterolemia associated with CVDs mainly results in atherosclerosis, and progression of such dysfunctionality of a blood vessel can lead to a blockage in blood flow, stroke, heart attack, peripheral arterial disease, ischemic heart disease, and others.2−4
To manage hypercholesterolemia and its associated consequential health outcomes, the protein target to inhibit is the main regulatory enzyme “3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase or Hmgcr”, which catalyzes the conversion of HMG-CoA to mevalonate as a rate-limiting catalytic step in the cholesterol biosynthesis pathway.5 The inhibition of HMG-CoA reductase lowers the expression of cholesterol synthesis in humans as well as in most animals by activating sterol regulatory element-binding protein (SREBP)-2, which upregulates this regulatory target and the low-density lipoprotein cholesterol (LDL-C) receptor, resulting in reduced cholesterol levels.6 Many nonclinical and human trials have proved that the risk of coronary complications could be minimized by halting the function of Hmgcr gene, a major cholesterol biosynthesis enzyme.7 Hypercholesterolemia can be treated with statins and other known drugs such as niacin’s and fibrates by inhibiting Hmgcr activity.8 Despite the fact that statins are widely used Hmgcr inhibitors, unfortunately their prolonged use are associated with some serious side effects such as hyperuricemia, nausea, diarrhea, gastric irritation, myositis, and abnormal liver function.9,10 These adverse impacts of synthetic pharmaceuticals have shifted the focus of researchers on plant-based alternatives to overcome these challenges. The isolation and use of medicinal compounds from plant sources to manage certain ailments in an accessible and cost-effective manner have represented the beneficial relationship of the plant environment and humans for decades.10
According to the World Health Organization, plant-based medicinal compounds are used by a large percentage of the world’s population for the primary treatment of various ailments.11,12 The medicinal plants are being researched, and the therapeutic characteristics of various isolated phytoconstituents with favorable nutritional value and pharmacological activity are being widely recorded.12 Various plants are utilized because of their prominent antioxidant, antithrombotic, anti-inflammatory, anti-atherogenic, and cardioprotective properties.11,12 Especially, the development and isolation of metformin, originally from natural product compounds found in the plant Galega officinalis, established the importance of medicinal phytochemicals.12
Since the last few decades, the whole plants, fruit, and roots of Withania coagulans Dunal (family: Solanaceae) have attracted attention due to their antidiabetic effects in animal models and human pilot trials.10,13−16 The fruit of W. coagulans contains a wide range of bioactive phytoconstituents with varying polarity, solubility, and chemical and physical characteristics. W. coagulans has been reported to have antifungal, emetic, antibacterial, anti-asthmatic, anti-inflammatory, diuretic, antimicrobial, anti-inflammatory, antitumor, hepato-protective, antihyperglycemic, antihyperlipidemic, anti-atherosclerotic, cardiovascular, immunosuppressive, free radical scavenging, and CNS depressant properties in the scientific literature.10−16 Interestingly, a number of various withanolides, including withaferin A, withanolide A, withanone, coagulin L, coagulanolide, ergostadiene, and sitosterol, were reported.10,13−16 According to phytochemical research, esterases, free amino acids, fatty oils, essential oils, and withanolides are the primary phytoconstituents of the fruit.13−16 The major phytoconstituents in W. coagulans fruit are withanolides, which are steroidal lactones with an ergostane skeleton.13−16 Therefore, the present study’s objective was to investigate and characterize the various compounds of W. coagulans as potential inhibitors of the HMG-CoA reductase enzyme of M. musculus through in silico analysis of their secondary metabolites.
2. Methodology
2.1. Gene’s Retrieval and Sequence Alignment
Protein-coding gene involved in lipid regulatory (regulation of total cholesterol) mechanisms in Mus musculus, that is, the Hmgcr gene, was retrieved from the UniProt (https://www.uniprot.org/) database. The key words used for the sequence retrieval were “M. musculus” and “lipid lowering enzyme”. The gene sequence was included and validated on the basis of the relevant records (annotation status), with information extracted from literature and curator-evaluated computational analysis. The gene record was also cross-validated with the NCBI accession number and gene identifier.17 Once the gene was selected, the FASTA format of the selected gene was also extracted from the NCBI databases. Protein BLAST (Basic Local Alignment Search Tool) was performed to generate alignments between the extracted protein sequence of M. musculus (as a “query”) and protein sequences within a database record (as “subject” sequences). The database chosen to search the respective set for the protein BLAST was the “Protein Data Bank” (PDB).17,18
2.2. Protein Modeling, 3D-Structural Validation, and Assessment
After the selection of the most potent protein-coding gene sequence, the 3D structure, active site identification, and ligand interaction information of proteins were retrieved through their reported PDB structure information database.18 However, in cases where no 3D-structural information or crystal structure was available for M. musculus, the main task was to perform ab initio homology modeling through the MODELLER programing method used for comparative modeling of protein 3D structures.19 It was done by considering template-based search through the NCBI’s pBLAST (protein–protein Basic Local Alignment Search Tool) server, and the MODELLER 3D model was evaluated.17 The overall strategy for this target-template modeling approach was based on searching for structures related to the Hmgcr gene, selecting the template, aligning the Hmgcr protein with the template, building the model, and then evaluating it.17,19
As a comparative and alternate modeling prediction strategy, we used the SWISS-MODEL tool to construct the 3D structure of a protein of interest from M. musculus.20 Threading or “ab initio” was also applied where the sequence similarity was low. UCSF Chimera was used to investigate the molecular characteristics and visualize the resulting protein structure interactively.21 DeepView/Swiss-PdbViewer was used for energy minimization of the obtained PDB structure of the Hmgcr gene.22 The model was validated through PROCHECK stereochemical assessment through an ERRAT quality factor, Ramachandran plot, Prosa-web (https://prosa.services.came.sbg.ac.at/prosa.php), and residual properties of the constructed model.23,24 The dihedral angles φ against ψ of possible conformations of amino acids in protein structure had also been studied in the Ramachandran plot. The SAVES (Structure Validation Server: https://saves.mbi.ucla.edu/) web server was used to know the probable structural errors and z-score.23,24 PDBsum was also used to investigate the protein’s secondary structural details.25
2.3. Structural Input of Lipid-Lowering Drugs and Identification of Druggable Protein-Binding Cavities
The 3D structures of the drugs used to inhibit the lipid-lowering rate regulatory enzyme were recruited as ligand molecules, and their 3D structures were downloaded from the PubChem compound library (https://pubchem.ncbi.nlm.nih.gov/).26 The identification of druggable protein-binding cavities was needed if the details of active site residues were not available. Therefore, to detect protein-binding cavities, the cavity binding blind docking (CB-dock) approach was used to generate consensus interaction cavities present in Hmgcr protein after docking with the lipid-lowering 3D drugs. Cavity-detection (CB)-guided blind docking investigated the homology model’s binding cavities using an automatic protein–ligand docking approach that identifies the binding cavities and sites. CB dock compared cavities and ranked them through a method called “CurPocket” with state-of-the-art protein–ligand binding site prediction methods using the benchmark set of COACH as prediction methods.27 With its novel curvature-based cavity detection approach, CB dock also calculates the center and size of the docking box of a putative cavity as a key parameter of the process. This method carefully optimizes and achieves a 70% success rate for the top-ranking poses whose root mean square deviation (rmsd) is within 2 Å from the X-ray pose.27
2.4. 3D Compound Library Compilation, Drug-Likeness Evaluation, and Energy Minimization from W. coagulans
Primarily, a 3D structure-based library comprising the compounds reported from W. coagulans was generated on the basis of an extensive literature search. The initial details of compounds of W. coagulans were taken from the literature.10,13−16 The 3D structures of selected compounds were downloaded from the PubChem compound database (https://pubchem.ncbi.nlm.nih.gov/).26 The structures were retrieved in SDF format originally and converted to PDB format using Open Babel software after visual inspection of each 3D compound.28
Drug-likeness evaluation was done through Pfizer’s rule of five, which was applied to all 3D compounds of W. coagulans.29 Following the assessment, only 3D derivatives of W. coagulans that passed all five physicochemical properties of Pfizer’s rule of five were chosen, as any single violation was considered an elimination aspect or treated as “fail”. In order to identify stable conformations, the process of energy minimization of compounds was performed to find a configuration space where all of the forces on the atoms are balanced. The drug-likeness properties were further computed and validated with the aid of OSIRIS Property Explorer (https://www.organic-chemistry.org/prog/peo/), which uses chemical structures for drug property prediction in terms of topological polar surface area (TPSA), c Log P calculation, log S calculation, molecular weight, fragment-based drug likeness, and drug score.30
2.5. Molecular Docking and Estimations of Inhibition Constant
AutoDock Vina software (version 4.2) was used to analyze the protein–drug interactions and estimate the molecular interactions of M. musculus protein with the selected compounds (ligand) molecules from lipid-lowering drugs as well as the screened compounds from W. coagulans.31 The cavity binding pattern of CB dock with the lowest free energy and rmsd value was chosen, and molecularly interacting targets were used as prominent residues in this cavity-guided auto docking approach.27 Polar hydrogen was added, and partial charges were assigned to the standard residue using the Gasteiger partial charge, which assumes all hydrogen atoms and was represented explicitly. The most-favorable binding interactions were estimated using the lowest predicted free energy of binding with the best molecular docking simulation pose.31 The inhibition constant (Ki) was obtained from the binding energy (ΔG) using the formula: Ki = exp(ΔG/RT), where R is the universal gas constant (1.985 × 10–3 kcal mol–1 K–1) and T is the temperature (298.15 K).
2.6. Visualization of Protein–Ligand Docked Complexes with Hmgcr Protein and the Top-Ranked Docked Compounds’ Toxicity Prediction
The interactions of the docked complexes were studied visually with the help of the Discovery Studio, and a 2D ligand–protein interaction plot was also drawn from this tool.32 The absorption, distribution, metabolism, excretion, and toxicity (ADMET) approach was used for in silico pharmacokinetic predictions of the selected compounds having higher binding affinities. The ADMET lab platform (http://admet.scbdd.com/home/index/) was used to access the ADMET properties.33 The assessment was carried out for each physiochemical property by submitting a canonical SMILE format of the individual compound.
2.7. MD Simulation
For the molecular dynamics (MD) simulation study, the Amber 20 package was used.34 The tleap module of Amber was used for the initial system setup. The protein–ligand complexes were immersed in an 8 Å TIP3P hydrated cubic box. For the neutralization of the residual charges in the system, counter ions were added. The MD systems underwent energy minimization, heating, density equilibration, and equilibration under periodic boundary conditions.35 The last production step of 50 ns as an NPT ensemble was completed at 310 K. After the 50 ns, MD simulation was successfully completed; rmsd, root mean square fluctuation (rmsf), and radius of gyration (Rog) studies were performed to measure the strength of the protein–ligand interaction.36
3. Results and Discussion
3.1. Gene Selection, Sequence Retrieval, and Alignment of Hmgcr Protein
Worldwide, CVDs are one of the leading causes for mortality.37 The underlying cause of this global health concern has been linked to elevated levels of total cholesterol, specifically LDL-C, as a major risk factor for acute myocardial infarction and other heart disease.2−4Hmgcr is the protein-coding gene in M. musculus linked to the rate-committing step for the biosynthesis of cholesterol and is related via a negative feedback mechanism.38 In this study, we investigate the cholesterol-lowering impact of the potential derivatives of W. coagulans in terms of the conversion of Hmgcr inhibition, one of the most significantly regulated enzymes in nature.39 This conversion in the cholesterol biosynthetic pathway is referred to as the main rate-limiting step in the field of drug discovery and development, as variations in the cholesterol synthesis follow changes in the enzyme’s activity and contribute to the management of various cholesterol-associated diseases.2−4 Therefore, investigating compounds that decrease blood cholesterol levels by blocking an enzyme required by the body to produce cholesterol is always considered a prolific outcome for the scientific community. To do so, the UniProt database was accessed using the key words “HMG-CoA reductase” and “M. musculus”, yielding a total of 17 gene sequences. From M. musculus, among the entire total, six of them were reviewed; the rest were unreview sequences, and filtering was used to select only reviewed sequences for the respective target search. After thorough assessment of each reviewed gene, we only selected the Hmgcr gene (UniProt accession number “Q01237”) with 887 amino acid residues and a mass of 97,040 Da. The same sequence was further validated from the NCBI protein database, which has accession ID “Q01237.3”, gene ID “15357”, and GI: 408360266.
The protein–protein BLAST (blastp: Basic Local Alignment Search Tool) algorithm was used to compare a protein query to a protein databank PDB. Overall, six sequences showed significant alignments, the top two being from Homo sapiens, with a percent identity of around 94% (E-value = 0) and template accession PDB-IDs of “IDQ8-A” and “2Q1L-A”. Afterward, 3D structures of aligned templates were downloaded from the PDB database Table 1. This regulatory protein has been shown to be active in the endoplasmic reticulum as well as proximal membranes, which catalyze the transformation of HMG-CoA to mevalonate, and is the orthologous gene for human HMG-CoA reductase.40,41 Therefore, opting for this protein has a clear rationale of relevance in terms of gene orthologs (which evolved in various species from a common and mutually ancestral gene) retaining a similar function throughout evolution.38,41
Table 1. Sequence Alignment (BLASTp: Protein–Protein Basic Local Alignment Search Tool) of Hmgcr Protein of M. musculus.
| S. No | PDB accession ID | Chain | Scientific name | Maximum score | Total score | Query cover (%) | E value | Identity (%) | Accession length |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1DQ8 | A | H. sapiens | 906 | 906 | 52 | 0 | 93.94 | 467 |
| 2 | 2Q1L | A | H. sapiens | 866 | 866 | 49 | 0 | 94.94 | 441 |
| 3 | 6HR7 | A | Methanothermococcus thermolithotrophicus DSM 2095 | 307 | 307 | 45 | 1.00 × 10–95 | 41.4 | 427 |
| 4 | 6HR8 | A | M. thermolithotrophicus DSM 2095 | 303 | 303 | 45 | 4.00 × 10–94 | 41.16 | 427 |
| 5 | 5WPJ | A | Streptococcus pneumoniae | 53.1 | 53.1 | 39 | 1.00 × 10–6 | 23.84 | 426 |
| 6 | 3QAE | A | S. pneumoniae R6 | 50.4 | 50.4 | 39 | 8.00 × 10–6 | 23.56 | 458 |
3.2. MODELLER-Based Structural Modeling and Validation of Hmgcr Gene
Computational 3D structure prediction through different homology modeling techniques has been explicitly used to establish a protein structure with 3D specification through available residue sequences for helpful purposes in molecular biology, such as protein hypotheses about drug design, predicting ligand binding cavities or sites, specifying a substrate, and gene functional annotation.42,45 It is considered to be the most accurate of the computational structure prediction methods.18,19 Once the target protein-template alignment is constructed, the MODELLER algorithm-based modeling approach was used to build a 3D model for the Hmgcr gene of M. musculus. MODELLER generates 3D models of the target sequence, with the best model selected based on the lowest value for the objective function.19 The model built with the lowest value of the MODELLER objective function, that is, the lowest DOPE assessment scores and/or the highest GA341 assessment score, was selected as the best model. Model 1 was chosen due to its DOPE score of −65241.76172, molpdf objective function of 8205.12988, and GA341 score of 1.000 (Supporting Information, Table S1).
Information regarding the stereochemically predicted protein’s 3D structure is a basic requirement for understanding the structural reliability of a protein’s function.23 Therefore, we performed the structural assessment of the top-ranked model, which showed 611 (77.8%) residues were in the most-favorable region, 126 (16.1%) residues were in additionally allowed regions, 25 (3.2%) residues were in the generously allowed region, and 23 (2.9%) residues were in the disallowed region. The maximum deviation was 19.2, the number of bad contacts was 69, and the bond length/angle was 14.5. According to the G-factor analyses, the dihedral score was −0.26, the covalent score was −0.71, and the overall score was −0.41. With an overall quality factor of 24.321, a planar group of the 3D model revealed that 99.6% were within the limit and 0.4% were highlighted (Supporting Information, Figure S1). Hence, the role of the predicted model for structural validation in terms of its accuracy serves as a critical factor for its further comprehensive molecular mechanistic investigations, like molecular docking or drug designing, as well as the functional prediction of the 3D protein model.43,45 Due to the lowest quality factor score and the presence of several residues in the disallowed region of the Ramachandran plot in all constructed 3D structures built by MODELLER, we used an alternative model prediction strategy called “SWISS-MODEL”-based prediction for better model prediction and structural accuracy.20
3.3. Alternate Structural Modeling, Validation, and Identification of Druggable Binding Cavities of Hmgcr
Accurate prediction of protein 3D structure is considered a significant matter in structural biology because a good resolution-based 3D protein model produced through computational methods can considerably reduce the time, labor, and cost of differential experimentation techniques.44,45 For consequential homology prediction and structural validation, we used an alternate homology modeling approach through the SWISS-MODEL, which is a fully automated protein structure modeling server.20 The target-template search strategy resulted in an overall set of 50 templates that correspond with our Hmgcr query protein sequence which were further validated and assessed for their utilization in this study (Supporting Information, Table S2). Only the top three build models with the highest percent template identity were chosen for further processing. We build a model with a template having PDB ID “2R4F-A” (substituted pyrazoles as hepatoselective HMG-CoA reductase inhibitors; Table 2). In the absence of experimentally determined protein 3D structures, homology modeling plays a cost-effective role in structure-based applications and the characterization of protein properties and functions.46 Therefore, an accurate 3D model of a protein has been constructed by assigning a conformation, which is typically based on an accurate sequence alignment approach followed by model generation and relative energy minimization of atoms.44−46
Table 2. Template Search and 3D Homology Model Generation of Hmgcr Protein of M. musculus through SWISS-MODEL.
| S. No | PDB accession ID | Scientific name | Chain | GMQE | QSQE | Identity (%) | Method | Resolution (Å) | Oligo state |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2R4F | Substituted pyrazoles as hepatoselective HMG-CoA reductase inhibitors | A | 0.5 | 0.56 | 94.94 | X-ray | 1.7 | Homotetramer |
| 2 | 3CD0 | Thermodynamic and structure-guided design of statin HMG-CoA reductase inhibitors | C | 0.5 | 0.58 | 94.94 | X-ray | 2.4 | Homotetramer |
| 3 | 1HW8 | Complex of the catalytic portion of human HMG-CoA reductase with compactin (also known as mevastatin) | B | 0.5 | 0.55 | 93.94 | X-ray | 2.1 | Homotetramer |
| 4 | 1HW8 | Complex of the catalytic portion of human HMG-CoA reductase with compactin (also known as mevastatin) | B | 0.49 | 0.55 | 94.48 | X-ray | 2.1 | Homotetramer |
| 5 | 1HWK | Complex of the catalytic portion of human HMG-CoA reductase with atorvastatin | A | 0.49 | 0.56 | 93.94 | X-ray | 2.2 | Homotetramer |
Once the 3D structure of a protein is modeled, it is always necessary to check the overall quality factor.44,47 In this manner, the quality factor of our top-ranked homology model was 97.28%, and the Verify3D score was 88.33% of the residues, which depicted an average 3D-1D score of >0.2. According to Ramachandran plot analysis, 93.4% of residues were in the most-favored region, 6.1% were in the additionally allowed region, and 0.6% were in the generously allowed region. No residues were in the disallowed region. The maximum deviation was 3.7, no bad contact was investigated, and the bond length/angle was 4.8. According to the G-factor analyses, the dihedral score was 0.08, the covalent score was 0.15, and the overall score was 0.02. A planar group of the 3D model showed that 96.7% were within the limit, and 3.3% is highlighted in Figure 1. Therefore, for further analysis, the estimation of the 3D model’s accuracy (validation and assessment) was a necessary step, as without undergoing the quality factor assessment and other essential analyses, we were unable to determine the significance of our generated 3D homology model for further computational work.47
Figure 1.
Detailed structural validation and assessment of 3D homology model of Hmgcr protein of M. musculus.
Besides this, energy minimization is a vital step for determining the appropriate molecular organization of atoms (in space), with the objective of establishing the lowest energy conformation of a molecule.44,47 We anticipated that due to the existence of any non-energetically favorable coordinates in the natively drawn 3D structure, the target was to perform energy minimization on the finalized top-ranked 3D model of the Hmgcr protein.47 After the energy minimization of Hmgcr protein, the final exhibited energy was −22102.396 kJ/mol, bonds were 430.65, angles were 1391.479, torsion was 1788.981, and electrostatic energy was −12233.70. The energy minimization of the homology model raised the Verify3D score to 91.43% and the overall quality factor to 98.23%, and the properties of the Ramachandran plot remain the same.
In general, statins, the first-line drugs that reduce cholesterol production, are the most commonly prescribed class of medications for the treatment and prevention of CVDs and other lipid disorders.48 In vitro studies revealed statins’ role by competitively inhibiting the rate-limiting enzyme in cholesterol biosynthesis, HMG-CoA reductase.48,49 Following that, the 3D structures of the lipid-lowering drugs atorvastatin (CID-60823), cerivastatin (CID-446156), fluvastatin (CID-446155), mevastatin (CID-64715), rosuvastatin (CID-446157), and simvastatin (CID-54454) were downloaded, visually inspected, and followed by a cavity-guided docking to identify the potential druggable pockets in the Hmgcr protein of M. musculus. Among the cavity binding energies estimated due to the interactions of various cholesterol-lowering therapeutic agents with the homology model of Hmgcr, the highest cavity grading energetic score was found in simvastatin (−7.5 kcal/mol), mevastatin, and atorvastatin (−7.4 kcal/mol), while cerivastatin had the lowest rank (−6.5 kcal/mol) among all (Table 3). Consensus-binding residues took into account the identification of amino acids with the most common appearance, the lowest binding scores, and the smallest rmsd range (Table 3). When the binding site information (usually in novel proteins) is lacking or not available, either the most likely binding sites are algorithmically predicted or a “blind docking” simulation is run to detect the possible druggable protein-binding site.27 Detection of potential novel binding cavities or residues on a particular protein’s surface (structure) and ranking them according to the ligand’s binding affinity (ligandability) will help to understand the role of proteins in typical biological or cellular processes, which are regulated through particular molecular interaction(s) or recognition(s) according to the available protein surfaces.27,43
Table 3. Identification of Druggable Protein Cavities and Amino Acids Interactions of Hmgcr Protein with Standardized Drugs Used to Manage Hypercholesterolemia.
| Drugs | Binding affinity (kcal/mol) | Amino acid interactions | Hydrogen-bond interactions |
|---|---|---|---|
| Atorvastatin | –7.4 | TYR516, VAL521, CYS526, VAL529, MET533, ILE535, ALA555, ALA762, ALA767, PRO812, GLN813 | MET533 |
| Cerivastatin | –6.5 | VAL537, ILE761, ALA762, PRO812, GLN813 | ILE761, GLN813 |
| Fluvastatin | –7.3 | TYR516, VAL521, CYS526, VAL529, ILE535, ILE761, PRO812 | ILE535 |
| Mevastatin | –7.4 | TYR516, TYR532, MET533, ILE535, PRO812 | TYR516, MET533, ILE535 |
| Rosuvastatin | –7.1 | TYR516, ILE535, ILE761 | TYR516, ILE535, ILE761 |
| Simvastatin | –7.5 | TYR516, TYR532, MET533, ILE535, VAL537, PRO812 | TYR516, MET533, ILE535 |
3.4. 3D Compounds of W. coagulans and Drug-Likeness Evaluation
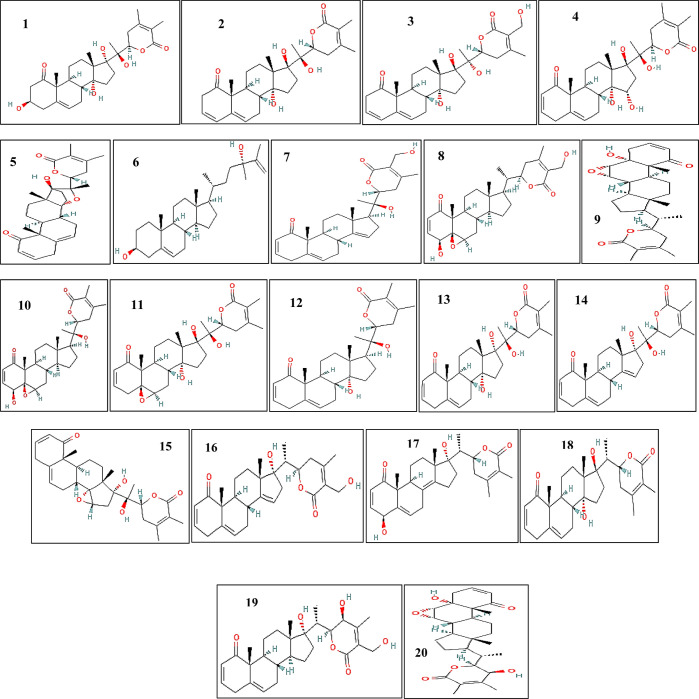
Emerging challenges to human health like managing CVD risk necessitate a coordinated effort to find both preventive and therapeutic approaches, placing natural products at the forefront of initiatives to develop novel treatments and lower disease transmission and associated mortality.37 The therapeutic efficacy of plant-based chemicals has long been recognized, leading to their dietary use as well as their being clinically recommended for the treatment and management of a wide range of illnesses.14,39 Despite the global advancements, many pathological conditions are still treated with plant-based medicines, or they are utilized as an alternative to modern pharmaceuticals.11 Plant-based derivatives or compounds will always be considered therapeutic agents in the management of various ailments because of their lesser side effects and easy accessibility.12 A total of 27 relevant compounds were downloaded in SDF format and converted into PDB file format. In order to explore pharmacokinetic and pharmacological qualities of medicinal compounds such as solubility, chemical stability, bioavailability, and distribution profile, we used the idea of drug-likeness assessment.29 Therefore, Pfizer’s rule of five was applied to all compounds, with 20 passing the drug-likeness criteria and the remaining 7 being excluded due to deviation (Supporting Information, Table S3; Figure 2). Drug-likeness evaluates qualitatively whether a molecule has the potential of becoming an oral drug in terms of bioavailability.30 The structure or physicochemical analysis of developing compounds regarded as oral drug candidates allowed for the determination of a compound’s drug likeness.30,50 The compounds that were excluded were 20 hydroxy-1-oxo-(22R)-witha-2,5,24-trienolide, coagulin D, coagulin L, withanolide S, withanolide C, withanolide E (6-alpha-hydroxy), and sitogluside. Afterward, the process of energy minimization was performed on the rest of the 20 compounds. The 3D structures of these 20 compounds were further assessed and validated with the aid of OSIRIS Property Explorer, as drug-like qualities including permeability, solubility, metabolic stability, and transporter actions are crucial for a drug candidate’s success in the future.30 They will have an impact on in vitro pharmacology as well as oral bioavailability, metabolism, clearance, and toxicity.50 In this manner, with the widespread acceptance of the concept of in silico toxicity risk assessment of the proposed compounds, we also performed the compounds’ (risk) assessment in terms of mutagenicity, tumorigenicity, irritating effects, reproductive effects, and drug scores, which, in general, exhibited the safest nature of all selected and screened compounds (Table 4).
Figure 2.
2D structures of W. coagulans compounds. 1: 3β hydroxyl-2,3-dihydrowithanolide F, 2: 17β hydroxywithanolide K, 3: ajugin E, 4: coagulanolide, 5: coagulin C, 6: ergosta 5,25-diene 3β,24-diol, 7: withacoagulin, 8: withaferin A, 9: withanolide B, 10: withanolide D, 11: withanolide E, 12: withanolide G, 13: withanolide J, 14: withanolide L, 15: withanolide M, 16: withanolide N, 17: withanolide O, 18: withanolide P, 19: withanolide Q, and 20: withanolide R.
Table 4. Drug-Likeness Assessment of W. coagulans Compoundsa.
| Compound | Mutagenicity scorea | Tumorigenicity scorea | Irritating effect scorea | Reproductive effect scorea | log S | c Log P | Solubility | TPSA | Drug likeness | Drug score |
|---|---|---|---|---|---|---|---|---|---|---|
| 3β hydroxyl-2,3-dihydrowithanolide F | 1 | 1 | 1 | 0.8 | 0.739 | 2.21 | –3.96 | 124.2 | 1.31 | 0.46 |
| 17β hydroxywithanolide K | 1 | 1 | 1 | 0.8 | 0.704 | 2.78 | –4.13 | 104 | –0.63 | 0.35 |
| Ajugin E | 1 | 1 | 1 | 0.8 | 0.798 | 1.86 | –3.62 | 124.2 | 1.65 | 0.5 |
| Coagulanolide | 1 | 1 | 1 | 0.8 | 0.78 | 1.93 | –3.73 | 124.2 | –0.08 | 0.4 |
| Coagulin C | 1 | 1 | 1 | 0.8 | 0.596 | 3.31 | –4.61 | 72.83 | –0.59 | 0.33 |
| Ergosta 5,25-diene 3β,24-diol | 1 | 1 | 1 | 1 | 0.299 | 6.74 | –5.85 | 40.46 | –12.72 | 0.16 |
| Withacoagulin | 1 | 1 | 1 | 1 | 0.677 | 3.4 | –4.26 | 83.83 | 1.24 | 0.56 |
| Withaferin A | 1 | 1 | 1 | 0.8 | 0.629 | 2.49 | –4.47 | 96.36 | 1.69 | 0.46 |
| Withanolide B | 1 | 0.8 | 0.8 | 0.8 | 0.505 | 3.42 | –4.98 | 76.13 | –1.04 | 0.18 |
| Withanolide D | 1 | 1 | 1 | 0.8 | 0.616 | 2.56 | –4.53 | 96.36 | 0.14 | 0.38 |
| Withanolide E | 1 | 1 | 1 | 0.8 | 0.726 | 1.77 | –4.03 | 116.5 | –0.41 | 0.37 |
| Withanolide G | 1 | 1 | 1 | 1 | 0.603 | 3.61 | –4.58 | 83.83 | –0.34 | 0.42 |
| Withanolide J | 1 | 1 | 1 | 0.8 | 0.704 | 2.78 | –4.13 | 104 | –0.2 | 0.37 |
| Withanolide L | 1 | 1 | 1 | 1 | 0.664 | 3.5 | –4.32 | 83.83 | –0.29 | 0.44 |
| Withanolide M | 1 | 1 | 1 | 0.8 | 0.687 | 2.49 | –4.21 | 96.36 | –0.21 | 0.37 |
| Withanolide N | 1 | 1 | 1 | 1 | 0.677 | 3.43 | –4.26 | 83.83 | 1.69 | 0.58 |
| Withanolide O | 1 | 1 | 1 | 1 | 0.661 | 3.68 | –4.33 | 83.83 | 0.85 | 0.52 |
| Withanolide P | 1 | 1 | 1 | 0.8 | 0.603 | 3.64 | –4.58 | 83.83 | –0.49 | 0.32 |
| Withanolide Q | 0.8 | 1 | 1 | 0.8 | 0.706 | 2.69 | –4.12 | 104 | 2 | 0.39 |
| Withanolide R | 1 | 0.8 | 0.8 | 0.8 | 0.604 | 2.57 | –4.58 | 96.36 | –0.88 | 0.2 |
No risk = 1 and moderate risk = 0.8.
3.5. In Silico Molecular Docking Experimentation and Estimations of Inhibition Constant
Molecular docking is mainly used to execute structure-based virtual screening and to aid in rationalizing ligand activity toward an interesting protein target.51 Docking experimentation-based energetic calculations commonly employ a grid representation strategy at the protein level, which includes pre-calculated charge force fields or potential energies required for drug–target interaction within the identified binding site specification.52 This grid representation approach, which is interestingly reliable than blind docking experimentation (binding mode runs), generates the significant conformation representing the optimal target’s binding cavity or sites with relevant potential energies.52 After obtaining the consensus-binding residues of our protein of interest, we used the AutoDock Vina tool to benchmark the grid box based on the target’s binding site information.31 The ligands we optimized for docking experiments were particularly explored for their accurate binding affinity, experimental binding pose, and active–inactive interactions with the Hmgcr protein of M. musculus.31 Captivatingly, the molecular docking of all 6 (cholesterol-lowering) drugs and 20 screened compounds with a 3D homology model of the Hmgcr protein of M. musculus showed significantly lower binding energies (Tables 5 and 6). Surprisingly, in comparison to all six (cholesterol-lowering) standardized drugs, we found higher binding affinities (ΔG) with very low inhibition constants (Ki) in all screened W. coagulans compounds, as shown in Table 6. Besides this, withanolide R (−10.77 kcal/mol; 12.68 nM), withanolide Q (−10.56 kcal/mol; 18.08 nM), and withanolide J (−10.52 kcal/mol; 19.29 nM) exhibited higher binding affinities and the lowest Ki with Hmgcr protein, compared to other screened compounds (Table 6 and Figure 3).
Table 5. Grid-Box (Residues Specific)-Based Docking Experimentation of Hmgcr Protein of M. musculus with Standardized Cholesterol-Lowering Drugsa.
| Drugs | ΔGbind (kcal/mol) | Inhibition constant (Ki) | ΔEvdw | ΔEele | ΔEMM | ΔE(unbound) | ΔE(torsional) | ΔE(total Internal) | ΔE(intermolecular) |
|---|---|---|---|---|---|---|---|---|---|
| Atorvastatin | –8.99 | 256.29 nM | –12.53 | –0.3 | –12.83 | –3.44 | 3.84 | –3.44 | –12.83 |
| Cerivastatin | –6.91 | 8.66 μM | –10.38 | –0.1 | –10.48 | –2.82 | 3.57 | –2.82 | –10.47 |
| Fluvastatin | –7.24 | 4.96 μM | –9.83 | –0.15 | –9.98 | –2.06 | 2.74 | –2.06 | –9.98 |
| Mevastatin | –8.56 | 531.02 nM | –10.71 | –0.04 | –10.75 | –2.02 | 2.2 | –2.02 | –10.76 |
| Rosuvastatin | –8.58 | 515.63 nM | –11.79 | –0.08 | –11.87 | –1.97 | 3.29 | –1.97 | –11.87 |
| Simvastatin | –8.66 | 450.92 nM | –10.78 | –0.07 | –1.99 | –1.99 | 2.2 | –1.99 | –10.85 |
ΔEvdw = van der Waals energy; ΔEele = electrostatic energy; ΔEMM = molecular mechanics.
Table 6. Grid-Box (Residues Specific)-Based Docking Experimentation of Hmgcr Protein of M. musculus with W. coagulans Compoundsa.
| Compounds | ΔGbind (kcal/mol) | Ligand efficiency | Inhibition constant (Ki) nM | ΔEvdw | ΔEele | ΔEMM | ΔE(unbound) | ΔE(torsional) | ΔE(total internal) | ΔE(intermolecular) |
|---|---|---|---|---|---|---|---|---|---|---|
| 3β hydroxyl-2,3-dihydrowithanolide F | –9.87 | –0.28 | 58.48 | –11.36 | –0.16 | –11.52 | –2.32 | 1.65 | –2.32 | –11.51 |
| Withanolide D | –10.29 | –0.3 | 28.7 | –11.24 | –0.15 | –11.39 | –1.35 | 1.1 | –1.35 | –11.39 |
| Withanolide L | –10 | –0.3 | 46.65 | –11.04 | –0.06 | –11.1 | –1.23 | 1.1 | –1.23 | –11.1 |
| Withaferin A | –10.32 | –0.3 | 27.28 | –11.6 | –0.09 | –11.69 | –1.49 | 1.37 | –1.49 | –11.69 |
| Withanolide E | –10.13 | –0.29 | 37.49 | –11.29 | –0.21 | –11.5 | –1.84 | 1.37 | –1.84 | –11.5 |
| Withacoagulin | –9.78 | –0.3 | 67.83 | –11.19 | 0.04 | –11.15 | –1.5 | 1.37 | –1.5 | –11.15 |
| Withanolide B | –10.51 | –0.32 | 19.86 | –11.24 | –0.09 | –11.33 | –1.22 | 0.82 | –1.22 | –11.33 |
| Withanolide J | –10.52 | –0.31 | 19.29 | –11.77 | –0.12 | –11.89 | –2.15 | 1.37 | –2.15 | –11.9 |
| Withanolide G | –10.36 | –0.31 | 25.56 | –11.36 | –0.09 | –11.45 | –1.14 | 1.1 | –1.14 | –11.46 |
| Withanolide P | –10.17 | –0.31 | 35.16 | –11.17 | –0.1 | –11.27 | –0.86 | 1.1 | –0.86 | –11.27 |
| Withanolide O | –10.44 | –0.32 | 22.43 | –11.44 | –0.09 | –11.53 | –0.43 | 1.1 | –0.43 | –11.53 |
| Withanolide N | –10.37 | –0.31 | 24.96 | –11.6 | –0.14 | –11.74 | –0.75 | 1.37 | –0.75 | –11.74 |
| Withanolide M | –9.63 | –0.28 | 87.67 | –10.69 | –0.04 | –10.73 | –2.22 | 1.1 | –2.22 | –10.73 |
| Coagulin C | –9.74 | –0.3 | 72.86 | –10.28 | –0.01 | –10.29 | –0.84 | 0.55 | –0.84 | –10.29 |
| Coagulanolide | –9.26 | –0.26 | 161.75 | –10.8 | –0.12 | –10.92 | –2.68 | 1.65 | –2.68 | –10.91 |
| 17β hydroxywithanolide K | –10.02 | –0.29 | 45.14 | –11.23 | –0.16 | –11.39 | –2.55 | 1.37 | –2.55 | –11.39 |
| Ajugin E | –10.04 | –0.29 | 43.37 | –11.84 | –0.13 | –11.97 | –2.13 | 1.92 | –2.13 | –11.97 |
| Withanolide R | –10.77 | –0.32 | 12.68 | –11.77 | –0.1 | –11.87 | –1.18 | 1.1 | –1.18 | –11.87 |
| Withanolide Q | –10.56 | –0.31 | 18.08 | –12.15 | –0.06 | –12.21 | –1.64 | 1.65 | –1.64 | –12.21 |
| Ergosta 5,25-diene 3β,24-diol | –9.29 | –0.31 | 155.65 | –11.15 | –0.06 | –11.21 | –0.15 | 1.92 | –0.15 | –11.21 |
ΔEvdw = van der Waals energy; ΔEele = electrostatic energy; ΔEMM = molecular mechanics.
Figure 3.
(A) Protein–ligand complexes showing the molecular interactions of the Hmgcr protein of M. musculus docked with the top-ranked three higher binding affinities compounds of W. coagulans. (B) 2D Ligand plot presenting the molecular interactions of the Hmgcr protein of M. musculus docked with the top-ranked three higher binding affinities compounds of W. coagulans. Color scheme: green color = conventional H-bonds; purple color = alkyl and π-alkyl bonds; red color = unfavorable acceptor–acceptor.
Due to statins’ competitive inhibition of HMG-CoA reductase, the liver produces less cholesterol.49 SREBPs are transported from the endoplasmic reticulum to the Golgi bodies, where they are broken down by proteases into active transcription factors as a result of the suppression of hepatic cholesterol production.40 The low-density lipoprotein (LDL) receptor and, most significantly, the HMG-CoA reductase are two of the genes that are expressed more when the SREBPs go to the nucleus.4,6,8,40 When HMG-CoA reductase is expressed more, hepatic cholesterol synthesis returns to normal, and when LDL receptor is expressed more, there are more LDL receptors on the plasma membrane of hepatocytes, which speeds up the clearance of LDL and very LDL (VLDL), which include apolipoprotein B and E.2−4,8 The decrease in plasma LDL-C and triglyceride levels is due to the enhanced clearance of LDL and VLDL.2−4,8 Statin therapy is ineffective in lowering LDL-C levels in patients with homozygous familial hypercholesterolemia, who lack all LDL receptors.48,49
ΔG (an independent variable) and Ki (a dependent variable) were used in the linear regression for both standard drugs and W. coagulans compounds. For Hmgcr protein interactions with W. coagulans compounds, the value of R2 equals 0.881, which means that 88.1% of the variability of Ki is explained by ΔG. The correlation (R) was equal to 0.9386, which indicated that there is a very strong direct relationship between ΔG and KiFigure 4A. When we compared the output of Hmgcr protein interactions with standardized cholesterol-lowering drugs, we found that R2 equals 0.6738, indicating that 67.4% of the variability of Ki is explained by ΔG). The correlation R equals −0.8209, which represents that there is a very strong inverse relationship between ΔG and KiFigure 4B. Statins are medications that lower serum lipid levels and thus lower the risk of CVDs and death. Researchers reported that the risk–benefit ratio of treating patients with statins is very favorable and has resulted in this class of drugs being widely utilized, so natural product compounds from W. coagulans have promising HMG-CoA reductase inhibitory activity compared to statins.48,48 This activity (in M. musculus) suggested that the compounds we selected from W. coagulans (withanolide R, J, and Q) are quite effective in decreasing cholesterol status through inhibiting the Hmgcr protein and will be useful in the management of CVDs.
Figure 4.
Linear regression analyses of docking score (ΔG—an independent variable and Ki—a dependent variable) computed with 3D homology model of Hmgcr protein of M. musculus docked with standardized cholesterol-lowering drugs (A) and compounds of W. coagulans (B).
3.6. ADMET Screening and MD Simulation Analysis
The overall in silico pharmacokinetics predictions of the three top-ranked (on the basis of binding affinity with Hmgcr protein) W. coagulans compounds exhibited the safest therapeutic properties with regard to the basic physicochemical characteristics of absorption, distribution, metabolism, excretion, and toxicity indices (Supporting Information, Table S4).
Persistent hypercholesterolemia (with elevated LDL-C and triglyceride levels) is the preliminary cause of atherosclerosis, including heart illnesses such as diabetic dyslipidemia and myocardial infarction.37 As a result of hypercholesterolemia, LDL-C accumulates pathologically in blood vessels, forming atherosclerotic plaques that are strongly associated with the emergence of CVDs.2−4,8 According to the oxidized LDL-C hypothesis, LDL-C oxidized by endothelial cells, macrophages, and smooth muscle cells of the artery wall plays a crucial role in the development of atherosclerosis.4,8 The molecular basis for understanding its role in atherogenesis begins with endothelial dysfunction that causes the formation of a lipid layer or fatty streak within the intima and the migration of leukocytes and smooth muscle cells into the vessel wall. This leads to the formation of foam cells and is consequently linked with the degradation of the extracellular matrix.2−4,8 In relation to this mechanism, the top-ranked docked compounds we investigated were withanolide R, withanolide J, and withanolide Q, which demonstrated a promising inhibitory impact in inhibiting the cholesterol’s biosynthesis linked regulatory enzyme, thus exhibiting an anti-atherogenic impact to manage CVD risk, similar to statins.48,49
The further authentication of our top-ranked docked complex (withanolide R) was done through the MD simulation strategy, which is based on a general model of the physics governing interatomic interactions.53 MD simulations predict how each atom in a protein or other molecular system will move over time, and protein–ligand complexes are found to be stable at least during the relevant time period (nanoseconds, ns).53 The rmsd as a function of time was calculated in this study to assess the stability of a biological complex (Figure 5). The rmsds of withanolide R and atorvastatin were compared and plotted in Figure 5A; withanolide R was more stable than atorvastatin. The rmsd of withanolide R revealed greater variations during 10–30 ns, but after 30 ns, the system gained stability until 50 ns. In contrast, atorvastatin initially showed large rmsd perturbations, especially between 20 and 25 ns. After that, the rmsd started to decline and remained stable until 50 ns. Residue flexibility indexing is a critical evaluation for understanding each residue’s involvement in various biological processes. According to Figure 5B, both the complexes showed a more consistent pattern of rmsf, with the exception of the regions between 25 and 75 for each complex Figure 5B. This region (25–75) of the protein’s flexibility displayed a distinct pattern, demonstrating the effect of specific residues on the protein’s internal dynamics.
Figure 5.
(A) rmsd plots of withanolide R (black color) and atorvastatin (red color). The time in nanoseconds is shown on the X-axis. (B) rmsf plot of withanolide R (black color) and atorvastatin (red color). The total number of residues is shown on the X-axis. (C) RoG of the withanolide R (black color) and atorvastatin (red color). The total number of frames is shown on the X-axis.
The radius of gyration (Rg) was determined as a function of time to comprehend the structural compactness. The withanolide R complex initially showed a high Rg value of up to 27 Å, but during the last frames, the complex showed a homogeneous pattern of Rg and the average Rg was observed as 25–25.5 Å throughout the simulation. In the case of atorvastatin, Rg slightly increased up to 26.5 A between 500 and 1500 frames, but it subsequently steadied once again until the last 2500 frames, and the average Rg was determined to be 25.4 as in Figure 5C.
4. Conclusions
This in silico study clearly demonstrated that the selected screened compounds of W. coagulans showed promising inhibitory activity against the Hmgcr protein of M. musculus deciphering their substantial propensity as HMG-CoA reductase inhibitors. This work also necessitates further study, that is, in vitro and in vivo trials on the same compounds, to support our findings and better understand the present inferences. The current study also suggested protein targets for a new multi-ligand approach to predict the efficacy of medicinal compounds from W. coagulans against various forms of hypercholesterolemia.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c07893.
3D model evaluation predictors for Hmgcr protein of M. musculus generated by MODELLER; stereochemical analysis of the top-ranked model of Hmgcr protein generated by SWISS-MODEL through PROCHECK webserver and SWISS-MODEL assessment indicators; details of W. coagulans compounds selected from PubChem database; in silico predicted pharmacokinetics profile of the three top (binding affinities based)-ranked compounds; and Ramachandran plot of the top rank 3D model generated through MODELLER of Hmgcr protein of M. musculus (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Brown A. J.; Coates H. W.; Sharpe L. J.. Cholesterol synthesis. Biochemistry of Lipids, Lipoproteins and Membranes; Elsevier, 2021; pp 317–355. [Google Scholar]
- Álvarez-López H.; Díaz-Aragón A.; Ruiz-Gastélum E. Knowing the basic mechanisms of lipid metabolism. Cardiovasc. Metab. Sci. 2021, 32, 147–152. 10.35366/100786. [DOI] [Google Scholar]
- Chandel N. S. Lipid metabolism. Cold Spring Harbor Perspect. Biol. 2021, 13, a040576. 10.1101/cshperspect.a040576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. H.; Garruti G.; Liu M.; Portincasa P.; Wang D. Q. Cholesterol and lipoprotein metabolism and atherosclerosis: recent advances in reverse cholesterol transport. Ann. Hepatol. 2017, 16, S27–S42. 10.5604/01.3001.0010.5495. [DOI] [PubMed] [Google Scholar]
- Feingold K. R. Maximizing the benefits of cholesterol-lowering drugs. Curr. Opin. Lipidol. 2019, 30, 388–394. 10.1097/mol.0000000000000631. [DOI] [PubMed] [Google Scholar]
- Xue L.; Qi H.; Zhang H.; Ding L.; Huang Q.; Zhao D.; Wu B. J.; Li X. Targeting SREBP-2-regulated mevalonate metabolism for cancer therapy. Front. Oncol. 2020, 10, 1510–1530. 10.3389/fonc.2020.01510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K.; Tam S.; Murray-Segal L.; Huynh K.; Meikle P. J.; Scott J. W.; Denderen B.; Chen Z.; Steel R.; LeBlond N. D.; Burkovsky L. A.; O’Dwyer C.; Nunes J. R. C.; Steinberg G. R.; Fullerton M. D.; Galic S.; Kemp B. E. Inhibition of Adenosine Monophosphate–Activated Protein Kinase–3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase Signaling Leads to Hypercholesterolemia and Promotes Hepatic Steatosis and Insulin Resistance. Hepatol. Commun. 2019, 3, 84–98. 10.1002/hep4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S.; Sun W.; Gao L.; Liu S. Therapeutic targets of hypercholesterolemia: HMGCR and LDLR. Diabetes, Metab. Syndr. Obes.: Targets Ther. 2019, 12, 1543–1553. 10.2147/dmso.s219013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreutzkamp B. Side effects of HMG-CoA Reductase inhibitors in placebo-controlled trials. Med. Monatsschr. Pharm. 2015, 38, 73–74. [PubMed] [Google Scholar]
- Lateef T.; Naeem S.; Qureshi S. A. In-silico studies of HMG-Co A reductase inhibitors present in fruits of Withania coagulans Dunal (Solanaceae). Trop. J. Pharm. Res. 2020, 19, 305–312. 10.4314/tjpr.v19i2.13. [DOI] [Google Scholar]
- Süntar I. Importance of ethnopharmacological studies in drug discovery: role of medicinal plants. Phytochem. Rev. 2020, 19, 1199–1209. 10.1007/s11101-019-09629-9. [DOI] [Google Scholar]
- Zhang L.; Song J.; Kong L.; Yuan T.; Li W.; Zhang W.; Hou B.; Lu Y.; Du G. The strategies and techniques of drug discovery from natural products. Pharmacol. Ther. 2020, 216, 107686. 10.1016/j.pharmthera.2020.107686. [DOI] [PubMed] [Google Scholar]
- Maurya R.; Akanksha J. Chemistry and pharmacology of Withania coagulans: an Ayurvedic remedy. J. Pharm. Pharmacol. 2010, 62, 153–160. 10.1211/jpp.62.02.0001. [DOI] [PubMed] [Google Scholar]
- Khan M. I.; Maqsood M.; Saeed R. A.; Alam A.; Sahar A.; Kieliszek M.; Miecznikowski A.; Muzammil H. S.; Aadil R. M. Phytochemistry, Food Application, and Therapeutic Potential of the Medicinal Plant (Withania coagulans): A Review. Molecules 2021, 26, 6881. 10.3390/molecules26226881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher S.; Choudhary M. I.; Saleem F.; Rasheed S.; Waheed I.; Halim S. A.; Azeem M.; Abdullah I. B.; Froeyen M.; Mirza M. U.; Ahmad S. Isolation of antidiabetic withanolides from Withania coagulans Dunal and their in vitro and in silico validation. Biology 2020, 9, 197. 10.3390/biology9080197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.; Sonawane T.; Pai S. An overview on pharmaceutical properties and biotechnological advancement of Withania coagulans. Adv. Tradit. Med. 2022, 22, 673–683. 10.1007/s13596-021-00558-7. [DOI] [Google Scholar]
- Sayers E. W.; Beck J.; Bolton E. E.; Bourexis D.; Brister J. R.; Canese K.; Comeau D. C.; Funk K.; Kim S.; Klimke W.; Marchler-Bauer A.; Landrum M.; Lathrop S.; Lu Z.; Madden T. L.; O’Leary N.; Phan L.; Rangwala S. H.; Schneider V. A.; Skripchenko Y.; Wang J.; Ye J.; Trawick B. W.; Pruitt K. D.; Sherry S. T. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2021, 49, D10–D17. 10.1093/nar/gkaa892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S. K.; Bhikadiya C.; Bi C.; Bittrich S.; Chen L.; Crichlow G. V.; Christie C. H.; Dalenberg K. D.; Costanzo L.; Duarte J. M.; Dutta S.; Feng Z.; Ganesan S.; Goodsell D. S.; Ghosh S.; Green R. K.; Guranović V.; Guzenko D.; Hudson B. P.; Lawson C. L.; Liang Y.; Lowe R.; Namkoong H.; Peisach E.; Persikova I.; Randle C.; Rose A.; Rose Y.; Sali A.; Segura J.; Sekharan M.; Shao C.; Tao Y.-P.; Voigt M.; Westbrook J. D.; Young J. Y.; Zardecki C.; Zhuravleva M. RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021, 49, D437–D451. 10.1093/nar/gkaa1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B.; Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinf. 2016, 54, 5.6.1–5.6.37. 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A.; Bertoni M.; Bienert S.; Studer G.; Tauriello G.; Gumienny R.; Heer F. T.; Beer T. A.; Rempfer C.; Bordoli L.; Lepore R.; Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Meng E. C.; Couch G. S.; Croll T. I.; Morris J. H.; Ferrin T. E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N.; Peitsch M. C.; Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30, S162–S173. 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- Ravikumar A.; Ramakrishnan C.; Srinivasan N. Stereochemical assessment of (Φ, Ψ) outliers in protein structures using bond geometry-specific Ramachandran steric-maps. Structure 2019, 27, 1875–1884. 10.1016/j.str.2019.09.009. [DOI] [PubMed] [Google Scholar]
- Azmi M. B.; Naeem U.; Saleem A.; Jawed A.; Usman H.; Qureshi S. A.; Azim M. K. In silico identification of the rare-coding pathogenic mutations and structural modeling of human NNAT gene associated with anorexia nervosa. Eat. Weight Disord. 2022, 27, 2725–2744. 10.1007/s40519-022-01422-6. [DOI] [PubMed] [Google Scholar]
- Laskowski R. A.; Jabłońska J.; Pravda L.; Vařeková R. S.; Thornton J. M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. 10.1002/pro.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Chen J.; Cheng T.; Gindulyte A.; He J.; He S.; Li Q.; Shoemaker B. A.; Thiessen P. A.; Yu B.; Zaslavsky L.; Zhang J.; Bolton E. E. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. 10.1093/nar/gkaa971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Grimm M.; Dai W. T.; Hou M. C.; Xiao Z. X.; Cao Y. CB-Dock: a web server for cavity detection-guided protein–ligand blind docking. Acta Pharmacol. Sin. 2020, 41, 138–144. 10.1038/s41401-019-0228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa N.; Hutchison G. R. Fast, efficient fragment-based coordinate generation for Open Babel. J. Cheminf. 2019, 11, 49. 10.1186/s13321-019-0372-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz M. D. Two decades under the influence of the rule of five and the changing properties of approved oral drugs. J. Med. Chem. 2019, 62, 1701–1714. 10.1021/acs.jmedchem.8b00686. [DOI] [PubMed] [Google Scholar]
- Osman W.; Ismail E. M.; Shantier S. W.; Mohammed M. S.; Mothana R. A.; Muddathir A.; Khalid H. S. In silico assessment of potential leads identified from Bauhinia rufescens Lam. as α-glucosidase and α-amylase inhibitors. J. Recept. Signal Transduct. Res. 2021, 41, 159–169. 10.1080/10799893.2020.1800734. [DOI] [PubMed] [Google Scholar]
- Nguyen N. T.; Nguyen T. H.; Pham T. N.; Huy N. T.; Bay M. V.; Pham M. Q.; Nam P. C.; Vu V. V.; Ngo S. T. Autodock vina adopts more accurate binding poses but autodock4 forms better binding affinity. J. Chem. Inf. Model. 2019, 60, 204–211. 10.1021/acs.jcim.9b00778. [DOI] [PubMed] [Google Scholar]
- Jejurikar B. L.; Rohane S. H. Drug designing in discovery studio. Asian J. Res. Chem. 2021, 14, 135–138. 10.5958/0974-4150.2021.00025.0. [DOI] [Google Scholar]
- Dong J.; Wang N. N.; Yao Z. J.; Zhang L.; Cheng Y.; Ouyang D.; Lu A. P.; Cao D. S. ADMETlab: a platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J. Cheminf. 2018, 10, 29. 10.1186/s13321-018-0283-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano M.; Galati S.; Ortore G.; Caligiuri I.; Rizzolio F.; Ceni C.; Bertini S.; Bononi G.; Granchi C.; Macchia M.; Poli G.; Tuccinardi T. Machine Learning-Based Virtual Screening for the Identification of Cdk5 Inhibitors. Int. J. Mol. Sci. 2022, 23, 10653. 10.3390/ijms231810653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadood A.; Ajmal A.; Junaid M.; Rehman A. U.; Uddin R.; Azam S. S.; Khan A. Z.; Ali A. Machine Learning-based Virtual Screening for STAT3 Anticancer Drug Target. Curr. Pharm. Des. 2022, 28, 3023–3032. 10.2174/1381612828666220728120523. [DOI] [PubMed] [Google Scholar]
- Singh R.; Pokle A. V.; Ghosh P.; Ganeshpurkar A.; Swetha R.; Singh S. K.; Kumar A. Pharmacophore-based virtual screening, molecular docking and molecular dynamics simulations study for the identification of LIM kinase-1 inhibitors. J. Biomol. Struct. Dyn. 2022, 2022, 1–15. 10.1080/07391102.2022.2101529. [DOI] [PubMed] [Google Scholar]
- Mensah G. A.; Roth G. A.; Fuster V. The global burden of cardiovascular diseases and risk factors: 2020 and beyond. J. Am. Coll. Cardiol. 2019, 74, 2529–2532. 10.1016/j.jacc.2019.10.009. [DOI] [PubMed] [Google Scholar]
- Engelking L. J.; Liang G.; Hammer R. E.; Takaishi K.; Kuriyama H.; Evers B. M.; Li W. P.; Horton J. D.; Goldstein J. L.; Brown M. S. Schoenheimer effect explained–feedback regulation of cholesterol synthesis in mice mediated by Insig proteins. J. Clin. Invest. 2005, 115, 2489–2498. 10.1172/jci25614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdavi A.; Bagherniya M.; Fakheran O.; Reiner Ž.; Xu S.; Sahebkar A. Medicinal plants and bioactive natural compounds as inhibitors of HMG-CoA reductase: A literature review. BioFactors 2020, 46, 906–926. 10.1002/biof.1684. [DOI] [PubMed] [Google Scholar]
- Johnson B. M.; DeBose-Boyd R. A. Underlying mechanisms for sterol-induced ubiquitination and ER-associated degradation of HMG CoA reductase. Semin. Cell Dev. Biol. 2018, 81, 121–128. 10.1016/j.semcdb.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher M. M.; DeBose-Boyd R. A. Posttranslational regulation of HMG CoA reductase, the rate-limiting enzyme in synthesis of cholesterol. Annu. Rev. Biochem. 2021, 90, 659–679. 10.1146/annurev-biochem-081820-101010. [DOI] [PubMed] [Google Scholar]
- Frye L.; Bhat S.; Akinsanya K.; Abel R. From computer-aided drug discovery to computer-driven drug discovery. Drug Discov. Today Technol. 2021, 39, 111–117. 10.1016/j.ddtec.2021.08.001. [DOI] [PubMed] [Google Scholar]
- Sabe V. T.; Ntombela T.; Jhamba L. A.; Maguire G. E.; Govender T.; Naicker T.; Kruger H. G. Current trends in computer aided drug design and a highlight of drugs discovered via computational techniques: A review. Eur. J. Med. Chem. 2021, 224, 113705. 10.1016/j.ejmech.2021.113705. [DOI] [PubMed] [Google Scholar]
- Adiyaman R.; McGuffin L. J. Methods for the refinement of protein structure 3D models. Int. J. Mol. Sci. 2019, 20, 2301. 10.3390/ijms20092301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman B.; Bradley P. Advances in protein structure prediction and design. Nat. Rev. Mol. Cell Biol. 2019, 20, 681–697. 10.1038/s41580-019-0163-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsamy G.; Soliman M. E. Homology modeling in drug discovery-an update on the last decade. Lett. Drug Des. Discov. 2017, 14, 1099–1111. 10.2174/1570180814666170110122027. [DOI] [Google Scholar]
- Haddad Y.; Adam V.; Heger Z. Ten quick tips for homology modeling of high-resolution protein 3D structures. PLoS Comput. Biol. 2020, 16, e1007449 10.1371/journal.pcbi.1007449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crismaru I.; Stoian A.; Bratu O. G.; Gaman M. A.; Stanescu A. M.; Bacalbasa N.; Diaconu C. C. Low-density lipoprotein cholesterol lowering treatment: the current approach. Lipids Health Dis. 2020, 19, 1–10. 10.1186/s12944-020-01275-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirtori C. R. The pharmacology of statins. Pharmacol. Res. 2014, 88, 3–11. 10.1016/j.phrs.2014.03.002. [DOI] [PubMed] [Google Scholar]
- Atanasov A. G.; Zotchev S. B.; Zotchev V. M.; Dirsch C. T.; Supuran C. T. Natural products in drug discovery: advances and opportunities. Nat. Rev. Drug Discovery 2021, 20, 200–216. 10.1038/s41573-020-00114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzi L.; Rastelli G. Molecular docking: shifting paradigms in drug discovery. Int. J. Mol. Sci. 2019, 20, 4331. 10.3390/ijms20184331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres P. H.; Sodero A. C.; Jofily P.; Silva-Jr F. P. Key topics in molecular docking for drug design. Int. J. Mol. Sci. 2019, 20, 4574. 10.3390/ijms20184574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Shi D.; Zhou S.; Liu H.; Liu H.; Yao X. Molecular dynamics simulations and novel drug discovery. Expet Opin. Drug Discov. 2018, 13, 23–37. 10.1080/17460441.2018.1403419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.