Abstract
To identify pan-ancestry and ancestry-specific loci associated with attempting suicide among veterans, we conducted a genome-wide association study (GWAS) of suicide attempts within a large, multi-ancestry cohort of U.S. veterans enrolled in the Million Veterans Program (MVP). Cases were defined as veterans with a documented history of suicide attempts in the electronic health record (EHR; N=14,089) and controls were defined as veterans with no documented history of suicidal thoughts or behaviors in the EHR (N=395,064). GWAS was performed separately in each ancestry group, controlling for sex, age and genetic substructure. Pan-ancestry risk loci were identified through meta-analysis and included two genome-wide significant loci on chromosomes 20 (p=3.64×10−9) and 1 (p=3.69×10−8). A strong pan-ancestry signal at the Dopamine Receptor D2 locus (p=1.77×10−7) was also identified and subsequently replicated in a large, independent international civilian cohort (p=7.97×10−4). Additionally, ancestry-specific genome-wide significant loci were also detected in African-Americans, European-Americans, Asian-Americans, and Hispanic-Americans. Pathway analyses suggested overrepresentation of many biological pathways with high clinical significance, including oxytocin signaling, glutamatergic synapse, cortisol synthesis and secretion, dopaminergic synapse, and circadian rhythm. These findings confirm that the genetic architecture underlying suicide attempt risk is complex and includes both pan-ancestry and ancestry-specific risk loci. Moreover, pathway analyses suggested many commonly impacted biological pathways that could inform development of improved therapeutics for suicide prevention.
INTRODUCTION
Death by suicide accounts for nearly 800,000 deaths worldwide each year, making it the second leading cause of death among young adults.1 In the U.S., the age- and sex-adjusted rate of death by suicide has increased by 33% since 1999.2 Among U.S. military veterans, the rate has increased even faster, jumping by nearly 50% since 2005.3 While heritability estimates range from 30–55%,4 the genetic basis of suicide and suicide attempts remains largely unknown. There have been more than 100 candidate-gene studies of suicide and suicidal behavior to date5—yet few, if any, candidate single nucleotide polymorphisms (SNPs) have been reliably associated with suicidal behavior.5 For this reason, there has been growing interest in using genome-wide association studies (GWAS) to identify novel genetic risk factors associated with suicide and suicide attempts. Multiple GWAS of suicide attempts have now been conducted,5–8 and several have identified genome-wide significant risk loci; however, only two associations have been independently replicated.6,8
A GWAS of suicide attempts among active-duty military personnel identified multiple genome-wide significant loci near Melanocortin 2 Receptor Accessory Protein 2 (MRAP2), which was subsequently replicated in an independent sample of U.S. veterans.6–7 More recently, Mullins and colleagues8 reported genome-wide significant associations between suicide attempts and the major histocompatibility complex and an intergenic locus on chromosome 7 within the International Suicide Genetics Consortium (ISGC; N=549,743) cohort. Notably, the Million Veteran Program (MVP) cohort9–10 was used to replicate the association between the chromosome 7 index SNP and suicide attempts (p=3.27×10−3); however, a GWAS of suicide attempts has not yet been conducted within the MVP cohort. Accordingly, the objective of the present research was to conduct a GWAS within the MVP cohort to identify pan-ancestry and ancestry-specific loci associated with risk for attempting suicide among U.S. military veterans.
SUBJECTS AND METHODS
Study Participants
The MVP study, which currently includes over 800,000 veteran participants from across the U.S., is one of the largest and most diverse biorepositories in the world.9–10 Moreover, electronic health records (EHR) make this cohort a powerful resource for investigating the genetic basis of a wide range of phenotypes. Recruitment and study procedures for MVP have been described in detail previously and entailed donating a blood sample, consenting to genetic analyses, linking one’s genetic information with the VA’s EHR, and completion of two optional surveys. 9–10 The present study, which involved secondary analysis of existing data collected through the primary MVP study, 9–10 was reviewed and approved by the Department of Veterans Affairs (VA) Central Institutional Review Board (IRB). All study participants provided informed consent as part of the larger MVP study.9–10
Development of the Suicide Attempt Phenotype
Three VA EHR sources were utilized to create a suicide attempt phenotype in the present study, including: (a) International Classification of Diseases (ICD9 and ICD10) codes for intentional self-harm; (b) suicide behavior reports from the VA’s Suicide Prevention Applications Network (SPAN) database;11 and (c) mental health survey responses from the VA’s Mental Health Assistant database indicating a history of attempting suicide (see Supplementary Phenotyping Methods and Supplementary Tables 1a–d for details). Veteran participants were classified as suicide attempt cases if their EHR contained one or more: ICD-9/ICD-10 self-injury codes; SPAN reports of suicide attempts; or mental health survey responses in which participants endorsed a history of attempting suicide. As can be seen Supplementary Table 1d, 60.3% of suicide attempt cases were identified by more than one source, 30.9% were identified by diagnostic codes only, 5.6% were identified by SPAN records only, and 3.2% were identified by mental health surveys only. Veteran participants were classified as controls if they had no documented lifetime history of suicide attempts or suicidal ideation based on qualifying ICD codes, suicide behavior reports, or mental health survey responses. Note that veterans who had a history of suicidal ideation, but not suicide attempts (N=36,732), were excluded from the present analyses to ensure that control participants did not have a history of suicidal thoughts or behaviors.
Code Availability
Please note that the code used to phenotype suicide attempts and suicidal ideation from VA EHR data in the present study are available through the VA’s Centralized Interactive Phenomics Resource (CIPHER) https://www.research.va.gov/programs/cipher.cfm(VAnetworkaccessonly)).
MVP Genotyping and Imputation
Genotyping methods and quality control (QC) for the MVP genotype data have been described previously.10 Briefly, DNA extracted from peripheral blood was genotyped on the MVP custom Axiom 1.0 array. Samples of questionable identity or with call rates below 98.5% were excluded. After phasing the chromosomes with EAGLE v 2.4, genotype data were imputed with Minimac v412 using the 1000Genomes p3v5 as the global reference panel. The present analysis used Release 3 of the imputed MVP dataset, excluding markers with a minor allele frequency (MAF) < 0.01 in the entire data set.
Statistical Methods
Genetic substructure.
We performed principal component analysis (PCA) using PLINK213 with the non-imputed genotypes within each of the four largest, mutually-exclusive racial groups assigned through a prior MVP study14 focused on harmonizing genetic ancestry and self-identified race/ancestry (HARE) within the MVP cohort: European-Americans, African-Americans, Hispanic-Americans, and Asian-Americans. The HARE approach to assign ancestry uses a combination of genetic markers and self-report to assign individuals to major ancestral groups.14 To further control for population substructure within ancestral group, we used 10 principal components (PC’s) for the European-Americans (lambdaGC=1.06 after PC adjustment), 6 for African-Americans (lambdaGC=1.03), 8 for Hispanic-Americans (lambdaGC=1.02), and 6 for Asian-Americans (lambdaGC=0.95).
Genetic Association and meta-analysis.
Ancestry-specific GWAS was performed using PLINK 2,13 controlling for age, gender and genetic PC’s. Meta-analysis was performed with the R package metafor,15 and the QE-test of heterogeneity of effect sizes16 was performed across the ancestral groups. GWAS results were summarized using Manhattan plots, along with LocusZoom plots17 for specific associated genomic regions.
Replication.
We used the ISGC8 and the Mid-Atlantic MIRECC cohorts7 to replicate top associations (p<10−6). The ISGC cohort was larger (N=549,743 total subjects, N=29,782 cases), but is primarily a civilian cohort, whereas the MIRECC cohort is smaller (N=2,423 total subjects, N=218 cases), but comprised entirely of U.S. military veterans, many of whom have seen combat and have histories of PTSD, depression, and suicide attempts. Direct replication of the top GWAS associations was performed for findings from the meta-analysis as well as for European-American and African-American ancestry-specific loci in the comparable ancestral groups in ISGC, using the original SNP, or a surrogate SNP with r2>0.5 or D’=1 identified with LDproxy, part of the LDlink suite of programs.18–19 Note that some MVP loci did not have proxy SNPs in the ISGC meeting those criteria. Due to the smaller size of the MIRECC cohort, this study was used as a target sample to test polygenic risk scores (PRS) generated from the MVP cohort. Using the program PRSice,20 the effect sizes from the MVP GWAS for the European-American and African-American groups were used to generate PRS to test for association with suicide attempts adjusting for age, sex and genetic PCs (15 for European-Americans, 3 for African-Americans) in the comparable ancestral groups in the MIRECC data set.7
Genetic correlation.
Cross-trait linkage disequilibrium score regression21 was used to estimate the genetic correlation between suicide attempts in the European-American subsets of the MVP and the ISGC. Additionally, we examined the genetic correlation between suicide attempts in the MVP European-American subset with five phenotypes relevant to suicide attempts including bipolar disorder,22 schizophrenia,23 major depression,24 posttraumatic stress disorder (PTSD),25 and sleep disorders,26 all of which were obtained from LD Hub.27
Pathway analysis.
Pathway analysis was performed with Over-Representation Analysis in the Web-Gestalt package.28 Genes were included from the following SNP designations: exonic, intronic, ncRNA exonic and ncRNA intronic. A single marker with the smallest p-value (< 0.05) was chosen to represent each gene from among the 10% most significant SNPs. The KEGG database was used for defining pathways, and the top 30 pathways for each GWAS analysis were reported.
RESULTS
Overview of Analyses
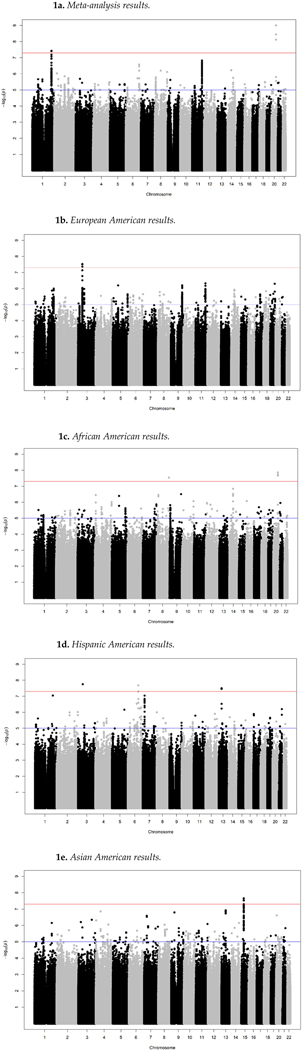
Clinical and genotype data from 409,153 individuals (including 14,089 cases) were examined in this study (Table 1). The four ancestral groups exhibited significantly different rates of suicide attempts (2.7% in Asian-Americans, 3.1% in European-Americans, 4.2% in Hispanic-Americans, 4.5% in African-Americans, p < 2.2e-16), which is consistent with national data that finds that African-Americans are more likely to attempt suicide than European Americans;29–30 whereas European-Americans are more likely than African Americans to die by suicide.30–31 Given these important differences in suicide attempt rates by ancestry, we elected to first conduct GWAS within each group and then to conduct a pan-ancestry meta-analysis. These analyses identified nine loci associated with suicide attempts with genome-wide significance (p<5×10−8; Figure 1; Table 2).
Table 1.
Sample characteristics.
| Controls | Cases | Standardized Mean Difference1 | |
|---|---|---|---|
|
| |||
| Total (%) | 395,064 (96.6) | 14,089 (3.4) | |
| Age (mean (SD)) | 62.84 (13.63) | 52.01 (12.77) | 0.820 |
| Age Group (%) | 0.814 | ||
| 18–29 | 9,192 (2.3) | 898 (6.4) | |
| 30–39 | 20,058 (5.1) | 1,811 (12.9) | |
| 40–49 | 31,109 (7.9) | 2,292 (16.3) | |
| 50–59 | 67,781 (17.2) | 4,855 (34.5) | |
| 60 and over | 266,710 (67.5) | 4,228 (30.0) | |
| Missing | 214 (0.1) | 5 (0.0) | |
| Male (%) | 363,773 (92.1) | 12,005 (85.2) | 0.218 |
| HARE Ancestry Group (%) | 0.170 | ||
| European-American | 287,370 (72.7) | 9,196 (65.3) | |
| African-American | 74,306 (18.8) | 3,507 (24.9) | |
| Asian-American | 4,082 (1.0) | 115 (0.8) | |
| Hispanic-American | 29,306 (7.4) | 1,271 (9.0) | |
| Self-Reported Race2 (%) | |||
| White (only) | 293,512 (74.3) | 9,544 (67.7) | 0.145 |
| Black (only) | 69,986 (17.7) | 3,339 (23.7) | 0.148 |
| Asian (Only) | 3,434 (0.9) | 93 (0.7) | 0.024 |
| Self-Reported Ethnicity (%) | 0.053 | ||
| Hispanic or Latino/a | 23,917 (6.1) | 1,031 (7.3) | |
| Not Hispanic or Latino/a | 362,546 (91.8) | 12,784 (90.7) | |
| Military Service3 (%) | |||
| September 2001 or later (%) | 42,128 (10.7) | 2,396 (17.0) | 0.184 |
| August 1990 to August 2001 (includes Persian Gulf War) (%) | 83,531 (21.1) | 4,788 (34.0) | 0.290 |
| May 1975 to July 1990 (%) | 90,036 (22.8) | 4,568 (32.4) | 0.217 |
| Prior Feb1975 (includes Vietnam, Korea, World War II) (%) | 267,917 (67.8) | 5,077 (36.0) | 0.671 |
Notes:
Standardized mean differences of 0.2, 0.5, and 0.8 can be interpreted as small, medium, and large effects sizes, respectively.
Self-reported race utilized a “mark all that apply format,” we report here results for participants who endorsed only one race category to facilitate comparison with the ancestral groups that were utilized, which were mutually exclusive.
Military service utilized a “mark all that apply format.” Thus, these categories are not mutually exclusive.
Figure 1. Manhattan plots summarizing the GWAS results for the pan-ancestry meta-analysis and ancestry-specific GWAS.
Notes: Red line indicates genome-wide (p<5X10−8) significant threshold. Figure la) Metaanalysis; Figure lb) European Americans; Figure lc) African Americans; Figure Id) Hispanic Americans; Figure le) Asian Americans.
Table 2.
SNP Associations with p<10−6 from Meta-Analysis and Ancestry-Specific Genome-Wide Association Studies.
| SNP | Chromosome: position | Alleles eff/alt | p-value | Odds Ratio (SE) | Annotation | Replication |
|---|---|---|---|---|---|---|
| * Meta-Analysis Results (14089 (3.4%) cases, 395064 (96.6%) controls) | ||||||
| rs56817213 | 20: 51818256 | A/G | 3.64×10−9 | 1.22 (0.03) | TSHZ2 | |
| rs72730526 | 1: 218026968 | A/T | 3.69×10−8 | 1.21 (0.03) | SPATA17 | 6.75×10−1 |
| rs10407501 | 19: 30569741 | A/G | 5.41×10−8 | 1.23 (0.04) | URI1, ZNF536 | |
| rs116165183 | 8: 138806806 | A/G | 7.25×10−8 | 1.35 (0.06) | LOC101927915, LOC401478 | |
| rs12883260 | 14: 56398379 | A/G | 1.62×10−7 | 1.11 (0.02) | LINC00520, PELI2 | 9.44×10−1 |
| rs6589377 | 11: 113355736 | A/G | 1.77×10−7 | 0.93 (0.01) | DRD2, TMPRSS5 | 7.97×10−4 |
| rs1107162 | 11: 113289037 | A/G | 2.16×10−7 | 0.93 (0.01) | DRD2 | 2.52×10−5 |
| rs28731324 | 14: 61747119 | C/G | 2.28×10−7 | 1.20 (0.04) | TMEM30B | |
| European American Results ( 9196 (3.1%) cases, 287370 (96.9%) controls) | ||||||
| rs4858820 | 3: 48484016 | T/A | 3.20×10−8 | 1.10 (0.02) | ATRIP | 1.73×10−1 |
| rs202061221 | 3: 48412264 | CG/C | 3.10×10−7 | 1.10 (0.02) | SPINK8, FBXW12 | 1.23×10−1 (rs56023037) |
| rs555562525 | 11: 113318196 | GA/G | 4.80×10−7 | 0.91 (0.02) | DRD2 | 1.89 × 10−4 (rs17601612) |
| rs376014007 | 19: 51031620 | C/T | 5.10×10−7 | 1.09 (0.02) | LRRC4B | 5.66×10−2 |
| rs360210 | 9: 127797945 | C/T | 6.39×10−7 | 0.92 (0.02) | SCAI | 3.94×10−2 |
| rs2174168 | 5: 62971367 | A/T | 6.44×10−7 | 1.08 (0.02) | IPO11, HTR1A | 3.50 ×10−1 (rs2365873) |
| African American Results (3507 (4.5%) cases, 74306 (95.5%) controls) | ||||||
| rs56817213 | 20: 51818256 | G/A | 1.38×10−8 | 1.22 (0.04) | TSHZ2 | |
| rs116165183 | 8: 138806806 | G/A | 2.90×10−8 | 1.37 (0.06) | LOC101927915, LOC401478 | |
| rs28731324 | 14: 61747119 | C/G | 1.46×10−7 | 1.21 (0.04) | TMEM30B | |
| rs536061513 | 9: 131007176 | C/A | 3.16×10−7 | 1.82 (0.12) | DNM1 | |
| rs34326041 | 8: 19193436 | T/G | 3.50×10−7 | 1.18 (0.03) | SH2D4A | 6.70×10−1 |
| rs3796820 | 4: 10097446 | A/G | 3.59×10−7 | 1.58 (0.09) | WDR1 | 4.72×10−2 |
| rs75861007 | 14: 61730115 | G/T | 3.60×10−7 | 1.32 (0.05) | SNORD112 | |
| rs114579897 | 5: 83946735 | T/G | 4.10×10−7 | 2.26 (0.16) | EDIL3, NBPF22P | 2.42 × 10−2 (rs78097367) |
| rs369284963 | 16: 7462413 | C/CA | 8.58×10−7 | 1.15 (0.03) | RBFOX1 | 9.98 ×10−2 (rs8050278) |
| rs61859846 | 10: 131294704 | C/A | 8.67×10−7 | 1.21 (0.04) | MGMT | 3.09×10−1 |
| rs56291711 | 4: 190043389 | T/C | 9.89×10−7 | 0.87 (0.03) | LINC02508, LINC01262 | 9.85.x10−2 (rs57050950) |
| Hispanic American Results (1271 (4.2%) cases, 29306(95.8%) controls) | ||||||
| rs34173987 | 3: 54892449 | G/A | 1.76×10−8 | 2.34 (0.15) | CACNA2D3 | |
| rs6907713 | 6: 120589671 | C/A | 2.08×10−8 | 15.67 (0.49) | MIR3144, TBC1D32 | |
| rs9596440 | 13: 51646178 | G/C | 3.17×10−8 | 2.73 (0.18) | GUCY1B2 | |
| Asian American Results (115 (2.7%) cases, 4082 (97.3%) controls) | ||||||
| rs199633759 | 15: 48822272 | A/G | 2.20×10−8 | 2.77 (0.18) | FBN1 | |
| rs77378519 | 15: 48831864 | C/T | 2.89×10−8 | 2.69 (0.18) | FBN1 | |
| rs57790277 | 15: 48855753 | T/C | 2.98×10−8 | 3.07 (0.20) | FBN1 | |
For the meta-analysis, the direction of effects were (EA AA HA AS): rs56817213 (- - - -), rs72730526 (+ + + +), rs10407501 (+ + + +), rs116165183 (- - + -), rs12883260 (+ - + +), rs6589377 (+ - + +), rs1107162 (- + - -), rs28731324 (+ - + -).
Pan-Ancestry GWAS Results
Meta-analysis across the four ancestral groups identified two genome-wide significant pan-ancestry loci on chromosomes 20 and 1 (Figure 1, Table 2). Locus zoom plots for genome-wide significant genomic regions are presented in Supplemental Figure 1. The top SNPs were close to Teashirt Zinc Finger Homeobox 2 (TSHZ2), a transcription factor linked to smooth muscle development32 and Spermatogenesis Associated Protein 7 (SPATA17). The direction (same risk allele) and magnitude of the effect sizes of these associations were similar across ancestry groups. Supporting this observation, the test for heterogeneity was not statistically significant for either locus. Five other loci were associated with suicide attempts at p<10−6 in the meta-analysis, including two SNPs near Dopamine Receptor D2 (DRD2), a G protein-coupled receptor located on postsynaptic dopaminergic neurons involved in reward-mediating pathways and addictive behavior.33 We also performed a gene-based analysis using FUMA34 on the meta-analysis results. Two genes were genome-wide significant for the gene-based tests, including SPATA17 on chromosome 1 and DRD2 on chromosome 11, providing further support for these meta-analytic signals.
Ancestry-Specific GWAS Results
We also analyzed association with suicide attempt risk within each ancestral group (Figure 1, Table 2). Among European-Americans, a genome-wide significant association was identified near ATR-Interacting Protein (ATRIP), which has been associated with Seckel syndrome, a rare Mendelian disorder characterized by microcephaly and mental retardation.35 Among African-Americans, one of the genome-wide significant SNPs from the meta-analysis (rs56817213) was also found to be genome-wide significant within this cohort. Among Hispanic-Americans, three genome-wide significant loci were identified, including one in Calcium Channel, Voltage-Dependent, Alpha-2/Delta Subunit 3 (CACNA2D3), which modulates presynaptic nerve function36 and is associated with pain sensitivity.37 Genome-wide significant associations were also identified near TBC1 Domain Family Member 32 (TBC1D32) and near Guanylate Cyclase 1 Soluble Subunit Beta 2 (GUCY1B2), a pseudogene, among Hispanic-Americans. Finally, among Asian-Americans, three genome-wide significant SNPs were identified near Fibrillin 1 (FBN1). Locus Zoom plots of these regions are provided in Supplemental Figure 1.
Replication
We attempted to replicate findings in the ISGC cohort8 (29,782 cases; 519,961 controls) for SNPs in Table 2, or high LD surrogates, when available. We observed strong replication of three SNPs identified in DRD2 in the overall meta-analysis and in European Americans. Two African-American loci were also replicated, including WDR1 (WD-repeat containing protein 1) and EDIL3 (EGF-like Repeats and Discoidin I-like Domains Containing Protein 3).
Polygenic Risk and Top Loci Follow-Up in an Independent Veteran Cohort
Results from the MVP GWAS were used to construct polygenic risk scores (PRSs). We then evaluated the association between these PRSs and suicide attempts in the MIRECC cohort.7 For African-Americans (N=1,245), the MVP PRS was significantly associated with suicide attempts (p=0.001), with the most significant association achieved using MVP p-values below 0.3 (PT=0.3; Supplemental Figure 3). This PRS explained slightly less than 2% of the phenotypic variability of suicide attempts within the MIRECC cohort. Among European-Americans (N=1,178), none of the MVP PRSs were significantly associated with suicide attempts.
Genetic Correlations
The SNP heritability of suicide attempts in the MVP was estimated to be 0.0125 (se 0.0022). Using the EA subset of the MVP as the reference, we found a large genetic correlation (rGC=0.86; p=1.30e-21) for suicide attempts between the MVP and ISGC data sets, despite ISGC being a largely civilian cohort. The LDSC intercept for this analysis was 0.0062 (se 0.0059), providing support that the two data sets were independent. We also estimated rGC between suicide attempts in the MVP European-American subset with several other psychiatric disorders, including bipolar disorder (rGC = 0.38; se=0.0641; p=4.5E-09), schizophrenia (rGC =0.46; se=0.0571; p=9.23E-16) major depression (rGC =0.63; se=0.0685; p=1.9E-20), and PTSD (rGC =0.57; se=0.073; p=5.59E-15). We observed a small correlation with sleep disorders (rGC = 0.096; se=0.0347 p=0.0057). All of the rGC estimates with suicide attempts remained significant after a Bonferroni correction for the number of phenotypes examined (corrected p = 0.05/5 = 0.01).
Pathway Analysis
Thirty pathways were significantly over-represented in the multi-ancestry meta-analysis (Table 3), including oxytocin signaling, glutamatergic synapse, axon guidance, calcium signaling, circadian entrainment, cortisol synthesis and secretion, dopaminergic synapse, and circadian rhythm (all FDR <0.05). There was striking consistency for the pathway analyses across the ancestral groups, including two that were significant across all five analyses: arrhythmogenic right ventricular cardiomyopathy (ARVC), and phospholipase D signaling, the latter of which mediates alcohol’s effect on ion channels38 and has been linked to Alzheimer’s disease.39
Table 3.
Top 30 False-Discovery Rate (FDR)-Significant Findings from the Pathway Analyses.
| Gene Set | Description | Meta-Analysis Ratio | European-Americans Ratio | African-Americans Ratio | Hispanic-Americans Ratio | Asian-Americans Ratio |
|---|---|---|---|---|---|---|
|
| ||||||
| hsa00604 | Glycosphingolipid biosynthesis | 4.98 | ||||
| hsa04710 | Circadian rhythm | 3.37 | ||||
| hsa04520 | Adherens junction | 3.32 | ||||
| hsa04924 | Renin secretion | 3.22 | 2.88 | 2.75 | ||
| hsa04540 | Gap junction | 3.06 | 2.66 | |||
| hsa04724 | Glutamatergic synapse | 3.01 | 2.33 | 2.67 | 2.99 | |
| hsa04921 | Oxytocin signaling | 2.95 | 2.31 | 2.56 | 2.25 | |
| hsa05412 | Arrhthmogenic righ ventricular cardiomyopathy | 2.90 | 2.82 | 2.75 | 3.16 | 3.09 |
| hsa04927 | Cortisol synthesis and secretion | 2.80 | 3.42 | 2.86 | ||
| hsa04713 | Circadian entrainment | 2.80 | 2.60 | 2.22 | 2.71 | |
| hsa04925 | Aldosterone synthesis and secretion | 2.65 | 2.44 | 2.70 | 2.37 | |
| hsa04360 | Axon guidance | 2.56 | 1.96 | 2.00 | ||
| hsa05414 | Dilated cardiomyopathy | 2.49 | 2.37 | 2.53 | 2.66 | |
| hsa04270 | Vascular smooth muscle contraction | 2.47 | 2.32 | 2.65 | ||
| hsa04020 | Calcium signaling | 2.45 | 2.22 | 2.08 | 2.13 | |
| hsa04070 | Phosphatidylinositol signaling | 2.41 | 2.37 | 2.46 | ||
| hsa04912 | GnRH signaling | 2.41 | ||||
| hsa04725 | Cholinergic synapse | 2.40 | 2.23 | 2.99 | ||
| hsa04072 | Phospholipase D signaling | 2.35 | 2.46 | 2.40 | 2.22 | 2.34 |
| hsa05410 | Hypertrophic cardiomyopathy | 2.34 | 2.74 | 2.68 | ||
| hsa04911 | Insulin secretion | 2.28 | ||||
| hsa04750 | Inflammation/transient receptor potential channels | 2.26 | 2.31 | |||
| hsa04022 | Cyclic guanosine monophosphate-protein kinase G signaling | 2.11 | 2.62 | |||
| hsa04024 | Cyclic adenosine monophosphate signaling | 2.10 | 2.36 | 1.91 | ||
| hsa04371 | Apelin signaling pathway | 2.07 | ||||
| hsa04728 | Dopaminergic synapse | 2.05 | ||||
| hsa04015 | Ras-associated protein-1 signaling | 2.03 | 1.82 | 2.15 | 1.89 | |
| hsa04261 | Adrenergic signaling cardiomyocytes | 1.97 | 2.17 | 2.49 | ||
| hsa04514 | Cell adhesion molecules | 1.97 | 2.01 | 2.14 | 2.26 | |
| hsa04934 | Cushing syndrome | 1.94 | 2.03 | |||
Notes: All values shown are significant at FDR < 0.05.
Association between Sleep Problems and Suicide Attempts within MVP
Given the overrepresentation of both the circadian entrainment and circadian rhythm pathways, we examined the association between suicide attempts and sleep problems (see Supplementary Phenotyping Methods for details) in the MVP cohort, using the MVP Lifestyle Survey,9 which was available for 53.7% (N=220,001) of the participants from the current cohort. We observed significant, clinically-meaningful differences between suicide attempt cases and controls on total number of sleep problems endorsed (overall sample: 3.4 vs 2.1, p < 0.001; standardized mean difference = 0.65; Supplementary Table 3). Notably, these differences were highly consistent with respect to both direction and magnitude of effect across all four ancestries (Supplemental Figure 4), providing strong support for the potential role of sleep disturbance in suicide attempt risk among veterans.
DISCUSSION
We report here findings from the largest GWAS of suicide attempts among U.S. veterans to date. Meta-analysis across four ancestral groups identified two genome-wide significant pan-ancestry loci on chromosomes 20 and 1. Both loci demonstrated homogeneity of effects across ancestral groups, suggesting that common genetic factors underlie risk for attempting suicide among veterans. A strong pan-ancestry signal at the Dopamine Receptor D2 locus was also identified (p<10−7) and subsequently replicated in a large, independent international cohort. Ancestry-specific genome-wide significant risk loci were detected for each ancestral group examined, and an MVP-based PRS significantly predicted suicide attempts in an independent cohort of African-American veterans. As expected, we also observed significant genetic correlations between suicide attempts and other psychiatric disorders, including bipolar disorder, schizophrenia, PTSD, and depression. Perhaps most notable was the magnitude of the genetic correlation for suicide attempts across the MVP and ISGC cohorts (GC=0.86, p=1.30−21), which suggests that similar genetic factors influence risk for suicide attempts across diverse environmental exposures (e.g., combat), providing strong support for consortium-based studies of genetic risk for suicide.
The strongest meta-analytic association identified in the present study was near TSHZ2, which is highly expressed in human cerebral cortex40 and has been previously associated with sleep duration41 and comorbid depression, alcohol dependence.42 Notably, the TSHZ2 locus was also genome-wide significant within African-Americans.
Strong associations were also observed between DRD2 and suicide attempts in both the pan-ancestry meta-analysis and the European-American cohort. Moreover, this association was also replicated within the ISGC cohort and was genome-wide significant in the meta-analytic gene-based analysis. DRD2 is a G protein-coupled receptor located on postsynaptic dopaminergic neurons that is highly expressed in human and murine basal ganglia. It is centrally involved in reward-mediating pathways and is consistently associated with risk for substance use disorders.43 DRD2 is also associated with risk for schizophrenia,44 attention-deficit/hyperactivity disorder (ADHD),45 and sleep duration,46 all of which are, in turn, risk factors for suicide and suicidal behavior.47–49 DRD2 has also been associated with suicidal behavior previously;50–52 however, the present findings provide the strongest support to date.
The genome-wide significant association between CACNA2D3 and suicide attempts in the Hispanic-American cohort also represents a promising avenue for future investigation. CACNA2D3 has been associated with bipolar disorder,53 depression,54 schizophrenia,55 autism spectrum disorder,56 nicotine dependence,57 and pain sensitivity.37
Within the European-American cohort, the only SNP that reached genome-wide significance was located on chromosome 3 near ATRIP, which encodes a key component of the DNA damage checkpoint. A recent GWAS of shared risk across psychiatric disorders identified a genome-wide significant locus near ATRIP;58 however, we note that this region on chromosome 3 exhibits considerable LD in European-Americans, making it challenging to discern whether ATRIP or another gene is driving this association.
Among Asian-Americans, three genome-wide SNPs were identified, all within the Fibrillin 1 gene (FBN1), in which mutations give rise to Marfan syndrome,59 a heritable connective tissue disorder. While the functional significance of FBN1 to suicide is presently unclear, a prior GWAS found a suggestive association between FBNI and bipolar disorder among Norwegian individuals, which was subsequently replicated in an Icelandic sample.60
Finally, we identified a number of pathways of high relevance to suicide risk. Oxytocin signaling plays a key role in social bonding and feelings of well-being, and lower serum oxytocin concentrations have been associated with suicidal intent and suicide attempts.61–62 Over-representation of the glutamatergic synapse pathway is also of great interest, as neuroimaging studies have identified glutamatergic dysfunction among patients with schizophrenia and other psychiatric disorders,63 and at least two postmortem studies have found that suicide decedents are more likely than controls to have increased glutamatergic gene expression in the prefrontal cortex.64–65 Moreover, the glutamatergic modulator ketamine has been found to produce rapid decreases in suicidal ideation.66
Multiple stress pathways were also overrepresented, including cortisol synthesis and secretion, as well as blood pressure regulating pathways (e.g., renin secretion, aldosterone synthesis and secretion). Also featured were the circadian entrainment and circadian rhythm pathways, which have been shown to be stimulated by DRD2. Disruptions of circadian rhythm profoundly affect mood, as CLOCK protein regulates dopaminergic transmission in the ventral tegmental area—the critical neurological reward circuits.67 Furthermore, many psychiatric disorders (e.g., schizophrenia, mood disorders) are characterized by impaired circadian-clock controlled functions, such as sleep and cortisol secretion.68 A genome-wide significant association with a functional variant in the CLOCK gene PER1 has recently been reported for suicide death.69 We also found clear evidence for increased sleep problems among veterans with a history of suicide attempts within the MVP cohort, providing direct support for sleep disturbance in suicide risk among veterans. Notably, another recent study identified both circadian rhythms and glutamatergic signaling as top biomarker candidates for mood disorders.70
Study Limitations
Out of necessity, suicide attempts were defined from VA EHR sources, including ICD codes, SPAN reports, and survey responses, rather than by rigorous diagnostic interviews. It is possible that some control participants had a history of attempts that were not recorded in the VA’s EHR (particularly those that occurred prior to military service); however, while this might decrease statistical power, it is unlikely to introduce false positive SNP associations. A second limitation concerns the lack of replication for many of our top findings. In several cases, genotyped markers within loci of interest (i.e., TSHZ2, CACNA2D3, FBNT1) were simply not available within the ISCG cohort; in other instances (e.g., SPATA17, ATRIP), appropriate markers were available, but no replication was observed. This could be due to false positive associations in the MVP study, or alternatively, could represent population differences between the two data sets, such as the veteran vs. civilian nature of the ascertainments or the increased diversity present in the MVP study. A notable exception to this pattern was for DRD2, where we were able to replicate all three of our top SNPs in the ISGC cohort.
Conclusions
We report here findings from the largest GWAS of suicide attempts among U.S. veterans to date. Meta-analysis across ancestral groups identified genome-wide significant loci near TSHZ2 and SPATA17. Ancestry-specific genome-wide significant loci were also detected for each ancestral group examined and included TSHZ2, ATRIP, CACNA2D3, and FBN1. A strong pan-ancestry signal at the DRD2 locus was also identified and subsequently replicated in the ISGC cohort. Taken together, our findings confirm that the genetic architecture underlying suicide attempt risk is complex and includes both pan-ancestry and ancestry-specific risk loci. Moreover, findings from the pathway analyses suggest a number of promising biological pathways that could inform development of improved therapeutics for suicide prevention.
Supplementary Material
Acknowledgements
This research is based on data from the Million Veteran Program, Office of Research and Development (ORD), Veterans Health Administration (VHA), and was supported by award #I01CX001729 from the Clinical Science Research and Development (CSR&D) Service of VHA ORD. J.C. Beckham was also supported by a Senior Research Career Scientist Award (#lK6BX003777) from CSR&D. Niamh Mullins was supported by a NARSAD Young Investigator Award from the Brain & Behavior Research Foundation. This publication does not represent the views of the Department of Veteran Affairs or the United States Government. We also thank and acknowledge MVP, the MVP Suicide Exemplar Workgroup, and the ISGC for their contributions to this manuscript. A complete listing of contributors from the MVP, MVP Suicide Exemplar Workgroup, and ISGC is provided in the Supplementary Materials.
This work was also supported in part by the joint U.S. Department of Veterans Affairs and US Department of Energy MVP CHAMPION program. This manuscript has been co-authored by UT-Battelle, LLC under contract no. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a nonexclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan, last accessed September 16, 2020). The GWAS summary statistics generated from this study will be available via dbGaP. The dbGaP accession assigned to the Million Veteran Program is phs001672.v1.p.
Footnotes
Conflicts of Interest
The authors have no conflicts of interest to declare at this time.
References
- 1.World Health Organization. Suicide in the world: Global estimates, 2019. https://www.who.int/publications/i/item/suicide-in-the-world. [Google Scholar]
- 2.Hedegaard H, Curtin S, Warner M. NCHS Data Brief No. 398. Feb 2021. In Data Briefs [Internet]. Available from: https://www.cdc.gov/nchs/products/databriefs/db398.htm#:~:text=This%20report%20highlights%20trends%20in,than%20the%20rate%20in%202018 [Google Scholar]
- 3.Department of Veterans Affairs Office of Mental Health and Suicide Prevention. 2020. National Strategy for Preventing Veteran Suicide [Internet]. Available from: https://www.mentalhealth.va.gov/suicide_prevention/docs/Office-of-Mental-Health-and-Suicide-Prevention-National-Strategy-for-Preventing-Veterans-Suicide.pdf
- 4.Voracek M, Loibl LM. Genetics of suicide: A systematic review of twin studies. Wien Klin Wochenschr. 2007;119(15–16):463–475. [DOI] [PubMed] [Google Scholar]
- 5.Mirkovic B, Laurent C, Podlipski MA, Frebourg T, Cohen D, Gerardin P. Genetic association studies of suicidal behavior: A review of the past 10 years, progress, limitations, and future directions. Front Psychiatry. 2016;7:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stein MB, Ware EB, Mitchell C, Chen CY, Borja S, Cai T, et al. Genome-wide association studies of suicide attempts in US soldiers. J Med Genet Part B. 2017;174(8):786–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimbrel NA, Garrett ME, Dennis MF, VA Mid-Atlantic Mental Illness Research, Education, and Clinical Center Workgroup, Hauser MA, Ashley-Koch AE, et al. A genome-wide association study of suicide attempts and suicidal ideation in U.S. military veterans. Psychiatry Res. 2018;269:64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mullins N, Kang J, Campos AI, Coleman JRI, Edwards AC, Galfalvy H, et al. Dissecting the shared genetic architecture of suicide attempt, psychiatric disorders and known risk factors. Biol Psychiatry. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaziano JM, Concato J, Brophy M, Fiore L, Pyarajan S, Breeling J, et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–223. [DOI] [PubMed] [Google Scholar]
- 10.Hunter-Zinck H, Shi Y, Li M, Gorman BR, Ji SG, Sun N, et al. Genotyping array design and data quality control in the Million Veteran Program. Am J Hum Genet. 2020;106(4):535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmire C, Stephens B, Morley S, Thompson C, Kemp J, Bossarte RM. VA Suicide Prevention Applications Network: A National Health Care System-Based Suicide Event Tracking System. Public Health Rep. 2016;131(6):816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Center for Statistical Genetics. Minimac4. Available from: https://genome.sph.umich.edu/wiki/Minimac4.
- 13.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang H, Hui Q, Lynch J, Honerlaw J, Assimes TL, Huang J, et al. Harmonizing Genetic Ancestry and Self-identified Race/Ethnicity in Genome-wide Association Studies. Am J Hum Genet. 2019. Oct 3;105(4):763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viechtbauer W. Conducting meta-analyses in R with the metafor package. Journal of Statistical Software. 2010;36(3):1–48. [Google Scholar]
- 16.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. [DOI] [PubMed] [Google Scholar]
- 17.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Machiela MJ, Chanock SJ. LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machiela MJ, Chanock SJ. LDassoc: An online tool for interactively exploring genome-wide association study results and prioritizing variants for functional investigation. Bioinformatics. 2018;34(5):887–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Euesden J, Lewis CM, O’Reilly PF. PRSice: Polygenic Risk Score Software. Bioinformatics. 2015;31(9):1466–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J; Schizophrenia Working Group of the Psychiatric Genomics Consortium, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51(5):793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schizophrenia Psychiatric Genome-Wide Association Study Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43(10):969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A,et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50(5):668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nievergelt CM, Maihofer AX, Klengel T, Atkinson EG, Chen CY, Choi KW, et al. International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat Commun. 2019;10(1):4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.UK Biobank. http://www.nealelab.is/uk-biobank.
- 27.Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, et al. LD Hub: A centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33(2):272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Research. 2017;45(W1):W130–W137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Center for Behavioral Health Statistics and Quality. 2019 National Survey on Drug Use and Health: Detailed Tables. Rockville, MD: Substance Abuse and Mental Health Services Administration. https://www.samhsa.gov/data/report/2019-nsduh-detailed-tables; 2020. [Google Scholar]
- 30.Suicide Prevention Resource Center. https://www.sprc.org/scope/racial-ethnic-disparities 2019.
- 31.Centers for Disease Control and Prevention National Center for Health Statistics. 1999–2019 Wide Ranging Online Data for Epidemiological Research (WONDER), Multiple Cause of Death files [Data file]. Retrieved from http://wonder.cdc.gov/ucd-icd10.html. 2021.
- 32.Jenkins D, Caubit X, Dimovski A, Matevska N, Lye CM, Cabuk F, et al. Analysis of TSHZ2 and TSHZ3 genes in congenital pelvi-ureteric junction obstruction. Nephrol Dial Transplant. 2010;25(1):54–60. [DOI] [PubMed] [Google Scholar]
- 33.Comings DE, Blum K. Reward deficiency syndrome: genetic aspects of behavioral disorders. Prog Brain Res. 2000;126:325–341. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017. Nov 28;8(1):1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogi T, Walker S, Stiff T, Hobson E, Limsirichaikul S, Carpenter G, et al. Identification of the first ATRIP-deficient patient and novel mutations in ATR define a clinical spectrum for ATR-ATRIP Seckel Syndrome. PLoS Genetics. 2012;8(11):e1002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoppa MB, Lana B, Margas W, Dolphin AC, Ryan TA. alpha2delta expression sets presynaptic calcium channel abundance and release probability. Nature. 2012;486(7401):122–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neely GG, Hess A, Costigan M, Keene AC, Goulas S, Langeslag M, et al. A genome-wide Drosophila screen for heat nociception identifies alpha2delta3 as an evolutionarily conserved pain gene. Cell. 2010;143(4):628–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chung HW, Petersen EN, Cabanos C, Murphy KR, Pavel MA, Hansen AS, et al. A molecular target for an alcohol chain-length cutoff. J Mol Biology. 2019;431(2):196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cruchaga C, Karch CM, Jin SC, Benitez BA, Cai Y, Guerreiro R, et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature. 2014;505(7484):550–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Human Protein Atlas. http://www.proteinatlas.org/.
- 41.Gottlieb DJ, O’Connor GT, Wilk JB. Genome-wide association of sleep and circadian phenotypes. BMC Med Genet. 2007. Sep 19;8 Suppl 1(Suppl 1):S9. doi: 10.1186/14712350-8-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edwards AC, Aliev F, Bierut LJ, Bucholz KK, Edenberg H, Hesselbrock V, et al. Genome-wide association study of comorbid depressive syndrome and alcohol dependence. Psychiatr Genet. 2012;22(1):31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopez-Leon S, Gonzalez-Giraldo Y, Wegman-Ostrosky T, Forero DA. Molecular genetics of substance use disorders: An umbrella review. Neuro Biobehav Rev. 2021;124:358–369. [DOI] [PubMed] [Google Scholar]
- 44.Gonzalez-Castro TB, Hernandez-Diaz Y, Juarez-Rojop IE, López-Narváez ML, Tovilla-Zárate CA, Genis-Mendoza A, et al. The role of C957T, TaqI and Ser311Cys polymorphisms of the DRD2 gene in schizophrenia: systematic review and meta-analysis. Behav Brain Functions. 2016;12(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan YQ, Qiao L, Xue XD, Fu JH. Association between ANKK1 (rs1800497) polymorphism of DRD2 gene and attention deficit hyperactivity disorder: A meta-analysis. Neurosci Lett. 2015;590:101–105. [DOI] [PubMed] [Google Scholar]
- 46.Cade BE, Gottlieb DJ, Lauderdale DS, Bennett DA, Buchman AS, Buxbaum SG, et al. Common variants in DRD2 are associated with sleep duration: The CARe Consortium. Hum Mol Genet. 2016;25(1):167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fazel S, Runeson B. Suicide. N Engl J Med. 2020;382(3):266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Septier M, Stordeur C, Zhang J, Delorme R, Cortese S. Association between suicidal spectrum behaviors and Attention-Deficit/Hyperactivity Disorder: A systematic review and meta-analysis. Neurosci Biobehav Rev. 2019;103:109–118. [DOI] [PubMed] [Google Scholar]
- 49.Harris LM, Huang X, Linthicum KP, Bryen CP, Ribeiro JD. Sleep disturbances as risk factors for suicidal thoughts and behaviours: A meta-analysis of longitudinal studies. Scientific Reports. 2020;10(1):13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johann M, Putzhammer A, Eichhammer P, Wodarz N. Association of the −141C Del variant of the dopamine D2 receptor (DRD2) with positive family history and suicidality in German alcoholics. Am J Med Genetics, Part B Neuropsychiatric Gen. 2005;132B(1):46–49. [DOI] [PubMed] [Google Scholar]
- 51.Suda A, Kawanishi C, Kishida I, Sato R, Yamada T, Nakagawa M,et al. Dopamine D2 receptor gene polymorphisms are associated with suicide attempt in the Japanese population. Neuropsychobiology. 2009;59(2):130–134. [DOI] [PubMed] [Google Scholar]
- 52.Genis-Mendoza AD, Lopez-Narvaez ML, Tovilla-Zarate CA, Sarmiento E, Chavez A, Martinez-Magaña JJ, et al. Association between polymorphisms of the DRD2 and ANKK1 genes and suicide attempt: A preliminary case-control study in a mexican population. Neuropsychobiology. 2017;76(4):193–198. [DOI] [PubMed] [Google Scholar]
- 53.Smedler E, Bergen SE, Song J, Landen M. Genes, biomarkers, and clinical features associated with the course of bipolar disorder. Eur Neuropsychopharm. 2019;29(10):1152–1160. [DOI] [PubMed] [Google Scholar]
- 54.Dunn EC, Wiste A, Radmanesh F, Almli LM, Gogarten SM, Sofer T, et al. Genome-wide association study (GWAS) and Genome-wide by environment interaction study (GWEIS) of depressive symptoms in African American and Hispanic/Latina women. Depress Anxiety. 2016;33(4):265–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang T, Zhu L, Ni T, Liu D, Chen G, Yan Z, et al. Voltage-gated calcium channel activity and complex related genes and schizophrenia: A systematic investigation based on Han Chinese population. J Psych Res. 2018;106:99–105. [DOI] [PubMed] [Google Scholar]
- 56.Liao X, Li Y. Genetic associations between voltage-gated calcium channels and autism spectrum disorder: A systematic review. Molecular Brain. 2020;13(1):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yin X, Bizon C, Tilson J, Lin Y, Gizer IR, Ehlers CL, et al. Genome-wide meta-analysis identifies a novel susceptibility signal at CACNA2D3 for nicotine dependence. American Journal of Medical Genetics, Part B Neuropsychiatric Genetics. 2017;174(5):557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schork AJ, Won H, Appadurai V, Nudel R, Gandal M, Delaneau O, et al. A genome-wide association study of shared risk across psychiatric disorders implicates gene regulation during fetal neurodevelopment. Nat Neurosci. 2019;22(3):353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Collod-Beroud G, Le Bourdelles S, Ades L, Ala-Kokko L, Booms P, Boxer M, et al. Update of the UMD-FBN1 mutation database and creation of an FBN1 polymorphism database. Human Mutation. 2003;22(3):199–208. [DOI] [PubMed] [Google Scholar]
- 60.Djurovic S, Gustafsson O, Mattingsdal M, Athanasiu L, Bjella T, Tesli M, et al. A genome-wide association study of bipolar disorder in Norwegian individuals, followed by replication in Icelandic sample. J Affect Disord. 2010;126(1–2):312–316. [DOI] [PubMed] [Google Scholar]
- 61.Jahangard L, Shayganfard M, Ghiasi F, Salehi I, Haghighi M, Ahmadpanah M, et al. Serum oxytocin concentrations in current and recent suicide survivors are lower than in healthy controls. J Psychiatr Res. 2020;128:75–82. [DOI] [PubMed] [Google Scholar]
- 62.Jokinen J, Chatzittofis A, Hellstrom C, Nordstrom P, Uvnas-Moberg K, Asberg M. Low CSF oxytocin reflects high intent in suicide attempters. Psychoneuroendocrinology. 2012;37(4):482–490. [DOI] [PubMed] [Google Scholar]
- 63.Li CT, Yang KC, Lin WC. Glutamatergic dysfunction and glutamatergic compounds for major psychiatric disorders: Evidence from clinical neuroimaging studies. Front Psych. 2018;9:767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gray AL, Hyde TM, Deep-Soboslay A, Kleinman JE, Sodhi MS. Sex differences in glutamate receptor gene expression in major depression and suicide. Mol Psychiatry. 2015;20(9):1057–1068. [DOI] [PubMed] [Google Scholar]
- 65.Zhao J, Verwer RWH, Gao SF, Qi XR, Lucassen PJ, Kessels HW, et al. Prefrontal alterations in GABAergic and glutamatergic gene expression in relation to depression and suicide. J Psychiatr Res. 2018;102:261–274. [DOI] [PubMed] [Google Scholar]
- 66.Witt K, Potts J, Hubers A, Grunebaum MF, Murrough JW, Loo C, et al. Ketamine for suicidal ideation in adults with psychiatric disorders: A systematic review and meta-analysis of treatment trials. Aus and New Zealand J Psychiatry. 2020;54(1):29–45. [DOI] [PubMed] [Google Scholar]
- 67.McClung CA, Sidiropoulou K, Vitaterna M, Takahashi JS, White FJ, Cooper DC, et al. Regulation of dopaminergic transmission and cocaine reward by the Clock gene. Proc Natl Acad Sci U S A. 2005;102(26):9377–9381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walker WH 2nd, , Walton JC, DeVries AC, Nelson RJ. Circadian rhythm disruption and mental health. Trans Psychiatry. 2020;10(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DiBlasi E, Shabalin AA, Monson ET, Keeshin BR, Bakian AV, Kirby AV, et al. Rare protein-coding variants implicate genes involved in risk of suicide death. Am J Med Genet B Neuropsychiatr Genet. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Le-Niculescu H, Roseberry K, Gill SS, Levey DF, Phalen PL, Mullen J, et al. Precision medicine for mood disorders: objective assessment, risk prediction, pharmacogenomics, and repurposed drugs. Mol Psychiatry. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.