Abstract
READ, the RIKEN Expression Array Database, is a database of expression profile data from the RIKEN mouse cDNA microarray. It stores the microarray experimental data and information, and provides Web interfaces for researchers to use to retrieve, analyze and display their data. The goals for READ are to serve as a storage site for microarray data from ongoing research in the RIKEN mouse encyclopedia project and to provide useful links and tools to decipher biologically important information. The gene information is based mainly on the fully annotated FANTOM database. READ can be accessed at http://read.gsc.riken.go.jp/. READ also provides a search tool [READ integrates gene expression neighbor (RINGENE)] for genes with similarities in expression profiling.
INTRODUCTION
Although the emerging technology of DNA microarrays enables us to observe patterns of gene expression at the genomic scale, there are no well-established schemes for analyzing a large amount of data at once. Biologists are sometimes at a loss with microarray experimental data, because the data set is very large and often lacks proper functional annotations for each gene. Microarray data are usually data-mined for similarities in expression profiling, and co-expressed genes are inferred as functionally related to each other or under the same gene cascades. To properly infer the functions of unknown genes from microarray data, it is important for other genes in the same cluster to be well annotated. To this end, we have developed a new annotation tool called FANTOM+, which provides us with curated functional annotation of full-length mouse cDNAs (1). Achieving the functional annotation of anonymous sequences by using gene expression data from microarrays requires computational methods for analyzing these data to be explored.
To overcome this hurdle, we have been developing a Web-based system called READ, the Riken Expression Array Database, for analyzing microarray data from RIKEN mouse cDNA clones. Currently, READ includes information from microarray experiments (not only log ratios of gene expression intensity, but also experimental details such as slide format, lot numbers, hybridization conditions and probe preparation) and the results of sequence analyses for all the nucleotide sequences arrayed on the microarray chips. The READ system can be queried by RIKEN cDNA clone identifier and by keywords in the database description. It can also be searched by the log-transformed ratio of gene expression intensity in all 49 embryonic and adult mouse tissues in the collection.
IMPLEMENTATION
Because the READ system is implemented on the Web, it is easily accessible via typical Web browsers, which enables the system to be accessed without any special software. All data are stored in a flat-file format (tab-delimitated text), and only information frequently referred to is put into a relational database management system (RDBMS), PostgreSQL (http://www.postgresql.org/). Perl script language (http://www.cpan.org/) is frequently used for formatting data, and the scripts are combined in UNIX shell scripts and used in preparing and updating data. An Apache Web server (http://www.apache.org/) with PHP hypertext preprocessor (http://www.php.net/) makes it easy to query the RDBMS by structured query language (SQL) via the Web without the need for specific Web interfaces. The system is built on a PC-UNIX server running RedHat Linux 6.2 and Kondara MNU/Linux 2.0. The RDBMS, Web server and client software used in the READ system are all freeware, allowing us to distribute the system with little restriction to the academic community.
GENE EXPRESSION DATA FROM MICROARRAYS
Currently, READ contains microarray data from 49 embryonic and adult mouse tissues in the RIKEN 19K set (2). All experiments are replicated, and the data are critically checked using Scanalyze image analysis software (3) followed by the preprocessing implementation for microarray (PRIM) quality filtering program (4). PRIM efficiently extracts reproducible data from the results of replicated experiments. These data are transformed into log ratios for further analyses. READ stores mainly this log-transformed, normalized ratio data and integrates the functional annotation of cDNA clones. Sequence data for cDNA clones in the RIKEN 19K set are regularly updated by extracting corresponding sequence data entries in the internal sequence database.
FUNCTIONAL ANNOTATION OF cDNA CLONES
In the analysis of microarray expression data, the grouping or clustering of genes by their expression similarities is usually used. Several methods of clustering have been developed, including hierarchical clustering and neural net clustering. After the genes are clustered, it is hypothesized that genes in the same cluster are functionally correlated, may interact with each other at the protein level or may be under the same gene cascade. To infer the functions of genes that are yet to be determined on the basis of these hypotheses, accurate functional annotation information for known genes is indispensable. At the moment, all of the clones printed on the array are those derived from the RIKEN full-length enriched mouse cDNA clones. Therefore, the functional annotation information is taken mainly from the FANTOM database. For cDNAs that are not included in the FANTOM set, a computational functional annotation system called functional inference descriptor (FIND) is used to assign non-curated functional annotations. The FIND system simulates the assignment of a brief description (RIKEN definition) of cDNA clones. These functional annotations are regularly extracted and then integrated into the READ system to facilitate microarray data analysis. Gene ontology (GO; 5) terms, computationally assigned in the FANTOM meeting (6), are also included in READ, allowing us to query by GO numbers and GO terms. More detailed functional annotation can be retrieved from the FANTOM Web server (http://fantom.gsc.riken.go.jp/).
SEARCHING READ
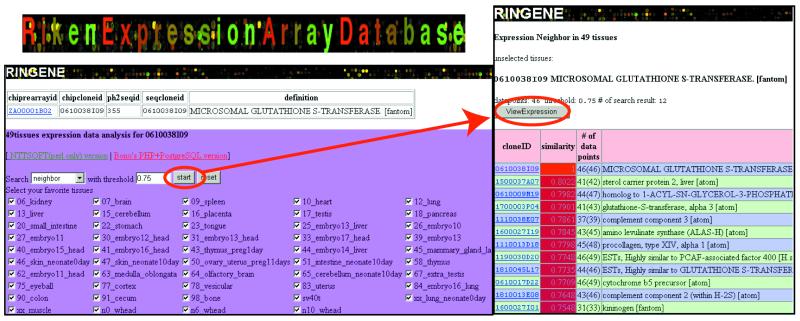
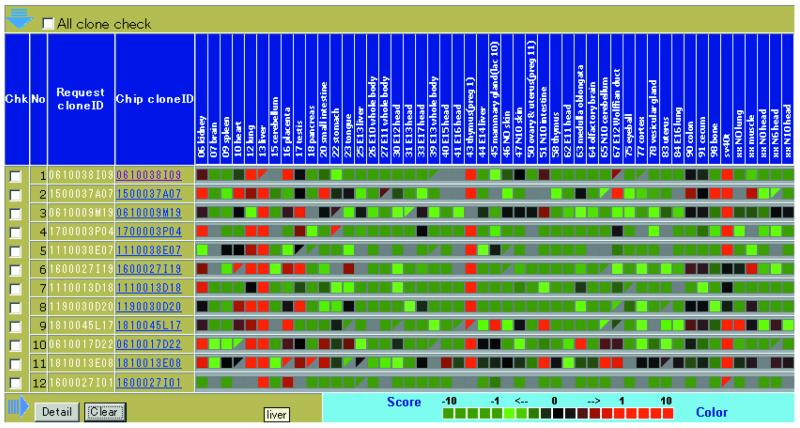
The READ system can be queried in the Web search form and by direct Web link from the TreeView application (3). A special configuration file for the direct Web link is available from the Web site. Currently, it is searchable by the RIKEN cDNA clone identifier on the chip and by keywords in the database description. It can also be searched by the log-transformed ratio of gene expression intensity in all 49 tissues. BLAST for searching a set of sequences on the chip (currently the RIKEN 19K microarray), called chipBLAST, is also available. Figure 1 shows the ‘READ integrates gene expression neighbor’ (RINGENE) query form and the result of a search. The special feature of this search tool is that similarities in the gene expression pattern of specified tissues can be dynamically calculated. Values of Pearson’s correlation coefficient, for correlations between the data of a specified clone identifier and those of all the other clones on the chip whose data are valid, are dynamically calculated to identify pairs with high correlation. Because the correlation coefficient is used to measure gene expression similarity, genes that have inverse correlation can also be queried in the RINGENE search. These inversely correlated genes could be regulated to run the program of living cells in the opposite direction. Figure 2 shows the expression profiling of the selected set of genes. This view appears by clicking the ‘ViewExpression’ button in the RINGENE view. A similarity search for gene expression patterns in specified tissues enables us to predict the function of genes, which is difficult to do just from a sequence similarity search.
Figure 1.
The RINGENE search form (left) and its result (right). The similarity in expression profile based on the Pearson’s correlation coefficient is queried. One can enter an arbitrary number (0.75 in this case) and select the tissues one would like to search for. A group of clones that shows the correlation efficient higher than the chosen value was extracted. By clicking the ‘ViewExpression’ button, the expression profile for each clone will be presented as shown in Figure 2.
Figure 2.
The expression profile of a clone set in 49 tissues chosen by the RINGENE program. The control for the microarray experiment was the RNA from 17.5 day whole embryos. Red depicts highly expressed genes, whereas green represents low expression. The color bar is shown at the bottom of the figure. Blank entries represent data missing after the PRIM filtration. All the experiments were done in duplicate and the triangles show cases where only a single result was extracted from the duplicates after the filtration.
DATA AVAILABILITY
Although users can interactively analyze all gene expression patterns on the Web, some users might prefer to download the data and analyze them locally. To do this, users can request a concatenated version of all data, including sequences, functional annotation of cDNA clones and log-transformed ratios of gene expression by sending an email to read@gsc.riken.go.jp.
FUTURE DIRECTIONS
Further microarray data from our group and other groups using the RIKEN mouse cDNA microarray will be deposited in the READ database and will be made publicly available. The READ system will include a laboratory information management system for microarray experiments capable of extensive use in the working research laboratory.
CITING READ
The following citation format is suggested when referring to READ: READ, Genome Exploration Research Group, Genomic Sciences Center, Yokohama Institute, RIKEN, Yokohama, Kanagawa, Japan (http://read.gsc.riken.go.jp/), and also citing the paper by Miki et al. (2).
Acknowledgments
ACKNOWLEDGEMENTS
We thank RIKEN Genome Exploration Research group members for the cDNA clones preparation and chip team members for the production of the expression profiling data. This study was supported by a Research Grant for the RIKEN Genome Exploration Research Project from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government and by ACT-JST (Research and Development for Applying Advanced Computational Science and Technology) of the Japan Science and Technology Corporation (JST).
REFERENCES
- 1.The RIKEN Genome Exploration Research Group Phase II Team and The FANTOM Consortium (2001) Functional annotation of a full-length mouse cDNA collection. Nature, 409, 685–690. [DOI] [PubMed] [Google Scholar]
- 2.Miki R., Kadota,K., Bono,H., Mizuno,Y., Tomaru,Y., Carninci,P., Itoh,M., Shibata,K., Kawai,J., Konno,H. et al. (2001) Delineating developmental and metabolic pathways in vivo by expression profiling using the RIKEN set of 18,816 full-length enriched mouse cDNA arrays. Proc. Natl Acad. Sci. USA, 98, 2199–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisen M.B., Spellman,P.T., Brown,P.O. and Botstein,D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA, 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kadota K., Miki,R., Bono,H., Shimizu,K., Okazaki,Y. and Hayashizaki,Y. (2001) Preprocessing implementation for microarray (PRIM): an efficient method for processing cDNA microarray data. Physiol. Genomics, 4, 183–188. [DOI] [PubMed] [Google Scholar]
- 5.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T. et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quackenbush J. (2000) Viva la revolution! A report from the FANTOM meeting. Nature Genet., 26, 255–256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Although users can interactively analyze all gene expression patterns on the Web, some users might prefer to download the data and analyze them locally. To do this, users can request a concatenated version of all data, including sequences, functional annotation of cDNA clones and log-transformed ratios of gene expression by sending an email to read@gsc.riken.go.jp.