Abstract
Early-life malnutrition increases adult disease risk in humans, but the causal changes in gene regulation, signaling, and metabolism are unclear. In the roundworm Caenorhabditis elegans, early-life starvation causes well-fed larvae to develop germline tumors and other gonad abnormalities as adults. Furthermore, reduced insulin/IGF signaling during larval development suppresses these starvation-induced abnormalities. How early-life starvation and insulin/IGF signaling affect adult pathology is unknown. We show that early-life starvation has pervasive effects on adult gene expression which are largely reversed by reduced insulin/IGF signaling following recovery from starvation. Early-life starvation increases adult fatty-acid synthetase fasn-1 expression in daf-2 insulin/IGF signaling receptor-dependent fashion, and fasn-1/FASN promotes starvation-induced abnormalities. Lipidomic analysis reveals increased levels of phosphatidylcholine in adults subjected to early-life starvation, and supplementation with unsaturated phosphatidylcholine during development suppresses starvation-induced abnormalities. Genetic analysis of fatty-acid desaturases reveals positive and negative effects of desaturation on development of starvation-induced abnormalities. In particular, the ω3 fatty-acid desaturase fat-1 and the Δ5 fatty-acid desaturase fat-4 inhibit and promote development of abnormalities, respectively. fat-4 is epistatic to fat-1, suggesting that arachidonic acid–containing lipids promote development of starvation-induced abnormalities, and supplementation with ARA enhanced development of abnormalities. This work shows that early-life starvation and insulin/IGF signaling converge on regulation of adult lipid metabolism, affecting stem-cell proliferation and tumor formation.
Keywords: L1 arrest, diapause, starvation, thrifty phenotype, DOHaD, phosphatidylcholine, arachidonic acid
Introduction
The Dutch Hunger Winter provides a tragic example of the consequences of early-life starvation on adult disease risk. Malnutrition in utero followed by replete nutrition during development results in decreased glucose tolerance, altered lipid metabolism, and higher incidence of cancer (Roseboom et al. 2001; Painter et al. 2006; Hughes et al. 2009). Numerous examples in humans and other animals demonstrate a similar correlation between early-life nutrient stress and adult pathology (Wadhwa et al. 2009; Wang et al. 2016; Hoffman et al. 2017). These observations suggest a critical period in early development when nutrient availability can persistently alter gene regulation and metabolism, potentially resulting in adult disease. However, signaling pathways and gene-regulatory mechanisms underlying such life-long effects are largely unknown.
The roundworm Caenorhabditis elegans has robust starvation responses, and the signaling and gene-regulatory pathways controlling its metabolism are widely conserved, making it a valuable animal model for developmental consequences of nutrient stress (Braeckman et al. 2009; Watts and Ristow 2017; Baugh and Hu 2020). When C. elegans larvae hatch in the absence of food, development remains arrested in the first larval stage (L1 arrest or L1 diapause; Baugh 2013). Larvae in L1 arrest continue foraging, and they initiate post-embryonic development after feeding. They can survive starvation during L1 arrest for weeks, but we found that many individuals develop germline tumors and other gonad abnormalities during early adulthood after being starved for a week as larvae and then fed ad libitum (Jordan et al. 2019). These abnormalities are pathological in that they limit reproductive success (Jordan et al. 2019). We also reported that reducing insulin/IGF signaling (IIS) with RNAi against the sole known insulin/IGF receptor (InsR) in C. elegans, daf-2, during larval development after starvation suppresses starvation-induced abnormalities in adults. Reducing DAF-2/InsR signaling promotes nuclear localization and activation of the transcription factor DAF-16/FoxO (Kenyon et al. 1993; Lin et al. 1997; Ogg et al. 1997; Henderson and Johnson 2001), and daf-16 is required for reduced IIS to suppress starvation-induced abnormalities (Jordan et al. 2019). Our companion manuscript shows that early-life starvation and IIS increase Wnt signaling to promote starvation-induced abnormalities (Shaul et al. co-submitted). However, other mechanisms by which early-life starvation promotes and daf-16 inhibits development of starvation-induced abnormalities are unknown.
Here we integrate transcriptomic and lipidomic analyses with genetic and dietary interventions to interrogate the consequences of early-life starvation and reduced IIS in C. elegans. We report that extended starvation during L1 arrest increases relative mRNA expression levels of a couple thousand genes in adults, and that these changes are largely reversed by reduced IIS during larval development. We confirmed this pattern of regulation for the fatty-acid synthetase (FASN) fasn-1/FASN, and we show that fasn-1 promotes development of starvation-induced abnormalities. Lipidomic analysis revealed that phosphatidylcholine levels are elevated in adults subjected to early-life starvation and that IIS regulates phosphatidylcholine metabolism. Dietary supplementation with unsaturated phosphatidylcholine during larval development suppressed starvation-induced abnormalities, suggesting functional relevance of phosphatidylcholine metabolism. Furthermore, genetic epistasis analysis and dietary supplementation suggest that arachidonic acid (ARA), or one of its derivatives, promotes starvation-induced abnormalities. This work connects early-life starvation and IIS to lipid metabolism in adults, elucidating developmental origins of adult disease.
Materials and methods
Nematode strains
The following worm strains were used in this study: N2 (Bristol), GG14 fasn-1(g14) I, IG348 fasn-1(fr8) I; frIs7 [nlp-29p::gfp + col-12p::DsRed] IV, RB1031 fat-4(ok958) IV, BX24 fat-1(wa9) IV, BX52 fat-4(wa14) IV; fat-1(wa9) IV, IG763 frEx266 [fasn-1p::gfp], VC788 fat-3(ok1126) IV, BX107 fat-5(tm420) V, BX106 fat-6(tm331) IV, and BX153 fat-7(wa36) V. Worms were fed Escherichia coli strains OP50 and HT115.
Gonad abnormalities assay
The assay was performed as described (Jordan et al. 2019). In brief, animals were given 72 h to develop after an egg-lay by well-fed parents (and well-fed for several generations prior). Eggs were released with sodium hypochlorite treatment, allowed to hatch and arrest development in virgin S-basal (no ethanol or cholesterol), and then recovered ad libitum on E. coli HT115 after 1 (control) or 8 (starved) days. After 72 h, animals were scored for gonad abnormalities as described (Jordan et al. 2019). It is important to note that HT115 was used for food in all abnormality assays since OP50 does not produce abnormalities at as high of a frequency.
RNAi
RNAi food was prepared and used for feeding on plates during recovery from starvation as described (Jordan et al. 2019). In brief, frozen, single-thaw aliquots of HT115 RNAi were seeded onto NGM plates containing 25 µg/ml carbenicillin and 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and allowed to grow at room temperature overnight.
mRNA-seq sample preparation
Wild-type starvation cultures were prepared as described above. Roughly 1,000 animals were recovered on two large NGM plates (25 µg/ml carbenicillin and 1 mM IPTG) seeded with empty vector (EV) or daf-2 RNAi HT115 E. coli. Worms were cultured at 20°C until egg-laying onset (∼52 h for control animals and ∼68 h for starved animals). Worms were quickly washed twice and flash frozen in liquid nitrogen.
RNA was extracted using 1 ml TRIzol Reagent (Invitrogen# 15596026) according to the manufacturer's protocol except 100 µl of acid-washed sand (Sigma# 27439) was added to each sample at the beginning of the protocol to aid homogenization. RNA was eluted in nuclease-free water and stored at −80°C until further use.
In the initial mRNA-seq experiment, libraries were prepared for sequencing using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs: E7530) with 500 ng of starting RNA per library and 12 cycles of polymerase chain reaction (PCR). Libraries were sequenced using Illumina HiSeq 4000 to obtain 50 bp single-end reads. In the subsequent fasn-1(g14) mRNA-seq experiment, libraries were prepared using the NEBNext Ultra II RNA Library Prep Kit for Illumina (New England Biolabs# E7770) starting with 1 µg total RNA per library and eight cycles of PCR. Libraries were sequenced using Illumina NovaSeq 6000 to obtain 50 bp paired-end reads.
mRNA-seq analysis
In the initial mRNA-seq experiment, Bowtie version 1.1.2 was used to map single-end reads to the WS210 of the worm genome with some WS220 features mapped to WS210 using the command –best –chunkmbs 256 -k 1 -m 2 -S (Langmead et al. 2009). HTSeq version 0.6.0 was used to count the reads (Anders et al. 2015). edgeR was used on count tables for differential expression analysis (Robinson et al. 2010). The original count table was filtered to contain those annotated as genes, reducing the number of genes to 22,581. Genes were further filtered by including only genes with counts per million (CPM) > 1 across three libraries. This resulted in 12,894 genes. edgeR exact test was used for four pair-wise comparisons, and edgeR GLM was used to find genes that changed across multiple conditions. The trimmed mean of M values (TMM) method was used to normalize for RNA composition. Tag-wise dispersions were used to test for differentially expressed genes.
In the subsequent fasn-1(g14) mRNA-seq experiment, Bowtie2 2.3.3.1 was used to map paired-end reads to the WS273 genome with the parameter k set to 1. HTSeq 0.12.4 was used to count reads. edgeR 3.28.1 was used to filter and normalize read counts and perform differential expression analysis. Detected genes were considered those with an expression level of at least one CPM in at least two libraries. Library sizes were recomputed after filtering out lowly expressed genes. The TMM method was used to normalize for RNA composition. Tag-wise dispersions were used to test for differentially expressed genes between wild-type and the fasn-1 mutant.
Gene ontology term enrichment analysis
Differentially expressed genes were intersected using “gplots” and “venn” in R. The intersection lists were analyzed for enrichment of Gene Ontology (GO) terms using Gorilla (Eden et al. 2009). The 12,891 detected genes (criteria detailed above) were used as a background set in the first mRNA-seq experiment. In the fasn-1 mRNA-seq experiment, a 13,757 gene background set was used. The most significantly enriched GO component terms are plotted in Figs. 1f and 2i. See Supplementary Files 2 and 4 for complete results.
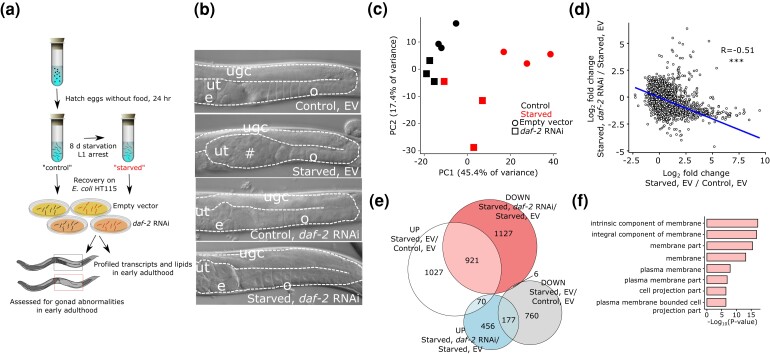
Fig. 1.
Early-life starvation and IIS have similar effects on adult gene expression. a) Diagram of the experimental design used throughout. Embryos are collected from well-fed mothers on the first day of egg laying and cultured without food they hatch and arrest development as L1-stage larvae (L1 arrest). “Control” animals are starved in L1 arrest for about 12 h, synchronizing the population, and “starved” animals were starved for 8 days. Both were recovered on EV or daf-2 double-stranded RNA-expressing (RNAi) E. coli HT115. Samples were collected for mRNA-seq, lipidomic, and phenotypic analyses at egg-laying onset. Because starved worms develop slower than controls, they were allowed an additional 16 h to reach egg-laying onset. b) Representative differential-interference contrast images of control and starved worms at egg-laying onset recovered with EV or daf-2 RNAi food. Eight days L1 starvation causes gonad abnormalities when worms are recovered on EV food, but recovery on daf-2 RNAi food suppresses abnormalities. Gonads are outlined with white dashed lines. ugc = undifferentiated germ cells, ut = uterus, o = oocytes, e = embryos, # = gonad abnormality. See Jordan et al. (2019) for additional characterization. c) The first two components from principal component analysis of mRNA-seq results are plotted. Adult samples were collected at egg-laying onset as depicted in (a). d) Log2 fold changes for all detected genes are plotted for the effect of starvation with EV and the effect of daf-2 RNAi in starved worms. ***P < 0.001; Pearson test. e) A Venn diagram of differentially expressed genes (FDR < 0.1) is plotted, intersecting the effects of early-life starvation and IIS on adult gene expression. f) GO component terms enriched (FDR < 10−3) among the 921 genes that are up in starved worms and down with daf-2 RNAi. Complete results including process and function GO terms can be found in Supplementary File 2.
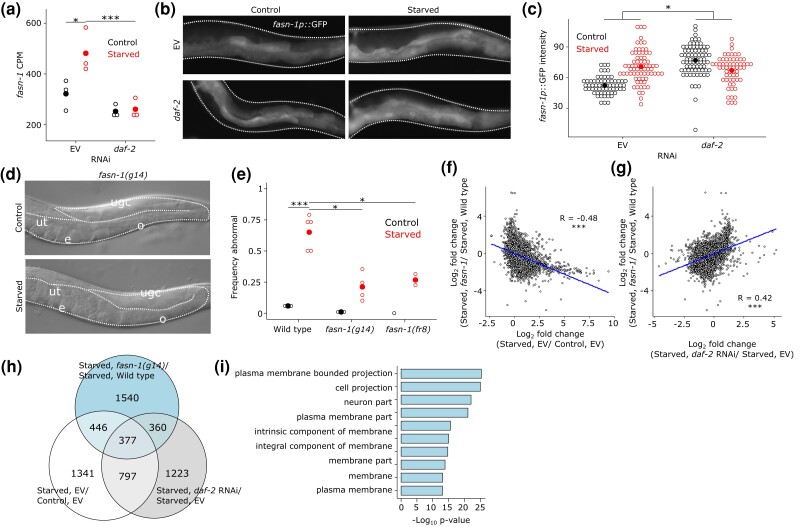
Fig. 2.
fasn-1/FASN mediates effects of early-life starvation and IIS on starvation-induced abnormalities and adult gene expression. a) fasn-1 mRNA-seq read CPM in control and starved worms recovered on EV or daf-2 RNAi food. *FDR < 0.05 and ***FDR < 0.001. b) Representative images of the posterior region of adult worms (72 h recovery from L1 arrest) expressing fasn-1p::gfp. Control and starved worms recovered on either EV or daf-2 RNAi food are shown. Worms are outlined with white-dotted lines. Expression is prominent in the intestine. c) Open dots represent the average intensity of fasn-1p::gfp in posterior intestine from two control replicates and three starved replicates. Closed dots indicate the mean. *Pinteraction < 0.05; two-way ANOVA. d) Representative images of control and starved fasn-1(g14) mutant worms. Gonads are outlined with white-dotted lines. ugc = undifferentiated germ cells, ut = uterus, e = embryos, and o = oocytes (compare with Fig. 1b). e) Suppression of starvation-induced gonad abnormalities with disruption of fasn-1. Open dots indicate the frequency per replicate of starved animals with gonad abnormalities after 8 days L1 arrest (starvation and recovery at 20°C) where at least 40 individual animals were scored. Closed dots indicate means. *P < 0.05, ***P < 0.001; unpaired t-test. f) Log2 fold changes for all detected genes are plotted for the effect of starvation on EV and the effect of fasn-1 mutation in starved worms. g) Log2 fold changes for all detected genes are plotted for the effect of daf-2 RNAi in starved worms and the effect of fasn-1 mutation in starved worms. f and g) ***P < 0.001; Pearson test. h) A Venn diagram of differentially expressed genes (FDR < 0.1) is plotted, intersecting the effects of fasn-1, early-life starvation, and IIS on adult gene expression. i) GO component terms enriched (FDR < 10−11) among the 2,724 differentially expressed genes in fasn-1(g14) compared with wild type (compare with Fig. 1f). Complete results including process and function GO terms can be found in Supplementary File 4.
fasn-1p::gfp microscopy and quantification
fasn-1p::gfp worms (Lee et al. 2010), cultured and recovered on RNAi as described, were mounted on 4% noble agar pads and paralyzed with 50 mM levamisole. Images of posterior intestines were captured using an AxioImager compound (Zeiss) microscope at 400× with a fixed exposure time. fasn-1p::gfp was quantified by outlining and assessing the average pixel intensity of the region of interest in ImageJ.
Lipidomics sample preparation
Animals were cultured and collected as described for mRNA-seq. Samples were normalized to ∼200 µl aqueous volume. About 200 µl of MeOH-containing stable-isotope-labeled internal standards were added to the samples; samples were sonicated, then 667 µl methyl tert-butyl ether (MTBE) was added to each sample for lipid extraction. Samples were shaken for 1 h at 50°C to extract lipids, followed by a 5-min centrifugation at 10,000 RCF to separate phases. Four hundred microliters of the MTBE phase were removed and then taken within a few microliters of dryness in a glass autosampler under a stream of nitrogen. Lipids were then resuspended in 100 µl 95%/5% v/v MeCN/water.
Targeted lipidomics analysis
Aliquots of each sample pool (2 µl) were injected several times using a Hydrophilic Interaction Liquid Chromatography-Tandem Mass Spectrometry (HILIC-MS/MS) “screening” method (Waters Acquity UPLC-Xevo TQS) which targets over 500 lipid species including the following classes: cholesterol (ChoE), DAG, TAG, SM, PC, LPC, PE, PG, PI. For those lipids which showed consistent signals after three injections, a single positive ion and a single negative ion injection targeting those signals was performed. The study pool QC was analyzed four times and each sample analyzed once with 2 μl by HILIC-MS/MS in each mode. Mobile phase A was 95%:5% v/v MeCN:water with 10 mM ammonium acetate, while mobile phase B was 50%:50% v/v MeCN:water with 10 mM ammonium acetate. The separation used a 2.1 mm × 100 mm BEH HILIC Amide column (Waters), with a flow rate of 0.6 ml/min and a column temperature of 45°C. Lipids were separated in the HILIC mode using a gradient from 0.1% to 20% mobile phase B in 2 min, a ramp to 80% B at 5 min, followed by 3 min re-equilibration at 0.1% B. Electrospray ionization in the positive mode (3.5 kV) or negative mode (2.0 kV) was used for sample introduction, and molecule-specific precursor and product ion MRM transitions were used to quantify lipids.
Skyline v4.2 was used to measure peak area for each MRM chromatogram, and to ratio each native analyte to an appropriate internal standard. The SPLASH Lipidomix mass spectrometry standard kit (Avanti: 330707) supplemented with deuterated free fatty-acid internal standards (d8-ARA, d5-DHA, d2-palmitic acid, and d14-palmitoleic acid) was used for peak area normalization.
The sum of all lipids in the class (LPC, PC, TAG, SM, and PG) was calculated, and the ratio of the maximum sum of all samples and each individual sample was calculated as a normalization factor. Each lipid was multiplied by this normalization factor to scale each individual lipid measurement to “total” lipid quantity prior to Log2 normalization and statistical analysis in R.
Phospholipid supplemented plate preparation
Stock solutions of egg phosphatidylcholine (Sigma: P3556), lysophosphatidylcholine (Sigma: L4129), phosphatidylethanolamine (Sigma: P7943), and crude phosphatidylinositol from soybeans (Sigma: P6636) were made with 100% EtOH, further diluted in S-basal, applied to NGM plates, allowed to dry for ∼2 h, and then seeded with HT115 E. coli. Specific PC species 20:0 (Avanti: 850368P), 20:4n-6 (Avanti: 850397C), 18:1n-7 (Avanti: 790626C), and 18:1n-9 (Avanti: 850375P) were applied similarly.
Diacylglycerol and choline supplementation
DAG 18:1 (Avanti: 800811O) was diluted in 100% EtOH, further diluted in S-basal, and applied to plates at a final concentration of 120 µM. Choline (Sigma: P7527) was diluted in water and added to NGM plates to make the final concentration 30 mM. Plates were allowed to dry for ∼2 h and then seeded with HT115 E. coli.
Free fatty-acid supplementation
Stock solutions (10 mM) of dihomo-gamma-linoleic acid (DGLA; Cayman: 90230), eicosatetraenoic acid (ETA; Matreya: 1042), ARA (Cayman: 90010), and eicosapentaenoic acid (EPA; Cayman: 90110) were prepared in DMSO after evaporation of proprietor's solvent. Stock solutions were diluted in EV HT115 E. coli to a final concentration of 600 µM and then seeded onto carbenicillin and IPTG plates. Plates were protected from light and bacterial lawns were allowed to grow overnight prior to being plated with wild-type animals starved in L1 arrest for 6 days.
Results
Early-life starvation has pervasive effects on adult gene expression
We characterized the effects of early-life starvation and reduced IIS during larval development on adult gene expression using mRNA-seq of whole worms. In our two-factor design (Fig. 1a), “starved” worms were cultured without food (E. coli) as L1-stage larvae (L1 arrest) for 8 days, and “control” worms were starved overnight for synchronization. Both populations were fed ad libitum with EV (negative control) or daf-2/InsR RNAi food (reduced IIS). RNAi was used rather than a daf-2 mutant so that the results would not be confounded by daf-2 function during starvation, instead disrupting daf-2 later in fed, developing larvae. Upon reaching adulthood, animals were collected for transcript profiling and scored for the presence of germ-cell tumors, differentiated uterine masses, and other gonad abnormalities (Fig. 1b; Jordan et al. 2019).
The results demonstrate similar effects of early-life starvation and IIS on adult gene expression. Principal component analysis revealed a major effect of L1 starvation in the first component and a secondary effect of IIS in the second component (Fig. 1c). Notably, the effect of early-life starvation on adult gene expression appeared smaller with daf-2 RNAi, consistent with suppression of starvation-induced abnormalities (Fig. 1b; Jordan et al. 2019). There was a significant negative correlation between the effects of starvation with EV and effects of daf-2 RNAi in starved worms (Fig. 1d), showing that reduced IIS during larval development suppresses starvation-induced gene expression changes. Notably, relative increases in expression caused by early-life starvation were more prominent than decreases. Expression of 2,018 (68%) of the 2,961 genes affected by starvation was increased [false-discovery rate (FDR) < 0.1; Supplementary File 1], and strikingly, it was reduced for 921 (46%) of those 2,018 genes when starved worms were recovered on daf-2 RNAi (P = 2.17e−274; Fig. 1e; Supplementary File 1). These results show that for most genes affected by extended L1 starvation, their relative expression is increased in adults, and that these increases are largely suppressed by reduced IIS during development. GO term analysis of genes displaying this pattern (up in previously starved worms and down with daf-2 RNAi) identified a variety of “functions” and “processes” apparently affected by early-life starvation (Supplementary File 2). However, enrichment of “component” terms was more coherent, with the most significant terms relating to plasma membranes (Fig. 1f). These results reveal that early-life starvation and IIS converge on the regulation of many genes in adult worms, and they suggest that membrane biology is affected, potentially contributing to the pathological effects of early-life starvation.
Early-life starvation up-regulates fasn-1/FASN which promotes starvation-induced abnormalities
Given strong enrichment of genes associated with plasma membranes among those affected by early-life starvation and IIS (Fig. 1f, Supplementary File 5), we sought to identify candidate lipid or membrane-related genes that may contribute to starvation-induced abnormalities. FASN controls rate-limiting conversion of carbohydrates into free fatty acids, which are either subjected to beta-oxidation to generate energy or incorporated into phospholipid bilayers. By affecting phospholipid bilayers, FASN can affect membrane structure, membrane permeability, immune response, and gene expression (Kimura et al. 2020). In addition, FASN is up-regulated in a variety of cancers and is a potential chemotherapeutic target (Flavin et al. 2010; Papaevangelou et al. 2018; Che et al. 2019). Notably, fasn-1/FASN, the sole C. elegans FASN homolog, displays increased expression in adult worms starved as L1 larvae (Fig. 2a). Furthermore, increased fasn-1 expression is dependent on daf-2/InsR during recovery, making it an ideal candidate. We attempted to confirm this pattern of regulation using a fasn-1p::gfp transcriptional reporter. This multicopy, extrachromosomal array-based reporter is expressed throughout the intestine in early adulthood, and its expression is significantly elevated by prior L1 starvation, except when larvae are recovered from starvation on daf-2 RNAi food (Fig. 2b, c). Reporter analysis suggested that fasn-1 expression is elevated by daf-2 RNAi in control conditions, which we did not see with mRNA-seq, but there are a number of caveats in quantitative reporter gene analysis that could account for this discrepancy. For example, the reporter may lack critical regulatory elements and mRNA stability could be affected by the non-native 3′ UTR. We believe that the salient features of these data are the increase in reporter expression following starvation without RNAi and absence of that increase with daf-2 RNAi. Together with the mRNA-seq data, these results suggest that fasn-1 transcription is persistently activated in worms subjected to early-life starvation in IIS-dependent fashion.
We sought to disrupt fasn-1/FASN function to test the hypothesis that it promotes starvation-induced abnormalities. Disrupting gene function with RNAi is better than mutants in this context since RNAi uncouples effects of function during starvation and development. However, fasn-1 RNAi resulted in larval growth arrest, and gonad abnormalities could not be assayed, so we instead tested viable fasn-1 hypomorphs. fasn-1 mutant alleles g14 and fr8 both reduced starvation-induced abnormalities (Fig. 2d, e). These results suggest that up-regulation of fasn-1 promotes starvation-induced abnormalities, supporting the hypothesis that early-life starvation alters adult lipid metabolism to cause pathology.
Given the large number of genes whose adult expression was affected by early-life starvation and IIS, we wondered if fasn-1 is one among many possible effectors or if it has a relatively substantial role in promoting starvation-induced abnormalities. To address this, we performed mRNA-seq of wild-type and a fasn-1 mutant at egg-laying onset in worms that had been starved for 8 days as L1 larvae. A total of 2,724 genes were differentially expressed (FDR < 0.1; Supplementary File 3), demonstrating widespread effects of fatty-acid synthesis on gene expression. Furthermore, there was a significant negative correlation between the effects of early-life starvation and mutation of fasn-1 on adult gene expression (Fig. 2f). In addition, there was a significant positive correlation between the effects of reduced IIS and mutation of fasn-1 on expression (Fig. 2g). Consistent with these correlations, 377 genes were significantly affected by starvation, IIS, and fasn-1 (P = 2.08e−64; Fig. 2h). Finally, GO component term enrichments for genes affected by fasn-1 are very similar to those for the genes that are up-regulated following starvation and down-regulated by daf-2/InsR RNAi, again highlighting membrane biology (compare Fig. 2i to 1f, Supplementary File 4). Together these results suggest that alteration of fasn-1 expression (and therefore activity) contributes substantially to the effects of early-life starvation and IIS on adult gene expression, consistent with it having a critical role in development of starvation-induced abnormalities.
Early-life starvation and insulin/IGF signaling affect adult phosphatidylcholine levels
We used targeted lipidomic analysis in whole worms to address our hypothesis that early-life starvation and IIS affect lipid metabolism in adults. We used the same experimental design as for mRNA-seq (Fig. 1a), except we also included double RNAi of daf-2/InsR and daf-16/FoxO. We previously showed the reliability of this particular perturbation in this context (Jordan et al. 2019), which is not confounded by disruption of gene function during starvation as would occur with mutations. We measured abundance of nine lipid classes including diacylglycerols, free fatty acids, lysophosphatidylcholines, phosphatidylcholines, phosphatidylethanolamines, phosphoglycerides, phosphatidylinositols, sphingomyelins, and triacylglycerols. Because there are many molecular species per class (with different hydrocarbon chain lengths and degrees of saturation), we initially pooled all species for each class for class-level analysis (Fig. 3a). Phosphatidylcholine, free fatty acid, lysophosphatidylcholine, phosphoglyceride, and triacylglyceride each displayed significant variance across all conditions in our data set, with diacylglycerol and sphingomyelin marginally significant. These results reveal pervasive effects of early-life starvation and IIS on adult lipid metabolism.
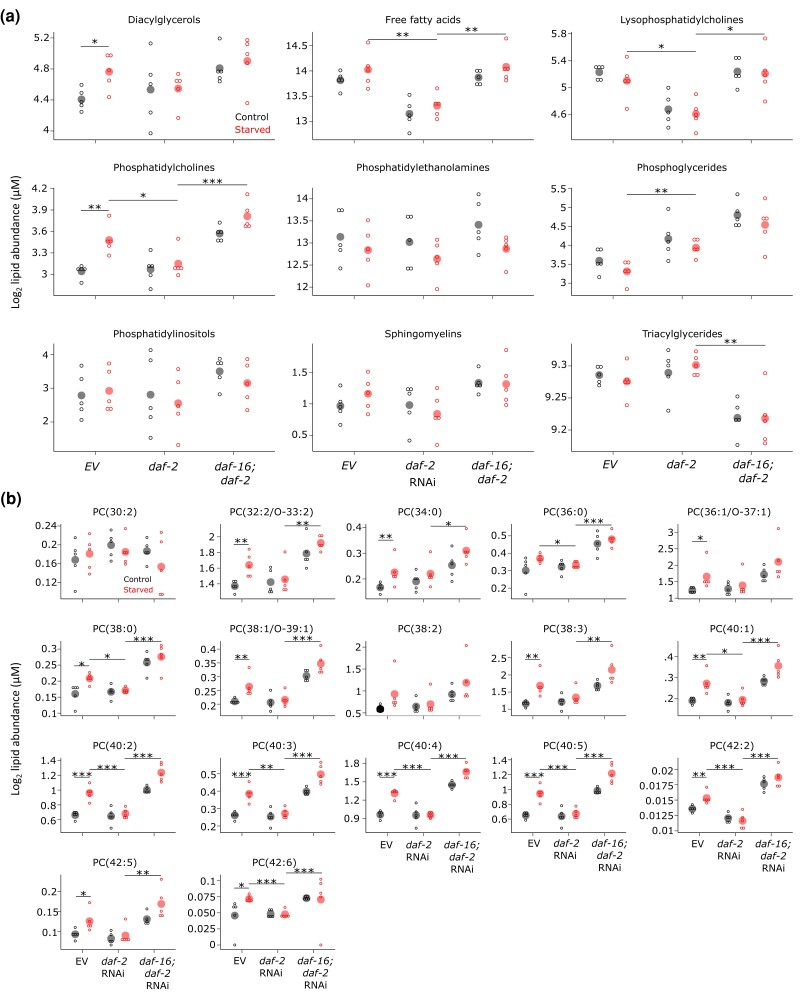
Fig. 3.
Early-life starvation and IIS have similar effects on the adult lipid profile including extensive effects on abundance of phosphatidylcholine species. a) Pooled lipid abundance for each class of targeted lipids is plotted for the six conditions assayed. Open dots indicate sum of all species within each class for a biological replicate. Closed dots indicate the mean. b) Abundance of individual phosphatidylcholine species is plotted. Open dots indicate abundance in an individual biological replicate. Closed dots indicate the mean. a and b) Lipid levels normalized by total lipid. *P < 0.05, **P < 0.01, ***P < 0.001; lines connecting two conditions indicate significance in an unpaired t-test. Five pair-wise tests were considered here: control vs starved in each RNAi condition, EV vs daf-2 in starved worms, and daf-2 vs daf-16; daf-2 in starved worms. See Supplementary File 6 for complete statistical analysis.
Continuing with class-level analysis, we dissected the effects of early-life starvation, daf-2/InsR, and daf-16/FoxO. Phosphatidylcholine showed the most significant effects between pairs of individual conditions (Fig. 3a). For example, phosphatidylcholine was significantly elevated in adults that had been starved as L1 larvae, and this increase was suppressed with daf-2 RNAi, mirroring the effect of reduced IIS on starvation-induced abnormalities and starvation-induced changes in gene expression (Fig. 1). In addition, daf-16 RNAi suppressed the effects of daf-2 RNAi, significantly increasing phosphatidylcholine abundance. This result suggests that reduced IIS acts through DAF-16/FoxO. Qualitatively similar patterns were observed among the other lipid classes (though in some cases, the signs are reversed), with lysophosphatidylcholine and free fatty acids displaying the most significance (Fig. 3a). These results suggest that early-life starvation and IIS converge on metabolism of phosphatidylcholine as well as lysophosphatidylcholine and free fatty acids in adults.
Individual species of each lipid class generally showed similar patterns of abundance as seen at the class level (Figs. 3b, Supplementary Figs. 1 and 2). Fourteen out of 17 individual phosphatidylcholine species were significantly increased by L1 starvation (Fig. 3b, Supplementary File 6). Remarkably, this effect of starvation was absent for all 14 with daf-2/InsR RNAi. Furthermore, the effects of daf-2 RNAi required daf-16 function, with 14 phosphatidylcholine species displaying increased levels with daf-16 RNAi. These results show that the patterns observed for the entire family of phosphatidylcholine species (Fig. 3a) are evident for most individual species, reflecting coherent behavior of the family. Also consistent with the results of class-level analysis, individual species of diacylglycerol and free fatty acids appeared to increase in abundance in adults starved as larvae, and species of lysophosphatidylcholine appeared to decrease (Supplementary Fig. 1). These effects were also apparently suppressed by daf-2 RNAi in daf-16/FoxO-dependent fashion. Notably, not all these individual effects were statistically significant, but many were (Supplementary File 6), and they display strong concordance within class, supporting the significance of these effects. These results affirm our conclusion from class-level analysis that early-life starvation and IIS converge on metabolic regulation of phosphatidylcholine, lysophosphatidylcholine, and free fatty acids.
Closer examination of individual phosphatidylcholine species suggested that unsaturated species are particularly affected by early-life starvation. All 8 phosphatidylcholine species with at least 40 carbons in their 2 fatty-acid tails were significantly increased in worms starved as larvae (Fig. 3b, Supplementary File 6). Notably, none of these species are fully saturated, though the identity of the two tails cannot be determined from these data, so individual tails may be saturated. Furthermore, six of these eight, and none of the other phosphatidylcholine species, showed a significant interaction between starvation and daf-2 RNAi in a stringent two-factor test (Supplementary File 6). Together these results suggest that early-life starvation and IIS affect metabolic regulation of long-chained, unsaturated phosphatidylcholine.
Unsaturated phosphatidylcholine supplementation suppresses starvation-induced abnormalities
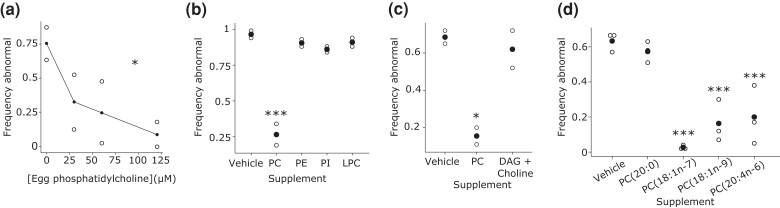
Lipidomic analysis suggested that unsaturated phosphatidylcholine is involved in the development of starvation-induced abnormalities. Dietary supplementation of previously starved worms during development with phosphatidylcholine extracted from chicken egg suppressed starvation-induced abnormalities in dose-dependent fashion (Fig. 4a). A similar effect was not seen with chicken-egg phosphatidylethanolamine or lysophosphatidylcholine, nor with soybean phosphatidylinositol (Fig. 4b), demonstrating specificity of phosphatidylcholine. Phosphatidylcholine can be synthesized from diacylglycerol and choline in the cytidine-diphosphate choline pathway (Lands 1958; Moessinger et al. 2014). Supplementation with diacylglycerol and choline had no effect on starvation-induced abnormalities (Fig. 4c) suggesting deficit in these precursors is not a cause of abnormalities. Phospholipids extracted from chicken eggs comprise a complex mixture, including saturated and unsaturated species. We therefore used individual, synthetic phosphatidylcholine species that varied in degrees of saturation for supplementation. Three different unsaturated phosphatidylcholine species suppressed starvation-induced abnormalities, but a saturated form did not (Fig. 4d). Notably, the saturated species had 20 carbons as did one of the unsaturated species (ARA), suggesting that desaturation, rather than the number of carbon atoms, is critical for suppression of starvation-induced abnormalities. However, differences in solubility or bioavailability with supplementation may also be factors. Since phosphatidylcholine levels were elevated in whole worms with starvation-induced abnormalities (Fig. 3a, b), it was surprising that phosphatidylcholine suppressed rather than enhanced starvation-induced abnormalities (see Discussion). Nonetheless, these results suggest a specific, functional role of unsaturated phosphatidylcholine in development of starvation-induced abnormalities.
Fig. 4.
Dietary supplementation of unsaturated phosphatidylcholine during larval development suppresses starvation-induced abnormalities. a) A chicken-egg phosphatidylcholine supplementation dose–response curve for gonad abnormalities in starved worms is plotted. *P < 0.05; one-way ANOVA. b) Frequency of gonad abnormalities in starved worms when supplemented with equimolar doses (120 µM) of different phospholipids is plotted. c) Frequency of gonad abnormalities in starved worms when supplemented with diacylglycerol and choline. d) Frequency of gonad abnormalities in starved worms when supplemented with phosphatidylcholine with specific fatty-acid tails. a–d) Open dots indicate a biological replicate where at least 40 individual animals were scored. Closed dots indicate the mean. b–d) *P < 0.05, ***P < 0.001; unpaired t-test vs vehicle. DAG, diacylglycerol; LPC, lysophosphatidylcholine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol.
Fatty-acid desaturation modifies starvation-induced abnormalities
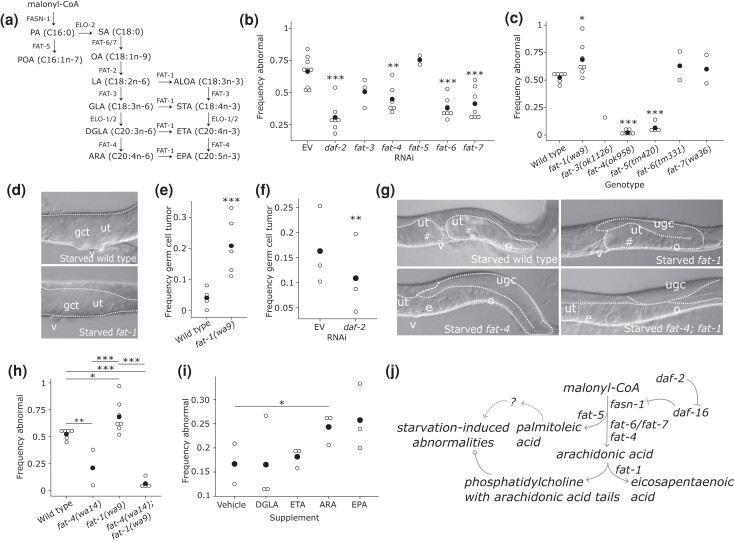
Lipidomic analysis and phospholipid supplementation suggest that unsaturated fatty-acid tails on phosphatidylcholine may be involved in the development of starvation-induced abnormalities. There are seven fatty-acid desaturases in C. elegans that catalyze largely distinct biochemical reactions (Fig. 5a; Watts and Ristow 2017), and we performed genetic analysis of them to test this hypothesis. There is an early branch in the pathway, where palmitic acid (C16:0; PA) is either elongated or desaturated by the Δ9 desaturase FAT-5. We found that fat-5 RNAi by feeding during larval development after L1 starvation did not have a significant effect on starvation-induced abnormalities (Fig. 5b). However, mutation of fat-5 suppressed abnormalities (Fig. 5c), potentially reflecting greater disruption of gene function than RNAi. This result suggests a role of the FAT-5 product palmitoleic acid (C16:1n-7; POA), its derivatives, or phospholipids containing either, in development of starvation-induced abnormalities. RNAi of either fat-6 or fat-7, both of which also encode a Δ9 desaturase but with different substrate specificity than FAT-5, also suppressed starvation-induced abnormalities (Fig. 5b), though mutation of fat-6 or fat-7 did not (Fig. 5c). Genetic redundancy could account for the lack of mutant phenotype, with RNAi potentially affecting both genes given extensive similarity in their sequences. Together these results suggest that both branches of the pathway from PA affect development of starvation-induced abnormalities. In addition, given the relatively early place in the metabolic pathway of FAT-5, FAT-6, and FAT-7 (Fig. 5a), these results are consistent with the fasn-1/FASN phenotype (Fig. 2d, e) and reduced flux through the pathway reducing penetrance of starvation-induced abnormalities.
Fig. 5.
Fatty-acid desaturation modifies starvation-induced abnormalities, implicating arachidonic acid in their development. a) A metabolic model of fatty-acid synthesis and desaturation is presented. Lipid names are abbreviated, and the number of carbon atoms and the state of saturation are given in parentheses. Enzymes responsible for each biochemical reaction are given alongside arrows. ALOA, alpha-linolenic acid; ARA, arachidonic acid; DGLA, dihomo-gamma-linolenic acid; EPA, eicosapentaenoic acid; ETA, eicosatetraenoic acid; GLA, gamma-linolenic acid; LA, linoleic acid; OA, oleic acid; PA, palmitic acid; POA, palmitoleic acid; SA, stearic acid; STA, stearidonic acid. b) Frequency of gonad abnormalities in starved worms recovered on RNAi food targeting the indicated gene. c) Frequency of gonad abnormalities in starved worms of the given mutant genotypes. d) Representative images of germ-cell tumors in starved wild-type animals and fat-1 desaturase mutants. e) Frequency of proximal germ-cell tumors in starved wild-type and fat-1(wa9) mutant worms. f) Frequency of proximal germ-cell tumors in starved fat-1(wa9) mutant worms recovered on daf-2 RNAi. g) Representative images of starved worms of the indicated genotype. Note that fat-4 desaturase mutation suppresses starvation-induced abnormalities, and that fat-4 is epistatic to fat-1. h) Frequency of gonad abnormalities in starved worms of the given mutant genotypes. i) Frequency of gonad abnormalities in wild-type worms starved for 6 days (rather than 8 days as elsewhere) and recovered on lawns supplemented with the given free fatty acids (600 µM). j) Working model describing how early-life starvation and IIS influence lipid metabolism to affect starvation-induced abnormalities. Arrowheads indicate enzymatic products and functional consequences, bars indicate suppression, and circles indicate complex interactions. Dotted lines indicate gaps in knowledge, while solid lines indicate well-known relationships and findings from this study. b, c, e, f, h, and i) Open dots indicate a biological replicate where at least 40 individual animals were scored. Closed dots indicate the mean. *P < 0.05, **P < 0.01, ***P < 0.001; unpaired t-test vs EV, wild type or vehicle. d and g) Gonads are outlined with white-dotted lines. gct = germ-cell tumor; # = gonad abnormality; ugc = undifferentiated germ cells; ut = uterus; o = oocytes; e = embryos; v = vulva.
Downstream of FAT-6 and FAT-7, the Δ6 desaturase FAT-3 catalyzes a couple of different reactions (Fig. 5a). RNAi and mutation of fat-3 suggest that disruption of its function suppresses starvation-induced abnormalities, but the results are not statistically significant (Fig. 5b, c). Further downstream, the sole known C. elegans Δ5 desaturase FAT-4 also catalyzes a couple of different reactions (Watts et al. 2002), and RNAi and mutation of fat-4 both suppressed starvation-induced abnormalities. This result suggests that the terminal, most unsaturated products of the pathway, ARA (C20:4n-6) and EPA (C20:5n-3), which cannot be synthesized in the fat-4 mutant (Watts et al. 2002), promote starvation-induced abnormalities. However, it is unclear whether one, the other, or both are involved. In addition, it could be that DGLA (C:20:3n-6) or eicosatetraenoic acid (C20:4n-3) actually suppresses starvation-induced abnormalities.
The ω3 desaturase FAT-1 catalyzes a variety of reactions in the pathway, including the conversion of ARA into EPA (Watts et al. 2002). A fat-1 RNAi clone was not available in either commercially available RNAi library, but mutation of fat-1 significantly increased penetrance of starvation-induced abnormalities (Fig. 5c). Furthermore, the abnormality phenotype was more severe, with the frequency of proliferative, non-differentiated proximal germ-cell tumors increased in particular (Fig. 5d, e), as opposed to gonad abnormalities in general (Fig. 1b; Jordan et al. 2019). Further, daf-2 RNAi suppressed this enhanced germ-cell tumor phenotype (Fig. 5f). fat-1 mutants cannot convert DGLA into ETA or ARA into EPA, and fat-1 mutants have elevated ARA levels (Watts et al. 2002). Together with the fat-4 phenotype, the fat-1 phenotype narrows the possible interpretations of these results, suggesting that ARA promotes and/or ETA suppresses starvation-induced abnormalities.
Given opposite phenotypes, we used epistasis analysis to refine our interpretations of how fat-4 and fat-1 affect development of starvation-induced abnormalities. fat-4 is clearly epistatic to fat-1, with suppression of starvation-induced abnormalities in a double mutant comparable to a fat-4 single mutant (Fig. 5g, h). This result supports the conclusion that the Δ5 fatty-acid desaturase FAT-4 promotes starvation-induced abnormalities, and it suggests that the FAT-4 product ARA is responsible, possibly in the context of phosphatidylcholine metabolism. Dietary supplementation with ARA, but not DGLA or ETA, significantly enhanced starvation-induced abnormalities in wild-type animals starved for 6 days (Fig. 5i; duration of starvation was reduced to facilitate detection of enhancement). Supplementation with EPA had variable effects that are not statistically significant. Together with epistasis analysis, these results suggest that ARA promotes development of starvation-induced abnormalities.
Discussion
We show that early-life starvation has widespread effects on adult gene expression and lipid metabolism resulting in developmental pathology in C. elegans. The key lipogenic enzyme-encoding gene fasn-1/FASN is one of many genes whose adult expression is increased by early-life starvation and IIS. We show that fasn-1 contributes substantially to the effects of early-life starvation and IIS on adult gene expression and that it promotes development of starvation-induced gonad abnormalities. We also report extensive effects of early-life starvation and IIS on adult lipid levels, with prominent effects on unsaturated phosphatidylcholine. Dietary supplementation with unsaturated phosphatidylcholine suppresses starvation-induced abnormalities, suggesting functional relevance of its altered abundance. We also show that disruption of fatty-acid desaturation generally suppresses abnormalities, except that mutation of the ω3 desaturase-encoding gene fat-1 enhances abnormality formation. Finally, epistasis analysis suggests that ARA, one of the products of the Δ5 desaturase-encoding gene fat-4, promotes formation of starvation-induced abnormalities, and supplementation with ARA enhances frequency of abnormalities. We speculate that this function occurs in the context of phosphatidylcholine metabolism, potentially via the Lands cycle and eicosanoid signaling (see below; Fig. 5j). This work is significant for connecting early-life starvation and IIS with lipid metabolism and adult pathology, which together with our companion manuscript (Shaul et al. co-submitted) elucidates developmental origins of adult pathology.
Early-life starvation and insulin/IGF signaling have similar effects on adult gene expression
Remarkably, early-life starvation affects expression of thousands of genes in adults. Reduction of IIS with daf-2/InsR RNAi largely reversed these gene expression changes, consistent with daf-2 RNAi suppressing starvation-induced abnormalities (Jordan et al. 2019). Genes related to membranes were particularly enriched among those affected, but the data suggest a variety of consequences of early-life starvation. Lipid metabolism directly impinges on membrane structure and function, and the sole C. elegans FASN fasn-1/FASN was among the genes up-regulated by early-life starvation and down-regulated by daf-2/InsR RNAi. Notably, Fasn expression is also increased in livers of mice subjected to fasting and refeeding with a high-carbohydrate diet (Cabrae et al. 2020), suggesting what we see in worms is conserved in mammals. We found that FASN-1 promotes development of starvation-induced abnormalities, and mRNA-seq analysis suggests that changes in its activity account for much of the effect of early-life starvation and IIS on adult gene expression. These results suggest that an increase in lipid synthesis in well-fed animals following early-life starvation drives developmental pathology.
We did not identify transcriptional regulatory mechanisms affecting fasn-1 or other genes affected by early-life starvation and IIS. We suspect that DAF-16/FoxO, the major transcriptional effector of IIS, represses fasn-1 transcription, given that daf-16 is required for disruption of daf-2/InsR to suppress starvation-induced abnormalities (Jordan et al. 2019). We did not test this hypothesis by mRNA-seq, but we did include daf-16 RNAi in our lipidomics experiment, revealing that the effects of daf-2 on abundance of various lipid classes is in fact daf-16 dependent. However, DAF-16 is thought to act primarily as an activator of its direct targets (Schuster et al. 2010), and its antagonism of the transcription factor PQM-1 has been proposed to be responsible for indirect repression of so-called DAF-16 class II targets (Tepper et al. 2013). Other mechanisms are also likely to underlie such extensive effects on adult gene expression. Notably, our companion manuscript suggests that Wnt signaling is up-regulated by early-life starvation and IIS, affecting expression of numerous Wnt target genes (Shaul et al. co-submitted). In addition, post-translational histone modifications could contribute to epigenetic regulation. There could also be some sort of persistent molecular or cellular damage following starvation that affects gene regulation into adulthood.
Early-life starvation and insulin/IGF signaling have similar effects on adult lipid profiles, suggesting a role of phosphatidylcholine
In addition to affecting adult gene expression, early-life starvation and IIS also have similar effects on abundance of several lipid classes in adults. Our lipidomic analysis provides direct evidence that lipid metabolism is affected by early-life starvation and IIS, corroborating our interpretation of mRNA-seq analysis (Fig. 1f). Phosphatidylcholine levels were particularly affected, revealing that IIS, and DAF-16/FoxO in particular, regulates phosphatidylcholine metabolism. Our data suggest that early-life starvation and IIS have additive effects on phosphatidylcholine levels, as if they act through independent regulatory mechanisms.
Dietary supplementation with unsaturated phosphatidylcholine suppressed starvation-induced abnormalities, suggesting a functional role of phosphatidylcholine in pathological consequences of early-life starvation. It could be that exogenous unsaturated phosphatidylcholine acts as a sink for reactive oxygen species (Kim et al. 2019), buffering pathological effects of early-life starvation. However, supplementation with chicken-egg phosphatidylethanolamine, which is also rich in unsaturated fatty-acid species and has free-radical-scavenging capacity (Sun et al. 2019), did not suppress abnormalities.
We were surprised that phosphatidylcholine supplementation suppressed abnormalities, since endogenous phosphatidylcholine levels were increased following early-life starvation. Nonetheless, we believe this result demonstrates functional specificity of unsaturated phosphatidylcholine in developmental pathology, since saturated phosphatidylcholine and multiple other lipid species did not have the same effect. However, it should be noted that the solubility of unsaturated phosphatidylcholine may limit its effectiveness as a dietary supplement in C. elegans. The phosphatidylcholine concentration used for supplementation could also be high compared with endogenous levels, though we used concentrations typical for lipid supplementations in this system (Deline et al. 2013). It is unclear where in the animal the high levels of endogenous phosphatidylcholine detected after early-life starvation are synthesized or accumulated. One possibility is that endogenous phosphatidylcholine levels are high following starvation because there is not enough of it where it is needed to support developmental fidelity. For example, high phosphatidylcholine levels could reflect accumulation in lipid droplets in the body cavity or elsewhere rather than membrane incorporation. It is also unclear how exogenous phosphatidylcholine is transported through the animal or how it affects lipid metabolism. It is possible that phosphatidylcholine supplementation provokes a homeostatic response that reduces phosphatidylcholine synthesis at sites where its production promotes pathology. In conclusion, it is difficult to reconcile our results from lipidomic analysis and phosphatidylcholine supplementation, so they should be interpreted cautiously. Nonetheless, we believe that each of these results is robust, valuable observations.
Might arachidonic acid–derived oxylipin signaling promote starvation-induced abnormalities?
Phosphatidylcholine is an abundant phospholipid and major constituent of cellular membranes. Phosphatidylcholine is synthesized from choline and diacylglycerol in the cytidine-diphosphate choline pathway, and the fatty-acid tails can be modified in the Lands cycle (Lands 1958; Moessinger et al. 2014). Fatty-acid tails are removed from phosphatidylcholine by phospholipase A2 in the Lands cycle, and if they are unsaturated they are prone to oxidation, producing signaling molecules known as oxylipins (Lands 1958; Balsinde et al. 2002; Shindou and Shimizu 2009; Wang and Tontonoz 2019). If the oxylipins are derived from polyunsaturated, 20 carbon fatty acids such as ARA, then they are known as eicosanoids (Dennis and Norris 2015). Oxylipins regulate a variety of processes including cellular proliferation and differentiation, inflammation, and signaling, and they are implicated in cancer (Dennis and Norris 2015; Gabbs et al. 2015).
We cautiously speculate that participation of unsaturated phosphatidylcholine in the Lands cycle leads to the production of eicosanoid signaling molecules which contribute to developmental pathology following early-life starvation. Multiple observations support this tentative hypothesis. We show that phosphatidylcholine levels, especially for species with long unsaturated fatty-acid tails, are increased by early-life starvation and decreased by reduced IIS. Abundance of the other lipids that participate in the Lands cycle (diacylglycerol, lysophosphatidylcholine, and free fatty acids) is also affected by early-life starvation and IIS. We also show that supplementation with unsaturated phosphatidylcholine, but not saturated phosphatidylcholine or other abundant phospholipids, suppresses starvation-induced abnormalities. We show that mutation of the fatty-acid desaturase fat-4, which is required to produce ARA (Watts et al. 2002), a source of potent eicosanoids, suppresses starvation-induced abnormalities. In contrast, fat-1 mutants, which accumulate ARA (Watts et al. 2002), have enhanced penetrance and severity of starvation-induced abnormalities. Finally, we show feeding worms exogenous ARA enhances starvation-induced abnormalities.
Lipid metabolism is complex, and lipids other than ARA that are affected by mutation of fat-4 or fat-1 could be involved. For example, ω−3 fatty acids modulate inflammation and have a controversial role in suppressing cancer (Hanson et al. 2020). Levels of the ω−3 fatty-acid ETA are increased in fat-4 mutants and decreased in fat-1 mutants (Watts et al. 2002), which suppress and enhance starvation-induced abnormalities, respectively. However, the fat-4; fat-1 double mutant should have reduced levels of ETA like the fat-1 mutant, but starvation-induced abnormalities are suppressed in the double mutant like the fat-4 mutant, suggesting ETA is not responsible for inhibition of starvation-induced abnormalities. We also did not detect an effect of ETA when animals were supplemented with it. In addition, the ω−6 fatty-acid DGLA promotes germ-cell death in C. elegans (Perez et al. 2020), suggesting it could inhibit starvation-induced abnormalities. Furthermore, the DGLA-derived oxylipin 15S-HETrE inhibits cell proliferation in mammals (Pham et al. 2004), and its abundance is presumably increased in fat-4 mutants. However, mutation of fat-1 increases DGLA levels (Watts et al. 2002), and fat-1 mutation enhances starvation-induced abnormalities, arguing against DGLA or 15S-HETrE inhibiting starvation-induced abnormalities. We also did not detect an effect of DGLA when animals were supplemented with it. Notably, EPA supplementation had variable, statistically insignificant effects on the penetrance of starvation-induced abnormalities, as if it may promote them. However, fat-1, which suppresses starvation-induced abnormalities, is required for EPA synthesis. These observations suggest that EPA is not required for starvation-induced abnormalities though it could possibly contribute to them.
As detailed above, we cautiously suggest that unsaturated tails of phosphatidylcholine are removed in the Lands cycle and oxidized to produce eicosanoid signaling molecules, affecting gene expression and germ-cell proliferation to promote developmental pathology following early-life starvation (Fig. 5j). It should be emphasized that this is speculation and that we do not have direct evidence in support of the eicosanoid hypothesis. However, we believe it is the best interpretation of our observations and is worthy of consideration. If true, then it raises a variety of important questions. For example, which enzymes are responsible for the production of eicosanoids (or are they produced by non-enzymatic oxidation), which eicosanoids are responsible, in which tissue(s) does eicosanoid signaling occur, what are the receptors for the signals, and how do they affect cellular physiology to drive organismal pathology? Future work is required to address the possibility that eicosanoid signaling promotes starvation-induced abnormalities in C. elegans, and we suggest that the role of phosphatidylcholine and ARA metabolism in other models of early-life nutrient stress should be investigated.
Supplementary Material
Acknowledgments
The authors thank the Duke University School of Medicine and the Center for Genomic and Computational Biology for use of the Sequencing and Genomic Technologies core resource, which provided RNA sequencing service. fasn-1p::gfp was kindly provided by Jonathan Ewbank. They also thank WormBase! Transcriptomics and metabolomics experiments were performed by the Duke Center for Genomic and Computational Biology.
Contributor Information
James M Jordan, Department of Biology, Duke University, Durham, NC 27708, USA.
Amy K Webster, Department of Biology, Duke University, Durham, NC 27708, USA; University Program in Genetics and Genomics, Duke University, Durham, NC 27708, USA.
Jingxian Chen, Department of Biology, Duke University, Durham, NC 27708, USA.
Rojin Chitrakar, Department of Biology, Duke University, Durham, NC 27708, USA.
L Ryan Baugh, Department of Biology, Duke University, Durham, NC 27708, USA; Duke Center for Genomic and Computational Biology, Duke University, Durham, NC 27708, USA.
Data availability
Both mRNA-seq data sets discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE178172. Raw and normalized lipidomic data are available in Supplementary File 5.
Supplemental material available at GENETICS online.
Funding
This work was supported by the National Institutes of Health (R01GM117408, L.R.B.) and Duke University's Office of the Vice Provost for Research. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Author contributions
J.M.J. and L.R.B. conceived and designed the experiments and wrote the manuscript. J.M.J. and R.C. conducted the experiments. J.M.J., A.K.W., and J.C. analyzed the data.
Note added in proof
See https://doi.org/10.1093/genetics/iyac173 for a related work by Shaul et al.
Communicating editor: B. Conradt
Literature cited
- Anders S, Pyl PT, Huber W. HTSeq – a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsinde J, Winstead MV, Dennis EA. Phospholipase A2 regulation of acrachidonic acid mobilization. FEBS Lett. 2002;531:2–6. doi: 10.1016/S0014-5793(02)03413-0 [DOI] [PubMed] [Google Scholar]
- Baugh LR. To grow or not to grow: nutritional control of development during Caenorhabditis elegans L1 arrest. Genetics. 2013;194:539–555. doi: 10.1534/genetics.113.150847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Hu PJ. Starvation responses throughout the Caenorhabditis elegans life cycle. Genetics. 2020;216:837–878. doi: 10.1534/genetics.120.303565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braeckman BP, Houthoofd K, Vanfleteren JR. Intermediary Metabolism. WormBook; 2009. p. 1–24. doi: 10.1895/wormbook.1.146.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrae R, Dubuquoy C, Cauzac M, Morzyglod L, Guilmeau S, Noblet B, Feve B, Postic C, Burnol AF, Moldes M. Insulin activates hepatic Wnt/beta-catenin signaling through stearoyl-CoA desaturase 1 and porcupine. Sci Rep. 2020;10:5186. doi: 10.1038/s41598-020-61869-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che L, Paliogiannis P, Cigliano A, Pilo MG, Chen X, Calvisi DF. Pathogenetic, prognostic, and therapeutic role of fatty acid synthase in human hepatocellular carcinoma. Front Oncol. 2019;9:1412. doi: 10.3389/fonc.2019.01412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deline ML, Vrablik TL, Watts JL. Dietary supplementation of polyunsaturated fatty acids in Caenorhabditis elegans. J Vis Exp. 2013;81:50879. doi: 10.3791/50879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis EA, Norris PC. Eicosanoid storm in infection and inflammation. Nat Rev Immunol. 2015;15:511–523. doi: 10.1038/nri3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinf. 2009;10:48–55. doi: 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavin R, Peluso S, Nguyen PL, Loda M. Fatty acid synthase as a potential therapeutic target in cancer. Future Oncol. 2010;6:551–562. doi: 10.2217/fon.10.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabbs M, Leng S, Devassy JG, Monirujjaman M, Aukema HM. Advances in our understanding of oxylipins derived from dietary PUFAs. Adv Nutr. 2015;6:513–540. doi: 10.3945/an.114.007732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson S, Thorpe G, Winstanley L, Abdelhamid AS, Hooper L, PUFAH group . Omega-3, omega-6 and total dietary polyunsaturated fat on cancer incidence: systematic review and meta-analysis of randomised trials. Br J Cancer 2020;122:1260–1270. doi: 10.1038/s41416-020-0761-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/S0960-9822(01)00594-2 [DOI] [PubMed] [Google Scholar]
- Hoffman DJ, Reynolds RM, Hardy DB. Developmental origins of health and disease: current knowledge and potential mechanisms. Nutr Rev. 2017;75:951–970. doi: 10.1093/nutrit/nux053 [DOI] [PubMed] [Google Scholar]
- Hughes LA, van den Brandt PA, de Bruine AP, Wouters KA, Hulsmans S, Spiertz A, Goldbohm RA, de Goeij AF, Herman JG, Weijenberg MP, et al. Early life exposure to famine and colorectal cancer risk: a role for epigenetic mechanisms. PLoS One. 2009;4:e7951. doi: 10.1371/journal.pone.0007951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan JM, Hibshman JD, Webster AK, Kaplan REW, Leinroth A, Guzman R, Maxwell CS, Chitrakar R, Bowman EA, Fry AL, et al. Insulin/IGF signaling and vitellogenin provisioning mediate intergenerational adaptation to nutrient stress. Curr Biol. 2019;29:2380–2388.e5. doi: 10.1016/j.cub.2019.05.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtlang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0 [DOI] [PubMed] [Google Scholar]
- Kim S-H, Kim B-K, Park S, Park S-K. Phosphatidylcholine extends lifespan via DAF-16 and reduces amyloid-beta-induced toxicity in Caenorhabditis elegans. Oxid Med Cell Longev. 2019;2019:2860642. doi: 10.1155/2019/2860642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura I, Ichimura A, Ohue-Kitano R, Igarashi M. Free fatty acid receptors in health and disease. Physiol Rev. 2020;100:171–210. doi: 10.1152/physrev.00041.2018 [DOI] [PubMed] [Google Scholar]
- Lands WEM. Metabolism of glycerolipides: a comparison of lecithin and triglyceride synthesis. J Biol Chem. 1958;231:883–888. doi: 10.1016/S0021-9258(18)70453-5 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Kniazeva M, Han M, Pujol N, Ewbank J. The fatty acid synthase fasn-1 acts upstream of WNK and Ste20/GCK-VI kinases to modulate antimicrobial peptide expression in C. elegans epidermis. Virulence 2010;1:113–122. doi: 10.4161/viru.1.3.10974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Moessinger C, Klizaite K, Steinhagen A, Philippou-Massier J, Shevchenko A, Hoch M, Ejsing CS, Thiele C. Two different pathways of phosphatidylcholine synthesis, the Kennedy Pathway and the Lands Cycle, differentially regulate cellular triacylglycerol storage. BMC Cell Biol. 2014;15:43. doi: 10.1186/s12860-014-0043-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Painter RC, De Rooij SR, Bossuyt PM, Osmond C, Barker DJ, Bleker OP, Roseboom TJ. A possible link between prenatal exposure to famine and breast cancer: a preliminary study. Am J Hum Biol. 2006;18:853–856. doi: 10.1002/ajhb.20564 [DOI] [PubMed] [Google Scholar]
- Papaevangelou E, Almeida GS, Box C, deSouza NM, Chung YL. The effect of FASN inhibition on the growth and metabolism of a cisplatin-resistant ovarian carcinoma model. Int J Cancer. 2018;143:992–1002. doi: 10.1002/ijc.31392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez MA, Magtanong L, Dixon SJ, Watts JL. Dietary lipids induce ferroptosis in caenorhabditis elegans and human cancer cells. Dev Cell. 2020;54:447–454.e4. doi: 10.1016/j.devcel.2020.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham H, Banerjee T, Ziboh VA. Suppression of cyclooxygenase-2 overexpression by 15S-hydroxyeicosatrienoic acid in androgen-dependent prostatic adenocarcinoma cells. Int J Cancer. 2004;111:192–197. doi: 10.1002/ijc.20245 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseboom TJ, van der Meulen JH, Ravelli AC, Osmond C, Barker DJ, Bleker OP. Effects of prenatal exposure to the Dutch famine on adult disease in later life: an overview. Mol Cell Endocrinol. 2001;185:93–98. doi: 10.1016/S0303-7207(01)00721-3 [DOI] [PubMed] [Google Scholar]
- Schuster E, McElwee JJ, Tullet JM, Doonan R, Matthijssens F, Reece-Hoyes JS, Hope IA, Vanfleteren JR, Thornton JM, Gems D. DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol Syst Biol. 2010;6:399. doi: 10.1038/msb.2010.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul NC, Jordan JM, Falsztyn IB, Baugh LR. Insulin/IGF-dependent Wnt signaling promotes formation of germline tumors and other developmental abnormalities following early-life starvation in Caenorhabditis elegans. Genetics. 2022:iyac173. doi: 10.1093/genetics/iyac173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindou H, Shimizu T. Acyl-CoA:lysophospholipid acyltransferases. J Biol Chem. 2009;284:1–5. doi: 10.1074/jbc.R800046200 [DOI] [PubMed] [Google Scholar]
- Sun N, Chen J, Bao Z, Wang D, An B, Lin S. Egg yolk phosphatidylethanolamine: extraction optimization, antioxidative activity, and molecular structure profiling. J Food Sci. 2019;84:1002–1011. doi: 10.1111/1750-3841.14512 [DOI] [PubMed] [Google Scholar]
- Tepper RG, Ashraf J, Kaletsky R, Kleemann G, Murphy CT, Bussemaker HJ. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell. 2013;154:676–690. doi: 10.1016/j.cell.2013.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhwa PD, Buss C, Entringer S, Swanson JM. Developmental origins of health and disease: brief history of the approach and current focus on epigenetic mechanisms. Semin Reprod Med. 2009;27:358–368. doi: 10.1055/s-0029-1237424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li C, Yang Z, Zou Z, Ma J. Infant exposure to Chinese famine increased the risk of hypertension in adulthood: results from the China Health and Retirement Longitudinal Study. BMC Public Health. 2016;16:435–446. doi: 10.1186/s12889-016-3122-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Tontonoz P. Phospholipid remodeling in physiology and disease. Annu Rev Physiol. 2019;81:165–188. doi: 10.1146/annurev-physiol-020518-114444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JL, Browse J. Genetic dissection of polyunsaturated fatty acid synthesis in Caenorhabditis elegans. PNAS. 2002;99:5854–5859. doi: 10.1073/pnas.092064799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JL, Ristow M. Lipid and carbohydrate metabolism in Caenorhabditis elegans. Genetics. 2017;207:413–446. doi: 10.1534/genetics.117.300106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Both mRNA-seq data sets discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE178172. Raw and normalized lipidomic data are available in Supplementary File 5.
Supplemental material available at GENETICS online.