Abstract
Eukaryotic genomes are large and complex, and gene expression can be affected by multiple regulatory elements and their positions within the dynamic chromatin architecture. Transposable elements are known to play important roles in genome evolution, yet questions remain as to how transposable elements alter genome structure and affect gene expression. Previous studies have shown that genome rearrangements can be induced by Reversed Ends Transposition involving termini of Activator and related transposable elements in maize and other plants. Here, we show that complex alleles can be formed by the rapid and progressive accumulation of Activator-induced duplications and rearrangements. The p1 gene enhancer in maize can induce ectopic expression of the nearby p2 gene in pericarp tissue when placed near it via different structural rearrangements. By screening for p2 expression, we identified and studied 5 cases in which multiple sequential transposition events occurred and increased the p1 enhancer copy number. We see active p2 expression due to multiple copies of the p1 enhancer present near p2 in all 5 cases. The p1 enhancer effects are confirmed by the observation that loss of p2 expression is correlated with transposition-induced excision of the p1 enhancers. We also performed a targeted Chromosome Conformation Capture experiment to test the physical interaction between the p1 enhancer and p2 promoter region. Together, our results show that transposon-induced rearrangements can accumulate rapidly and progressively increase genetic variation important for genomic evolution.
Keywords: enhancer, RET, complex rearrangement, composite insertion, transposable elements, macrotransposons, Plant Genetics and Genomics
Using a convenient red kernel marker, Sharma and Peterson isolate a series of maize alleles containing Complex Chromosomal Rearrangements induced by the Ac transposable element. They show that these alleles were generated by the rapid and progressive accumulation of gene rearrangements that change the number and position of a known enhancer sequence. Their results demonstrate the remarkable capability of TEs to rapidly change genome structure and alter gene expression.
Introduction
Enhancers are cis-regulatory DNA sequences that interact with their target promoters to stimulate transcription. These elements work independently of orientation and can be present near the promoter they influence, or they could act from large distances. Enhancers can be promiscuous or promoter-specific, affecting genes/promoters nearby, or passing over some genes to affect 1 further away (Kvon et al. 2014). With the discovery of cis elements working from a distance, it became clear that the functionality of the genome is not only dependent on the linear DNA sequence but also on the spatial arrangement of the chromatin (Doğan and Liu 2018; Krumm and Duan 2019). The eukaryotic nucleus has chromatin grouped into compartments of higher and lower transcriptional activity, called A and B compartments, respectively (Lieberman-Aiden et al. 2009). Furthermore, chromatin topologically associated domains (TADs) with boundaries controlled by cohesin and CCCTC-binding factor (CTCF) play important roles in the regulation of gene expression (Dekker et al. 2013; Nora et al. 2013). CTCF is conserved within bilaterian phyla, whereas plants lack CTCF or an ortholog (Heger et al. 2012). Hi-C maps do show compartmentalization and TADs in most plant genomes tested (Doğan and Liu 2018). Enhancers function within these domains, and structural rearrangements that change the TAD boundaries or position of an enhancer relative to their target genes can lead to dysregulation and disease (Lupiáñez et al. 2015; Bompadre and Andrey 2019). With only a few well-studied examples of enhancers in plants (Weber et al. 2016), little is known about the mechanism of their interactions with target promoters.
The maize p1 and p2 genes encode Myb-homologous regulators of the flavonoid biosynthetic pathway to produce red phlobaphene pigments in floral organs (Dooner et al. 1991; Grotewold et al. 1994). The p1 gene is responsible for pigmentation in kernel pericarp, cob, and silk, while p2 is expressed in anther and silk (Zhang et al. 2000; Goettel and Messing 2009). The striking red kernel phenotype specified by p1 alleles has been used as a convenient indicator of gene expression since the earliest genetical studies in maize (Emerson 1917). The p1 gene was one of the first loci shown to carry Activator (Ac) transposable element insertions (Barclay and Brink 1954; Greenblatt and Brink 1962; Lechelt et al. 1989). The presence of 1.2-kb enhancer region near the p1 gene was inferred from early Ac insertional mutagenesis studies in the allele P1-rr4B2 (Athma et al. 1992; Moreno et al. 1992). The enhancer activity of this region was confirmed by both transient and stable transformations using the GUS reporter gene (Sidorenko et al. 1999; Sidorenko et al. 2000). In addition to increasing the rate of transcription, the p1 enhancer sequence has been shown to participate in p1 paramutation (Sidorenko and Peterson 2001). The P1-rr4B2 allele has a direct duplication of 1,269-bp fragment, which is a part of larger 5.2-kb direct repeats, which flank the p1 gene. The 3′ 5.2-kb direct repeat contains 2 copies of the 1.2-kb repeat, whereas the 5′ 5.2-kb direct repeat contains 1 full-length 1.2-kb direct repeat, and a stable Ds-like element inserted into the upstream copy. The p1 enhancer is a 405-bp fragment (fragment 15), which lies within the 1,269-bp direct duplication (Zhang and Peterson 2005a). For simplicity, following text refers to the 2 copies of fragment 15 as a single enhancer and the figures reflect the duplicate nature of the enhancer.
The maize P1-rr11 allele has insertions of Ac and fAc (fractured-Ac) elements in the p1 gene and produces a red pericarp phenotype. A derivative null allele p1-wwB54 has white pericarp and white cob, and contains a deletion of the upstream enhancer and exons 1 and 2 of the p1 gene (Yu et al. 2011). The p1-wwB54 allele retains p1 exon 3, the downstream enhancer, and the Ac and fAc elements in reversed orientation, separated by only 331 bp of DNA. These Ac/fAc termini can undergo frequent Reversed Ends Transposition (RET) events (Zhang and Peterson 2004; Huang and Dooner 2008; Zhang et al. 2009; Yu et al. 2011), forming a variety of alleles including deletions (Zhang and Peterson 2005b; Zhang et al. 2006), duplications (Zhang et al. 2013), Composite Insertions (CIs; Zhang et al. 2014; Su et al. 2018, 2020), and inversions (Zhang and Peterson 2004; Yu et al. 2011; Sharma et al. 2021). Here, we show how repeated RET events in maize can induce the formation of complex gene structures containing multiple rearrangements such as inversions, CIs, duplications and/or deletions. Moreover, these new structures alter the number and position of p1 enhancer elements, thereby affecting expression of the neighboring p2 gene.
Methods
Genetic screening and PCR
Nearly 4,000 plants of genotype p1-wwB54 heterozygous with the null allele p1-ww[4Co63] were grown and pollinated with p1-ww[4Co63]. Ears were screened for kernels with red pericarp to obtain RET-induced rearrangement alleles. In p1-wwB54, we see approximately 1 in 8 ears with a single red kernel and 1 in 40 ears with a multikernel red sector. All 5 of the cases presented here were derived from single kernel events. Pericarp is a maternal tissue derived from the ovary wall, sharing a common lineage with the egg progenitor cells. The size and heritability of red pericarp sectors depend on the stage of ear and kernel development at which an activating mutation occurred. Mutations occurring earlier in development produce larger sectors, and the underlying mutation can be more frequently transmitted. Because recovery of any mutant allele will depend on the segregation of that allele to the egg cell at meiosis, only half of the events seen as single kernel sectors will be transmitted to the next generation (Anderson and Brink 1952). Red kernels were selected and planted, and in case of transmission, the resulting plants would produce whole red ears. Kernels from these red ears were sown, and genomic DNA was extracted from seedling leaves by a modified CTAB method (Saghai-Maroof et al. 1984). To detect structural rearrangements (Su et al. 2020; Sharma et al. 2021), PCR was performed under standard conditions using Promega GoTaq Green Master Mix and primers specifically designed for the p1-wwB54 sequence (Supplementary Table 1). The models of RET predict that in rearrangements arising from the activity of Ac and fAc elements, each breakpoint will be expected to border a transposon involved in the reaction. Ac casting (Singh et al. 2003; Wang and Peterson 2013) and inverse-PCR (iPCR; Ochman et al. 1988) techniques were used to locate these breakpoints adjacent to Ac and fAc, respectively. For visualizing PCR products, a high-efficiency agarose gel electrophoresis method was used (Sharma and Peterson 2021). PCR amplicons were sequenced by the Iowa State University DNA Sequencing Facility.
Southern blotting
To confirm the internal structure of the complex rearrangements, digests using BglII, KpnI, HpaI, and EcoRI restriction enzymes and their combinations were performed (data not shown for EcoRV and some combinations). For double digests, digestions were performed in 2 steps, 1 enzyme at a time. Whole-genome DNA was digested and loaded on an agarose gel (0.7–0.8%) run for 24–26 h under 35–40 V for adequate separation of fragments. The DNA was transferred to a membrane for 24 h, followed by probing the membranes with fragment 15 located within the p1 gene enhancer (Zhang and Peterson 2005a).
RT-PCR
Pericarps were peeled from kernels 20 days after pollination (DAP) and flash-frozen in liquid nitrogen. Two biological replicates (pericarps from 2 siblings) were pooled to extract RNA. RNA was isolated using Purelink Plant RNA Reagent, treated with New England Biolabs DNase I to remove gDNA, and then reverse transcribed to cDNA using Invitrogen SuperScript II Reverse Transcriptase kit. Protocols recommended by the product suppliers were used. Two technical replicates were used for each sample in reverse transcription reaction. Finally, PCR was performed on the cDNA using primers specific to exons 1 and 3 of p2 to amplify the p2 gene transcript (Supplementary Table 2). Primers specific to the GAPDH gene were used as an internal control (Supplementary Table 2).
Chromosome conformation capture
A plant-specific 3C protocol was used with modifications (Louwers et al. 2009). Pericarps were peeled on ice from ∼40 developing kernels at 15 DAP totaling approximately 700 mg of tissue and added directly to a 2% formaldehyde solution made with a nuclei isolation buffer. The pericarps were fixed at room temperature for 1 h in a vacuum chamber under 11.7 psi pressure. For digestion, the sample was equally divided into 2 tubes, 150 units of BglII were added to each and kept overnight at 37°C while on rotation. An additional 50 U of BglII were added to each tube the next morning and incubated for 2 h. Slow rotations of 60 rpm were used on all steps requiring rotation. For DNA precipitation, samples were stored at −20°C. S-adenosyl-methionine decarboxylase (Sam) gene was used as an internal control in qPCR. As a control for primer efficiency differences in PCR amplification, the target regions were amplified and digested with BglII and re-ligated (Tolhuis et al. 2002). The DNA concentration was determined and mixed in equimolar amounts to make the control template containing all possible ligation products of the loci of interest (p1/p2 and Sam). qPCR was performed using SybrGreen master mix and supplier recommended protocols.
Results
Southern blot analysis using fragment 15 (Zhang and Peterson 2005a) allowed us to identify alleles with multiple p1 enhancers (Supplementary Fig. 1). In these alleles, rearrangement endpoints were found using Ac casting (Singh et al. 2003; Wang and Peterson 2013) and/or inverse-PCR (iPCR; Ochman et al. 1988) techniques. By evaluating the Southern blot results (Supplementary Figs. 1–3) together with the breakpoint sequences found with PCR techniques, the structures of 5 different rearrangement alleles were deduced in accordance with existing models of RET (Zhang and Peterson 2004; Huang and Dooner 2008; Zhang et al. 2009; Yu et al. 2011). The presence of 8-bp target site duplications (TSDs) at the 2 endpoints of each rearrangement structure confirmed their origin from Ac transposition (Supplementary Table S3). The structures of some cases could not be completely determined and thus are not considered here.
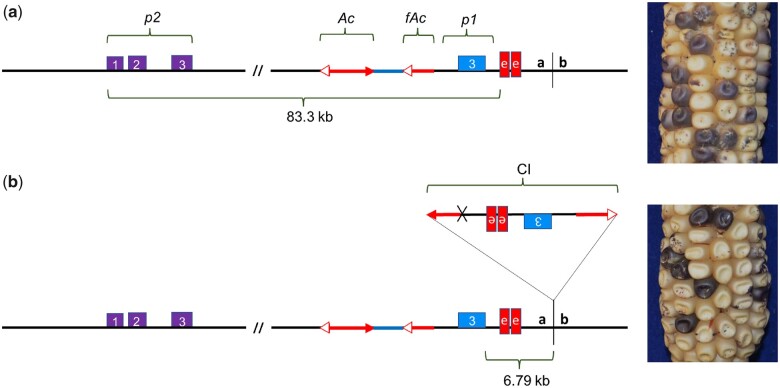
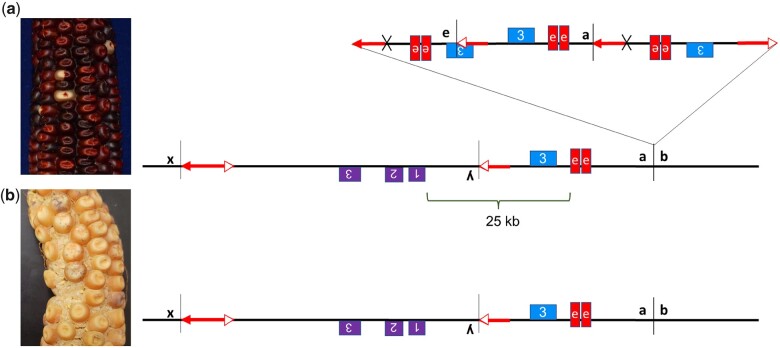
The null allele p1-wwB54 has a white kernel pericarp phenotype, and the p2 gene is not expressed in the pericarp (Sharma et al. 2021). The p1-wwB54 allele retains the p1 enhancer downstream of p1 exon 3; as described above, there are 2 nearby copies of the fragment 15 enhancer, indicated by the 2 red boxes in Fig. 1a (Zhang and Peterson 2005a). Structural rearrangements that move the p1 gene enhancer closer to the p2 gene can induce expression of the p2 gene in the pericarp and produce a red kernel pericarp phenotype. Secondary rearrangements are possible in many cases due to the continued presence of Ac/fAc termini and their ability to interact (Fig. 1a). Some of the rearrangements previously studied are deletions (Zhang et al. 2006), inversions (Sharma et al. 2021), and Composite Insertions (Su et al. 2020). The model for the formation of Composite Insertion (CI) through DNA re-replication has been described previously in detail (Zhang et al. 2014; Su et al. 2020). Briefly, a CI arises when a transposon pair along with flanking DNA moves during replication from an already replicated part to a nonreplicated region and gets re-replicated.
Fig. 1.
a) p1-wwB54: purple boxes represent the p2 gene with numbered exons 1, 2, and 3. The blue box is exon 3 of the p1 gene. Red boxes indicate 2 copies of fragment 15. Red arrows are Ac (2 arrowheads) and fAc (single arrowhead) elements. b) p1-wwB54-CI contains a CI at the site labeled a/b 6.79-kb downstream of p1 exon 3. This CI contains 1 copy of the enhancer region and exon 3 and is also present in 5 other rearrangement alleles. Corresponding pictures on the right show colorless pericarp phenotype in both alleles. The solid and spotted purple color is due to r1-m3::Ds activation in aleurone caused by Ac-induced excision of Ds.
At some point during the propagation of p1-wwB54, a new CI inserted into a site 6.79-kb downstream of the p1 exon 3 (Fig. 1, site a/b). This CI consists of 1,391 bp of Ac 5′ side sequence joined with 9,022 bp of fAc and flanking region containing p1 exon 3 and the p1 enhancer region. These 2 CI fragments are fused at a 4-bp sequence overlap (Supplementary Table 3), suggesting they were joined by microhomology-mediated end joining (McVey and Lee 2008). This CI insertion created a new allele termed p1-wwB54-CI (Fig. 1b), which contains 2 p1 enhancers located at 83.35 and 92.4 kb from the p2 5′ end. Similar to p1-wwB54, the p1-wwB54-CI allele has colorless pericarp. The lack of p2 expression in both alleles indicates that 1 or 2 copies of the enhancer cannot act on p2 from this distance. Because the kernel pericarp phenotype of p1-wwB54-CI is not significantly different from the parental p1-wwB54, the presence of p1-wwB54-CI remained undetected in our materials until we characterized 5 independent derivative alleles. These alleles all contained a CI with the same internal structure, inserted at the same position (Fig. 1, site a/b), and flanked by the same 8-bp TSD as that of p1-wwB54-CI (Supplementary Table 3). We conclude that the 5 mutant alleles were all derived from p1-wwB54-CI. The 5 complex rearrangements describe here appeared within 5 maize generations since the first isolation of p1-wwB54 (Yu et al. 2011).
Structures of complex rearrangements
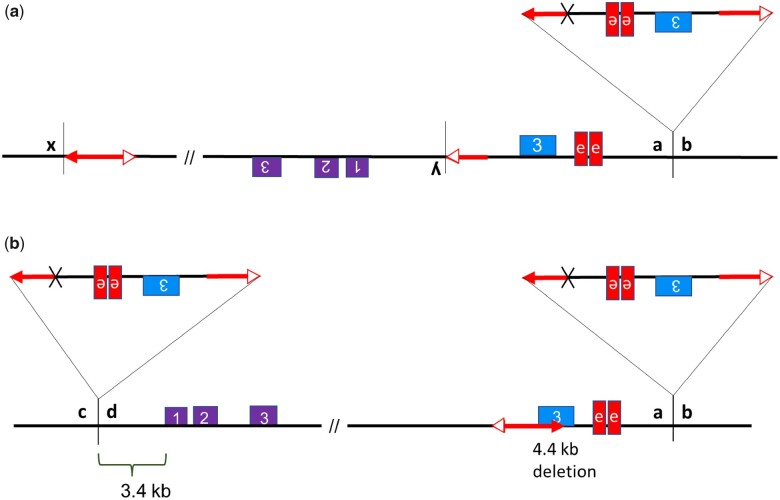
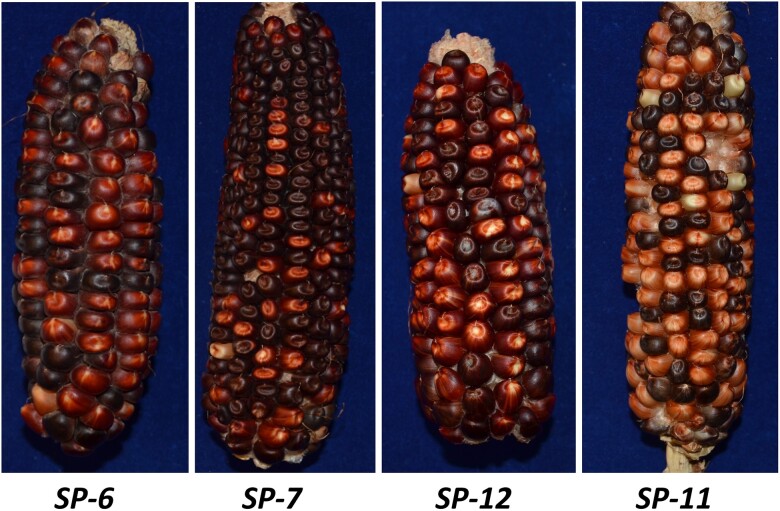
Five cases labeled SP-6, SP-7, SP-12, SP-11, and SP-97 with independent rearrangement structures were isolated. These cases contain combinations of rearrangements such as inversions, CIs, and deletions. All 5 cases share the same CI downstream of the p1 enhancer with p1-wwB54-CI. In addition, SP-6, SP-7, and SP-12 have an inversion, each with a unique endpoint near p2 (Fig. 2a). The target site x/y in SP-6, SP-7, and SP-12 is 199, 257, and 3,041 bp away from the p2 transcription start site (TSS), respectively, making the distances between p2 TSS and p1 enhancers from 4.9 to 16.8 kb. Target site x/y in each case was found to have flanking 8-bp target site duplications (Supplementary Table S3). In allele SP-6, the rearrangement structure leaves only 207 bp of promoter region sequence upstream of p2. Compared to p1-wwB54-CI, the inversions reduced the distance between p2 and the p1 enhancers in these cases. We expect SP-11 (Fig. 2b) to have originated from a progenitor that had the same structure as Fig. 1b. The CI structure with Ac and fAc terminal sequences at its ends is known to be able to transpose (Su et al. 2020). If the CI excises from its p1 location after being replicated and then inserts into a yet unreplicated region, the resulting allele will retain 2 copies of the CI. SP-11 has the common CI at the p1 downstream site, and a copy of the same CI is present near p2 (Fig. 2b). In addition to the movement/duplication of the CI, SP-11 has a deletion toward p1 exon 3, which could result from the RET event in which the Ac/fAc pair inserted into p1 exon 3, leading to deletion of the 4.4-kb fragment containing fAc and part of p1 exon 3 (Fig. 2b). With the 2 CIs carrying 1 copy of the p1 enhancer each, SP-11 has a total of 3 p1 enhancers. The enhancer in the CI inserted near p2 is at about 8.1 kb, and the 2 distant enhancers are at 78.95 and 88 kb from p2. SP-6, SP-7, and SP-12 ears have a dark red pericarp phenotype, whereas the SP-11 ear is fainter red in comparison (Fig. 3). The lighter red phenotype could possibly indicate that the 2 distant enhancers are not involved in p2 activation in SP-11 same as in p1-wwB54-CI. RT-PCR results show that these 4 alleles with red pericarp have active p2 gene expression in pericarp tissue (Supplementary Fig. 4). The p2 expression in pericarp was confirmed by sequence information (Sharma et al. 2021).
Fig. 2.
a) SP-6, SP-7, and SP-12: these alleles have unique x/y inversion endpoints and share the same a/b insertion site for the common CI containing a copy of the p1 enhancer. b) SP-11 has a copy of the same CI inserted near p2 and an additional 4.4-kb deletion in p1.
Fig. 3.
Ears of alleles with red pericarp phenotype. SP-6, SP-7, and SP-12 have darker red pericarp phenotypes compared to SP-11. The dark purple color in some kernels is due to anthocyanin pigmentation of kernel aleurone induced by Ds excision from the r1-m3::Ds tester allele.
Progressive rearrangements in SP-97
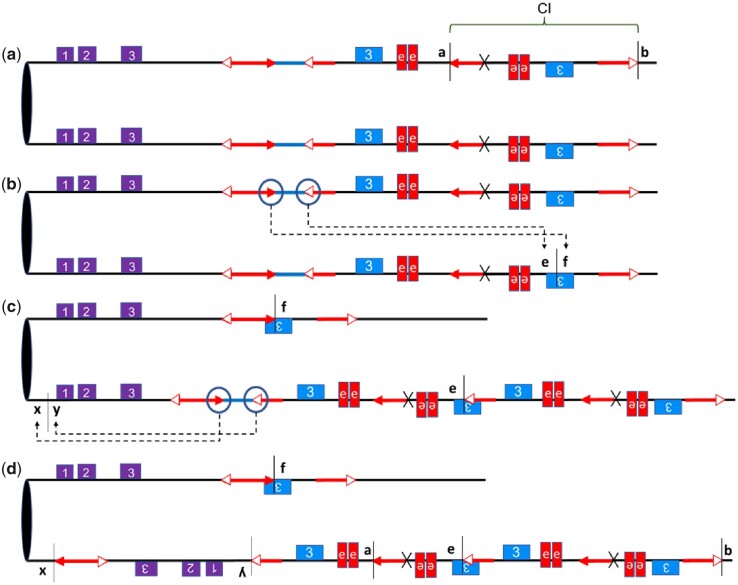
The ability of multiple Ac/fAc elements to interact and transpose leaves many possibilities for the formation of different complicated structures. This potential is exemplified in the fifth allele SP-97, which has a complex structure with 4 p1 enhancers and a dark red phenotype. Figure 4 describes a model for the origin of SP-97. We hypothesize that it originated after the common CI at the a/b site in p1-wwB54 (Fig. 4a, same as Fig. 1b); the Ac and fAc elements present at their original position underwent reversed ends transposition into the sister chromatid at site e/f (Fig. 4b). This resulted in a tandem direct duplication of the 17.73-kb region containing 2 copies of the enhancer, bringing the total to 4 copies (Fig. 4c). The Ac and fAc elements in the recipient chromatid are also capable of generating rearrangements. A subsequent RET event with insertion of Ac/fAc pair at target site x/y 20.3 kb from p2 caused an inversion that brought p2 closer to the 4 enhancers leading to activation in the pericarp (Fig. 4, c and d). The size of the inverted fragment is 98.5 kb (x to y in Fig. 4d). The final structure in Fig. 4d lower chromatid is the SP-97 allele containing a CI, a duplication, and an inversion. It would take these 3 RET events to form the final structure of SP-97 leading to the red pericarp phenotype selected. The event consisting formation of the CI occurred prior to other 2 since it is present in other alleles as well. The duplication and inversion could have occurred later in a single or 2 different generations. The inversion would occur at last reducing the distance between the p2 gene and the p1 enhancers. To confirm this structure, endpoints a/b and x/y were sequenced and found to contain the flanking 8-bp target site duplications (Supplementary Table 3). The new junction created by the duplication at target site e/f (Fig. 4, b and c) was also sequenced (Supplementary Table 3). The distance of the 4 enhancers from p2 is about 25, 34, 41.5, and 50.5 kb.
Fig. 4.
Model for the origin of allele SP-97: a) ancestral allele, which has a structure similar to p1-wwB54-CI. The diagram shows sister chromatids and the CI downstream of the p1 enhancer at site a/b. b) RET: Ac and fAc pair move from 1 chromatid and insert into the e/f target site in the sister chromatid, causing duplication and reciprocal deletion in the sister chromatids. c) RET: The Ac/fAc pair on the other chromatid underwent an inversion toward p2 inserting at target site x/y. d) The lower chromatid is SP-97, which contains 4 copies of the p1 enhancer, an inversion, a CI, and a duplication.
If the dark red phenotype results from multiple copies of the p1 enhancer acting on p2, then the absence of some of these enhancers should affect the phenotype. To test this hypothesis, we examined ears produced by different alleles for the loss-of-function colorless kernels. SP-97 had some colorless/faint red kernels on the ears with dark red kernel pericarp (Fig. 5a), which gave rise to the stable mutant called SP-97M1, which has a very faint red pericarp phenotype (Fig. 5b). SP-97 has 4 copies of the p1 enhancer near p2. Three of the 4 copies are within a structure that ends with Ac 5′ and 3′ terminal sequences, potentially forming a 28-kb large macrotransposon, which might be capable of transposition (Huang and Dooner 2008; Su et al. 2020). We analyzed the structure of SP-97M1 and found that the entire macrotransposon structure containing 3 copies of the p1 enhancer has been excised; the excision site retains a modified 8-bp TSD as a macrotransposon footprint (Supplementary Table S3). The resulting SP-97M1 allele is left with only 1 copy of the p1 enhancer at 25 kb from p2. SP-97M1 still has some p2 activity in the pericarp (Fig. 6). Similarly, loss-of-function mutants from SP-7 and SP-12 (called SP-7M1 and SP-12M1, respectively) were also found to have a light red pericarp phenotype (Supplementary Fig. 5). In both cases, the CI excised out, leaving only a single copy of the p1 enhancer near the p2 gene. An 8-bp TSD footprint was also sequenced in SP-7M1 (Supplementary Table 3). The lighter red phenotype in all 3 mutants shows that the darker red phenotype was a result of multiple copies of the enhancer present close to the p2 gene.
Fig. 5.
Phenotype (left) and structure (right). a) SP-97 has a dark red kernel pericarp. The final structure of SP-97 from Fig. 4 is consistent with a 28-kb macrotransposon at site a/b. b) SP-97M1 has a light red kernel pericarp. The large macrotransposon excised out, leaving only 1 copy of the enhancer in the resulting allele at 25 kb from p2.
Fig. 6.
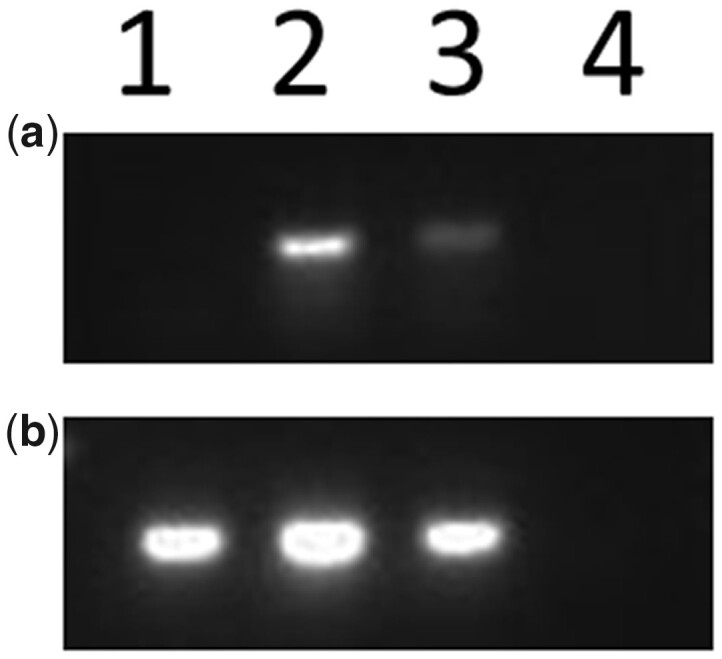
RT-PCR using RNA extracted from pericarp tissue and reverse transcribed to cDNA. Agarose gel image showing results of RT-PCR with primers from a) p2 exons 1 and 3, b) GAPDH as an internal control. Lane 1, p1-wwB54-CI; lane 2, SP-97; lane 3, SP-97M1; lane 4, negative control. p1-wwB54-CI lacks p2 expression; SP-97 and SP-97M1 have p2 expression.
Enhancer–promoter interaction
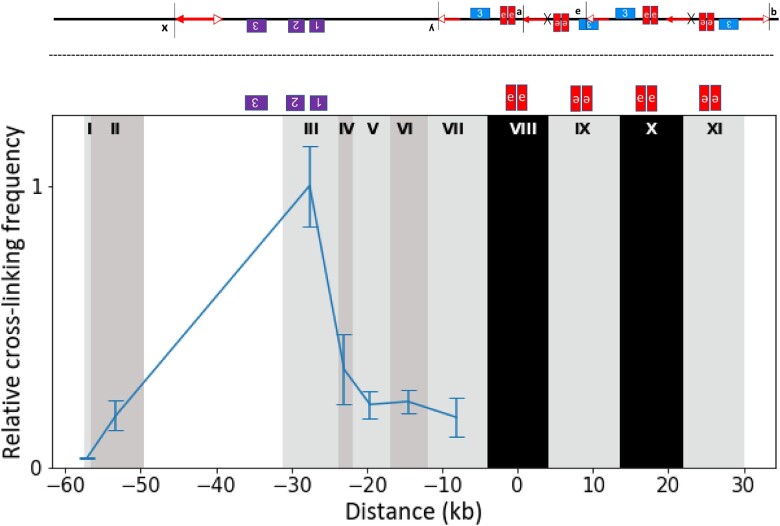
If the p1 enhancer is in fact activating p2 expression in the pericarp, we may be able to detect physical interactions between the p1 enhancer and p2 promoter. We tested for such interactions in Chromosome Conformation Capture (3C) experiments (Louwers et al. 2009) performed in the SP-97 allele, which was chosen for its dark red pericarp color. First, fresh pericarp tissue was cross-linked and intact nuclei were isolated. Then, the cross-linked nuclei were digested with BglII, the DNA re-ligated, and the crosslinks reversed. The resulting 3C DNA consists of new DNA molecules formed by the ligation of the BglII ends of DNA fragments from interacting loci. These new molecules were tested using primers specific to BglII fragments around p1/p2 region (Supplementary Table 4). Due to the presence of many repetitive retroelement sequences inserted within a 100-kb region around p2, many BglII fragments could not be tested. We were able to test 7 fragments (Fig. 7, labeled I to VII) located within a ∼60-kb region encompassing the p2 gene for interactions with the p1 enhancer. Due to the complex duplication in the SP-97 allele, the p1 enhancer is present on 4 BglII fragments labeled VIII to XI (Fig. 7). It is unknown whether all 4 copies of the p1 enhancer may interact with p2, so we tested the interaction of the p1 enhancer closest to p2. A primer specific to fragment VIII (and X) was used as an anchor and tested against primers specific to fragments I to VII. As shown in Fig. 7, the experiment detects a strong interaction peak in fragment III containing the p2 promoter region and lower interaction frequency in other nearby fragments. This result suggests that the interaction is specific between the p1 enhancer and the p2 gene; if there were no specific interactions and only random passive interactions, 1 would expect the highest interaction frequency with the fragment nearest the enhancer (fragment VII) and a declining frequency at further distances. The comparison of 7 different fragments from both sides of the p2 gene provides a level of internal control for the experiment. Additional 3C experiments testing other sites and alleles could further define the interaction, but such experiments are beyond the scope of this study.
Fig. 7.
(Top) The structure of SP-97 from Fig. 4d. (Bottom) Relative cross-linking frequency at p1/p2 locus in SP-97. The vertical shaded columns indicate the location of BglII fragments, numbered with Roman numerals. The anchor fragments are shaded in black. The blue line shows the relative crosslinking frequency of fragments tested against the anchors. Error bars indicate the standard error of the mean of 3 samples. The y-axis is cross-linking frequency, and the x-axis is the distance in kb. Location of p1 enhancers and p2 gene is shown above the graph. Fragments VIII and X have the same sequence and orientation, whereas fragments IX and XI are structurally different and in the opposite orientation. This is due to the duplication event in the origin of SP-97 (Fig. 4, b and c).
Discussion
Progressive rearrangements form complex alleles
Complex Chromosomal Rearrangements (CCRs) are defined in the human genetics literature as structures consisting of more than 2 breakpoints and involving 2 or more chromosomes (Pellestor et al. 2011). CCRs are clinically important as they are involved in a number of abnormal phenotypes such as recurrent miscarriages (Giardino et al. 2009), mental retardation (Batanian and Eswara 1998), and congenital malformations (Vermeulen et al. 2004). In addition to disease-causing variation, CCRs are important for the formation of loci with adaptive benefits in both animals and plants. Fibrinogen locus in humans, which is a major clotting factor (Kant et al. 1985), and the sh2-R allele of the maize shrunken-2 locus which gave rise to the sweetcorns (Kramer et al. 2015; Hu et al. 2021) are 2 examples of alleles originating from multiple rearrangements leading to a complex structure. The different p2 alleles we present here are also examples of CCRs in plants, except for the involvement of a single chromosome in their formation. We show that a pair of DNA transposable elements can form CCRs by recurrent alternative transposition events, leading to the formation of complex alleles that can alter gene expression.
Previous studies have shown that alternative transposition of Ac/Ds elements can generate genomic rearrangements (Zhang et al. 2009; Yu et al. 2011). Even a single rearrangement event can have a dramatic effect on gene expression (Sharma et al. 2021). In this study, we used the red pericarp phenotype as an indicator of p2 gene activation. We screened for red kernels to identify rearrangements caused by the movement of a pair of DNA elements (Ac/fAc). We show that these elements remain active and capable of undergoing transpositions causing progressive rearrangements. We identified CCRs with p2 activity, but it is possible for other CCRs to occur without activation of p2. So, although we describe only 5 CCRs among 4,000 ears, the total number of CCRs is likely much higher. The 5 CCR alleles described here consist of multiple rearrangement events leading to the activation of the p2 gene due to the presence of the p1 enhancer in close proximity. In addition to the enhancer being able to interact with the target gene, the loss of function in SP-97M1, SP-7M1, and SP-12M1 alleles with a decrease in the number of copies of the enhancer indicates that multiple enhancers work together to induce the comparatively darker red phenotype in SP-97, SP-7, and SP-12.
RET-induced DNA re-replication can generate CIs that act as Ac-macrotransposons (Zhang et al. 2014; Su et al. 2020). Ac-macrotransposons have been found in several different maize lines highlighting their role in genome evolution (Wang et al. 2022). The SP-11 allele is an example of the reinsertion of a 10.3-kb CI containing the p1 enhancer near the p2 gene. The retention of the macrotransposon at its original location together with the insertion of a second copy shows that these macrotransposons can increase in number, as previously proposed for Ac element transposition (Greenblatt and Brink 1962; Chen et al. 1987). In addition to the 10.3-kb macrotransposon present in all the cases discussed in our results, we present the example of SP-97, which contains a large macrotransposon of 28-kb size. The mutant SP-97M1 shows that this macrotransposon is able to excise. Ac/fAc termini in reverse or direct orientation have been known to cause chromosome breaks at a frequency inversely proportional to the distance between the interacting termini (Dooner and Belachew 1991; Yu et al. 2010). Both SP-12 and SP-97 were found capable of chromosome breakage, although breakage is much more frequent in SP-97 (Supplementary Fig. 6), likely due to the presence of numerous closely spaced Ac/fAc termini.
Effects of multiple enhancers
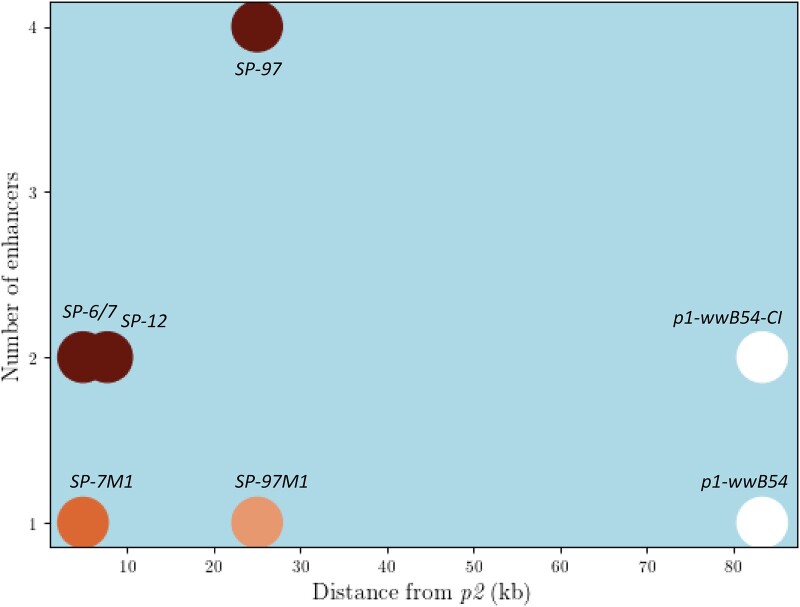
There is clearly a limited distance up to which the p1 enhancer can influence p2 gene expression. The 2 cases which have the p1 enhancers more than 80 kb away from p2, the parental p1-wwB54 allele with 1 p1 enhancer and p1-wwB54-CI with 2 p1 enhancers, both have colorless pericarp. But all alleles with active p2 expression in the pericarp have at least 1 enhancer within 25 kb of p2, indicating that the distance at which a single enhancer can no longer interact with p2 is somewhere between 25 and 80 kb. Among cases with a single p1 enhancer, the pericarp color gets lighter with an increase in distance (Fig. 8), indicating that the interaction is distance dependent. A positive correlation between gene expression and enhancer proximity has also been reported at a genomic scale (Downes et al. 2021) and a single locus (Nolis et al. 2009). Although enhancers are known to work from large distances, these long-range interactions are enabled by facilitating mechanisms such as chromatin looping and transcription factors that help the enhancer to reach the target promoter (Deng et al. 2012; Bartman et al. 2016). In addition to these facilitating mechanisms, an enhancer might have an intrinsic range in which it can interact with its target promoter. In a 2009 study using transgenic HeLa cells, Nolis et al. show the IFN-β enhancer activates transcription only up to a distance of 560 bp but with the addition of binding sites for Sp1 and CCAAT enhancer binding protein transcription factors, the range increases to at least 2,325 bp. In our case, SP-97M1 has a lighter red phenotype with the single enhancer at 25 kb, and SP-97 has a darker red phenotype with additional enhancers at distances of 34, 41.5, and 50.5 kb, which are all larger than 25 kb (Figs. 4 and 5), suggesting that presence of multiple enhancers can increase the maximum distance at which productive enhancer–promoter interactions can occur. If increasing the enhancer number strengthens the enhancer–promoter interaction, it is possible that it could also disrupt the 3D chromatin structure (Chakraborty et al. 2022).
Fig. 8.
Relationship between enhancer number, distance from p2, and pericarp phenotype. The circles represent different alleles and their pericarp color, the x-axis is the distance of the nearest enhancer in kb from p2, and the y-axis is the number of enhancers. The darker red phenotype seems to have a positive correlation with the number of enhancers and a negative correlation with the distance from the target gene. Additional factors may affect intensity of pericarp color.
Long-range transcriptional cis-regulatory elements, including enhancers, are widespread in the maize genome (Ricci et al. 2019). A hepta-repeat present at 100-kb upstream from the b1 gene and distal cis-element present at 140-kb upstream of the BX1 gene are candidate enhancer sequences for their respective target genes. In both cases, alleles containing multiple copies of these sequences have higher expression compared to their single copy counterparts (Louwers et al. 2009; Zheng et al. 2015). Here, we show that alleles containing multiple copies of the known p1 enhancer tend to exhibit darker red pericarp, while alleles with a single p1 enhancer copy near p2 specify lighter red phenotype (Fig. 8). Importantly, SP-97 with 4 enhancers has significantly greater pericarp color than its single enhancer derivative SP-97M1 (Fig. 6). A similar correlation between enhancer number and expression has been reported in transgenic grape and tobacco plants (Li et al. 2004). Having multiple copies of an enhancer is known to increase expression in mammalian systems as well (Downes et al. 2021). Although a change in enhancer number and position can cause misexpression (Will et al. 2017), in some contexts having multiple enhancers can be advantageous as it can provide robustness against disease-causing mutations (Osterwalder et al. 2018; Wang and Goldstein 2020). In summary, we show that RET-induced rearrangements can change enhancer copy number and position, fueling cis-regulatory variation vital for genomic evolution.
Acknowledgments
We thank Terry Olson and Libuse Brachova for technical assistance. We thank Harry T. Horner, Mohan Gupta, Allison Birnbaum, Erica Unger-Wallace, and Weijia Su for their assistance and contributions to the project.
Funding
This research is supported by the United States Department of Agriculture (USDA) National Institute of Food and Agriculture Hatch projects IOW05282 and IOW05669 and by State of Iowa funds.
Contributor Information
Sharu Paul Sharma, Department of Genetics, Development and Cell Biology, Iowa State University, Ames, IA 50011, USA.
Thomas Peterson, Department of Genetics, Development and Cell Biology, Iowa State University, Ames, IA 50011, USA; Department of Agronomy, Iowa State University, Ames, IA 50011, USA.
Data Availability
Maize genetic stocks are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplementary material is available on figshare: https://doi.org/10.25386/genetics.19944359.
Literature cited
- Athma P, Grotewold E, Peterson T. Insertional mutagenesis of the maize P gene by intragenic transposition of Ac. Genetics. 1992;131(1):199–209. 10.1093/genetics/131.1.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RE, Brink RA. Kernel pattern in variegated pericarp maize and the frequency of self-colored offspring. Am J Bot. 1952;39(9):637–644. 10.2307/2438369. [DOI] [Google Scholar]
- Barclay PC, Brink RA. The relation between modulator and activator in maize. Proc Natl Acad Sci U S A. 1954;40(12):1118–1126. 10.1073/pnas.40.12.1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartman CR, Hsu SC, Hsiung CC, Raj A, Blobel GA. Enhancer regulation of transcriptional bursting parameters revealed by forced chromatin looping. Mol Cell. 2016;62(2):237–247. 10.1016/j.molcel.2016.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batanian JR, Eswara MS. De novo apparently balanced complex chromosome rearrangement (CCR) involving chromosomes 4, 18, and 21 in a girl with mental retardation: report and review. Am J Med Genet. 1998;78(1):44–51. [DOI] [PubMed] [Google Scholar]
- Bompadre O, Andrey G. Chromatin topology in development and disease. Curr Opin Genet Dev. 2019;55:32–38. 10.1016/j.gde.2019.04.007 [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Kopitchinski N, Eraso A, Awasthi P, Chari R, Rocha PP. High affinity enhancer-promoter interactions can bypass CTCF/cohesin-mediated insulation and contribute to phenotypic robustness. bioRxiv. 10.1101/2021.12.30.474562, 2022. [DOI]
- Chen J, Greenblatt IM, Dellaporta SL. Transposition of Ac from the P locus of maize into unreplicated chromosomal sites. Genetics. 1987;117(1):109–116. 10.1093/genetics/117.1.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14(6):390–403. 10.1038/nrg3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149(6):1233–1244. 10.1016/j.cell.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doğan ES, Liu C. Three-dimensional chromatin packing and positioning of plant genomes. Nature Plants. 2018;4(8):521–529. [DOI] [PubMed] [Google Scholar]
- Dooner HK, Belachew A. Chromosome breakage by pairs of closely linked transposable elements of the Ac-Ds family in maize. Genetics. 1991;129(3):855–862. 10.1093/genetics/129.3.855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner HK, Robbins TP, Jorgensen RA. Genetic and developmental control of anthocyanin biosynthesis. Annu Rev Genet. 1991;25(1):173–199. 10.1146/annurev.ge.25.120191.001133 [DOI] [PubMed] [Google Scholar]
- Downes DJ, Beagrie RA, Gosden ME, Telenius J, Carpenter SJ, Nussbaum L, De Ornellas S, Sergeant M, Eijsbouts CQ, Schwessinger R, et al. High-resolution targeted 3C interrogation of Cis-regulatory element organization at genome-wide scale. Nat Commun. 2021;12(1):531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson RA. Genetical studies of variegated pericarp in maize. Genetics. 1917;2(1):1–35. 10.1093/genetics/2.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardino D, Corti C, Ballarati L, Colombo D, Sala E, Villa N, Piombo G, Pierluigi M, Faravelli F, Guerneri S, et al. De novo balanced chromosome rearrangements in prenatal diagnosis. Prenat Diagn. 2009;29(3):257–265. 10.1002/pd.2215 [DOI] [PubMed] [Google Scholar]
- Goettel W, Messing J. Change of gene structure and function by non-homologous end-joining, homologous recombination, and transposition of DNA. PLoS Genet. 2009;5(6):e1000516. 10.1371/journal.pgen.1000516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt IM, Brink RA. Twin mutations in medium variegated pericarp maize. Genetics. 1962;47(4):489–501. 10.1093/genetics/47.4.489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E, Drummond BJ, Bowen B, Peterson T. The Myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell. 1994;76(3):543–553. [DOI] [PubMed] [Google Scholar]
- Heger P, Marin B, Bartkuhn M, Schierenberg E, Wiehe T. The chromatin insulator CTCF and the emergence of metazoan diversity. Proc Natl Acad Sci U S A. 2012;109(43):17507–17512. 10.1073/pnas.1111941109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Colantonio V, Müller BSF, Leach KA, Nanni A, Finegan C, Wang B, Baseggio M, Newton CJ, Juhl EM, et al. Genome assembly and population genomic analysis provide insights into the evolution of modern sweet corn. Nat Commun. 2021;12:1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JT, Dooner HK. Macrotransposition and other complex chromosomal restructuring in maize by closely linked transposons in direct orientation. Plant Cell. 2008;20(8). 10.1105/tpc.108.060582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant JA, Fornace AJ, Saxe D, Simon MI, McBride OW, Crabtree GR. Evolution and organization of the fibrinogen locus on chromosome 4: gene duplication accompanied by transposition and inversion. Proc Natl Acad Sci U S A. 1985;82(8):2344–2348. 10.1073/pnas.82.8.2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer V, Shaw JR, Senior ML, Hannah LC. The Sh2-R allele of the maize Shrunken-2 locus was caused by a complex chromosomal rearrangement. Theor Appl Genet. 2015;128(3):445–452. [DOI] [PubMed] [Google Scholar]
- Krumm A, Duan Z. Understanding the 3D genome: emerging impacts on human disease. Semin Cell Dev Biol. 2019;90:62–77. 10.1016/j.semcdb.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, Kazmar T, Stampfel G, Yáñez-Cuna JO, Pagani M, Schernhuber K, Dickson BJ, Stark A. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature. 2014;512(7512):91–95. 10.1038/nature13395 [DOI] [PubMed] [Google Scholar]
- Lechelt C, Peterson T, Laird J, Chen SL, Dellaporta E, Dennis WJ, Peacock P Starlinger . Isolation and molecular analysis of the maize P locus. Mol General Genet. 1989;219:225–234. 10.1007/BF00261181 [DOI] [PubMed] [Google Scholar]
- Li ZT, Jayasankar S, Gray DJ. Bi-directional duplex promoters with duplicated enhancers significantly increase transgene expression in grape and tobacco. Transgenic Res. 2004;13(2):143–154. 10.1023/B:TRAG.0000026074.11859.77 [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louwers M, Bader M, Haring M, van Driel R, de Laat W, Stam M. Tissue- and expression level-specific chromatin looping at maize B1 epialleles. Plant Cell. 2009;21(3):832–842. 10.1105/tpc.108.064329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louwers M, Splinter E, van Driel R, de Laat W, Stam M. Studying physical chromatin interactions in plants using chromosome conformation capture (3C). Nat Protoc. 2009;4(8):1216–1229. 10.1038/nprot.2009.113 [DOI] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161(5):1012–1025. 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M, Lee SE. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24(11):529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno MA, Chen J, Greenblatt I, Dellaporta SL. Reconstitutional mutagenesis of the maize P gene by short-range Ac transpositions. Genetics. 1992;131(4):939–956. 10.1093/genetics/131.4.939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolis IK, McKay DJ, Mantouvalou E, Lomvardas S, Merika M, Thanos D. Transcription factors mediate long-range enhancer-promoter interactions. Proc Natl Acad Sci U S A. 2009;106(48):20222–20227. 10.1073/pnas.0902454106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Dekker J, Heard E. Segmental folding of chromosomes: a basis for structural and regulatory chromosomal neighborhoods? BioEssays. 2013;35(9):818–828. 10.1002/bies.201300040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120(3):621–623. 10.1093/genetics/120.3.621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder M, Barozzi I, Tissières V, Fukuda-Yuzawa Y, Mannion BJ, Afzal SY, Lee EA, Zhu Y, Plajzer-Frick I, Pickle CS, et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature. 2018;554(7691):239–243. 10.1038/nature25461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellestor F, Anahory T, Lefort G, Puechberty J, Liehr T, Hédon B, Sarda P. Complex chromosomal rearrangements: origin and meiotic behavior. Hum Reprod Update. 2011;17(4):476–494. 10.1093/humupd/dmr010 [DOI] [PubMed] [Google Scholar]
- Ricci WA, Lu Z, Ji L, Marand AP, Ethridge CL, Murphy NG, Noshay JM, Galli M, Mejía-Guerra MK, Colomé-Tatché M, et al. Widespread long-range Cis-regulatory elements in the maize genome. Nature Plants. 2019;5(12):1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA Spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci U S A. 1984;81(24):8014–8018. 10.1073/pnas.81.24.8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SP, Peterson T. Rapid detection of transposon-induced genome rearrangements. Methods Mol Biol. 2021;2250. 10.1007/978-1-0716-1134-0_13 [DOI] [PubMed] [Google Scholar]
- Sharma SP, Zuo T, Peterson T. Transposon-induced inversions activate gene expression in the maize pericarp. Genetics. 2021;218(2):iyab062. 10.1093/GENETICS/IYAB062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko L, Peterson T. Transgene-induced silencing identifies sequences involved in the establishment of paramutation of the maize P1 gene. Plant Cell. 2001;13(2):319–335. 10.1105/tpc.13.2.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko L, Li X, Cocciolone SM, Chopra S, Tagliani L, Bowen B, Daniels M, Peterson T. Complex structure of a maize Myb gene promoter: functional analysis in transgenic plants. Plant J. 2000;22(6):471–482. 10.1046/j.1365-313X.2000.00750.x [DOI] [PubMed] [Google Scholar]
- Sidorenko L, Li X, Tagliani L, Bowen B, Peterson T. Characterization of the regulatory elements of the maize P-rr gene by transient expression assays. Plant Mol Biol. 1999;39(1):11–19. 10.1023/A:1006172815663 [DOI] [PubMed] [Google Scholar]
- Singh M, Lewis PE, Hardeman K, Bai L, Rose JKC, Mazourek M, Chomet P, Brutnell TP. Activator mutagenesis of the pink Scutellum1/Viviparous7 locus of maize. Plant Cell. 2003;15(4):874–884. 10.1105/tpc.010249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Sharma SP, Peterson T. Evolutionary impacts of alternative transposition. Origin Evol Biodivers. 2018;113–130. 10.1007/978-3-319-95954-2_7 [DOI] [Google Scholar]
- Su W, Zuo T, Peterson T. Ectopic expression of a maize gene is induced by composite insertions generated through alternative transposition. Genetics. 2020;216(4):1039–1049. 10.1534/genetics.120.303592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell. 2002;10(6). 10.1016/S1097-27655 [DOI] [PubMed] [Google Scholar]
- Vermeulen S, Menten B, van Roy N, van Limbergen H, de Paepe A, Mortier G, Speleman F. Molecular cytogenetic analysis of complex chromosomal rearrangements in patients with mental retardation and congenital malformations: delineation of 7q21.11 breakpoints. Am J Med Genet. 2004;124A(1):10–18. 10.1002/ajmg.a.20378 [DOI] [PubMed] [Google Scholar]
- Wang D, Yu C, Zhang J, Peterson T. Excision and reinsertion of Ac macrotransposons in maize. Genetics. 2022;221(4):iyac067. 10.1093/genetics/iyac067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Peterson T. Isolation of sequences flanking Ac insertion sites by Ac casting. Methods Mol Biol. 2013;1057. 10.1007/978-1-62703-568-2_8 [DOI] [PubMed] [Google Scholar]
- Wang X, Goldstein DB. Enhancer domains predict gene pathogenicity and inform gene discovery in complex disease. Am J Hum Genet. 2020;106(2):215–233. 10.1016/j.ajhg.2020.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B, Zicola J, Oka R, Stam M. Plant enhancers: a call for discovery. Trends Plant Sci. 2016;21(11):974–987. 10.1016/j.tplants.2016.07.013 [DOI] [PubMed] [Google Scholar]
- Will AJ, Cova G, Osterwalder M, Chan W-L, Wittler L, Brieske N, Heinrich V, de Villartay J-P, Vingron M, Klopocki E, et al. Composition and dosage of a multipartite enhancer cluster control developmental expression of Ihh (Indian Hedgehog). Nat Genet. 2017;49(10):1539–1545. 10.1038/ng.3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Zhang J, Peterson T. Genome rearrangements in maize induced by alternative transposition of reversed Ac/Ds termini. Genetics. 2011;188(1):59–67. 10.1534/genetics.111.126847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Zhang J, Pulletikurti V, Weber DF, Peterson T. Spatial configuration of transposable element Ac termini affects their ability to induce chromosomal breakage in maize. Plant Cell. 2010;22(3):744–754. 10.1105/tpc.109.070052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Peterson T. Comparisons of maize Pericarp Color1 alleles reveal paralogous gene recombination and an organ-specific enhancer region. Plant Cell. 2005a;17(3):903–914. 10.1105/tpc.104.029660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Peterson T. Transposition of Reversed Ac element ends generates chromosome rearrangements in maize. Genetics. 2004;167(4):1929–1937. 10.1534/genetics.103.026229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Peterson T. A segmental deletion series generated by sister-chromatid transposition of Ac transposable elements in maize. Genetics. 2005b;171(1):333–344. 10.1534/genetics.104.035576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yu C, Pulletikurti V, Lamb J, Danilova T, Weber DF, Birehler J, Peterson T. Alternative Ac/Ds transposition induces major chromosomal rearrangements in maize. Genes and Development. 2009;23(6):755–765. 10.1101/gad.1776909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang F, Peterson T. Transposition of reversed Ac element ends generates novel chimeric genes in maize. PLoS Genet. 2006;2(10):e164. 10.1371/journal.pgen.0020164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zuo T, Peterson T. Generation of tandem direct duplications by reversed-ends transposition of maize Ac elements. PLoS Genet. 2013;9(8):e1003691. 10.1371/journal.pgen.1003691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zuo T, Wang D, Peterson T. Transposition-mediated DNA Re-replication in maize. eLife. 2014;3:e03724. 10.7554/eLife.03724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Chopra S, Peterson T. A segmental gene duplication generated differentially expressed Myb-homologous genes in maize. Plant Cell. 2000;12(12):2311–2322. 10.1105/tpc.12.12.2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, McMullen MD, Bauer E, Schön C-C, Gierl A, Frey M. Prolonged expression of the BX1 signature enzyme is associated with a recombination hotspot in the benzoxazinoid gene cluster in Zea mays. J Exp Bot. 2015;66(13):3917–3930. 10.1093/jxb/erv192 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Maize genetic stocks are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplementary material is available on figshare: https://doi.org/10.25386/genetics.19944359.