Abstract
The Developmental Origins of Health and Disease hypothesis postulates that early-life stressors can predispose people to disease later in life. In the roundworm Caenorhabditis elegans, prolonged early-life starvation causes germline tumors, uterine masses, and other gonad abnormalities to develop in well-fed adults. Reduction of insulin/insulin-like growth factor (IGF) signaling (IIS) during larval development suppresses these starvation-induced abnormalities. However, molecular mechanisms at play in formation and suppression of starvation-induced abnormalities are unclear. Here we describe mechanisms through which early-life starvation and reduced IIS affect starvation-induced abnormalities. Transcriptome sequencing revealed that expression of genes in the Wnt signaling pathway is upregulated in adults starved as young larvae, and that knockdown of the insulin/IGF receptor daf-2/InsR decreases their expression. Reduction of Wnt signaling through RNAi or mutation reduced starvation-induced abnormalities, and hyperactivation of Wnt signaling produced gonad abnormalities in worms that had not been starved. Genetic and reporter-gene analyses suggest that Wnt signaling acts downstream of IIS in the soma to cell-nonautonomously promote germline hyperproliferation. In summary, this work reveals that IIS-dependent transcriptional regulation of Wnt signaling promotes starvation-induced gonad abnormalities, illuminating signaling mechanisms that contribute to adult pathology following early-life starvation.
Keywords: early-life stress, starvation, DOHaD, insulin, Wnt, diapause, L1 arrest
Introduction
Early-life adversity can predispose an individual to a variety of diseases later in life (Gillman 2005; Suzuki 2018). Collectively, the ways in which developmental perturbations cause disease are referred to as Developmental Origins of Health and Disease (DOHaD). Nutrient stress during early development can exert substantial impacts on adult physiology. For instance, human populations exposed to malnutrition in utero experience higher rates of cardiovascular disease, obesity, diabetes, and breast cancer in adulthood (Roseboom et al. 2001; Painter et al. 2006; Barker 2007; Fentiman et al. 2007; Walker and Ho 2012). However, the regulatory mechanisms through which early-life malnutrition causes disease later in life are unclear.
The nematode Caenorhabditis elegans takes about 3 days to develop from embryo to adult in laboratory conditions (Corsi et al. 2015), facilitating its use as a model for DOHaD. Additionally, C. elegans reversibly arrest development in the first larval stage (L1 arrest or L1 diapause) upon hatching in the absence of food (Baugh 2013). Worms can survive L1 arrest for weeks, and they recover and initiate postembryonic development upon feeding. However, worms subjected to extended L1 arrest (∼1 week) followed by recovery with replete food develop gonad abnormalities, including both hyperproliferative germ-cell tumors and teratoma-like uterine masses (collectively referred to as gonad abnormalities or starvation-induced abnormalities), which decrease their reproductive success (Jobson et al. 2015; Jordan et al. 2019, co-submitted).
There is a single known insulin/insulin-like growth factor (IGF) receptor in C. elegans, daf-2/InsR, which signals through a conserved phosphatidylinositol-3-kinase (PI3K) transduction pathway to antagonize the forkhead transcription factor DAF-16/FoxO (Murphy and Hu 2013). We showed that reducing IGF signaling (IIS) with daf-2 RNAi during larval development after extended L1 arrest suppresses starvation-induced abnormalities, and suppression depends on daf-16/FoxO (Jordan et al. 2019). Our companion manuscript shows that early-life starvation and IIS affect adult lipid metabolism to promote starvation-induced abnormalities (Jordanet al. co-submitted). However, other mechanisms through which IIS and daf-16/FoxO affect starvation-induced abnormalities are unknown.
The widely conserved Wnt signaling pathway regulates important developmental processes in both vertebrates and invertebrates, including cell-fate specification, establishment of the anterior–posterior axis, regulation of neural development, and organogenesis (Komiya and Habas 2008). In many cases, Wnt signaling maintains stem-cell niches (Zhan et al. 2017; Nabhan et al. 2018). However, excessive Wnt activity at these sites can cause dysregulated stem-cell division eventually resulting in tumor formation (Zhan et al. 2017; Nabhan et al. 2018). Wnt signaling also promotes epithelial-to-mesenchymal transition, a common characteristic of metastatic and invasive cancers (Basu et al. 2018). Consequently, overactive Wnt signaling has been linked to many cancers, including colorectal cancer, leukemia, melanoma, breast cancer, and lung cancer (Zhan et al. 2017).
In C. elegans, Wnt signals are responsible for induction of cell polarity, embryonic cell-fate specification, vulval precursor cell-fate specification and neuronal development (Eisenmann et al. 1998). Wnt signals also maintain the proliferative state of stem-cell-like seam cells in larvae (Joshi et al. 2010; Gorrepati et al. 2015). The canonical Wnt signaling pathway is initiated when a Wnt ligand binds to a Frizzled receptor (Sawa and Korswagen 2013). This inhibits the degradation of the transcription factor β-catenin by a destruction complex consisting of Axin, APC, CK1, and GSK3β. β-Catenin can then enter the nucleus, bind T-cell factor (TCF), and activate the expression of Wnt-target genes (Sawa and Korswagen 2013). Notably, the C. elegans genome encodes multiple homologs of Wnt, Frizzled, and β-catenin.
Interestingly, FoxO proteins and IIS can affect Wnt signaling. Direct binding between β-catenin and FoxO has been shown to enhance FoxO activity while competitively inhibiting β-catenin activity in worms and mammalian cells (Essers et al. 2005; Hoogeboom et al. 2008). Insulin signaling also increases Wnt signaling in mouse hepatocytes that have undergone starvation and refeeding (Cabrae et al. 2020).
We describe signaling mechanisms linking early-life starvation and IIS to formation of germline tumors and other developmental abnormalities in adults. Gene expression analysis suggests that early-life starvation and IIS promote Wnt signaling through transcriptional regulation of pathway components, and genetic analysis shows that Wnt signaling contributes to starvation-induced abnormalities. This work elucidates how early-life starvation causes adult pathology, advancing understanding of the ways in which developmental homeostasis is affected by early-life nutrient availability and IIS.
Materials and methods
Worm maintenance
Worms were maintained under standard laboratory conditions at 20°C unless otherwise noted. Animals were passaged regularly to avoid starvation or dietary restriction for at least three generations prior to performing experiments.
Strains used in this study
N2 (Bristol), RB763 cwn-1(ok546) II., MT5383 lin-44(n1792) I., RB1162 cfz-2(ok1201) V., MT8904 lin-17(n3091) I., CB3303 mig-1(e1787) I., HS2329 osEx397 [cwn-1p::cwn-1::Venus], JK3025 gld-1(q485) I/hT2 [bli-4(e937) let-? (q782) qIs48] (I; III)., KN53 hsp-16.2pro:: ΔNTbar-1, LRB406 gld-1(q485) I/hT2 [bli-4(e937) let-? (q782) qIs48] (I; III); hsp-16.2pro:: ΔNTbar-1, NL2098 rrf-1(pk1417) I., LRB424 cwn-1(ok546) II; gld-1(q485) I/hT2 [bli-4(e937) let-? (q782) qIs48] (I; III)., NL2550 ppw-1(pk2505) I., BX107 fat-5(tm420) V., EU308 +/szT1 [lon-2(e678)] I; mom-1(or10) dpy-6(e14)/szT1 X.
Starvation and recovery assay
Caenorhabditis elegans embryos were obtained through sodium hypochlorite treatment, washed three times with virgin S-basal (no ethanol or cholesterol), and allowed to hatch and enter L1 arrest on a tissue culture roller drum at approximately 22°C (Hibshman et al. 2021). Worms were cultured for 1 or 8 days after sodium hypochlorite treatment before plating on lawns of E. coli HT115 bacteria. Animals were incubated at 20°C for 72 or 90 h, as indicated, before scoring gonad abnormalities.
Scoring gonad abnormalities and large mitotic tumors
Adult worms were selected at random and placed onto 4% noble agar pads containing 2.4 mM sodium azide. The animals were observed using Differential Interference Contrast (Nomarski) microscopy on a Zeiss AxioImager compound microscope at 200× magnification. Worms were scored on a binary scale based on the presence of any gonad abnormality (germ-cell tumor or uterine mass, abnormal) or complete absence of gonad abnormalities (normal) (Jordanet al. co-submitted.). For assessment of large mitotic tumors in gld-1 mutant worms, worms were scored on a binary scale based on the presence of enlarged masses of germ cells in the gonad (tumor) or complete absence of masses (no tumor). For assessment of gonad abnormalities in mom-1(or10)/+ worms (strain EU308), Dpy hermaphrodites and Lon males were excluded from scoring.
Gonad imaging
Worms were imaged with an AxioCam camera on a Zeiss AxioImager compound microscope using Zen software. Images were taken at 200× or 400× magnification as indicated. Gonads were outlined using Adobe Photoshop software, and images were cropped using Inkscape.
Heat shock
For experiments involving heat-shock activation of bar-1, worms were starved and recovered as described above. After 36 h of recovery, worms were incubated at 34°C (heat shock) or 20°C (no heat shock) for 1 h in an incubator. Worms were then returned to 20°C and scored 90 h after the start of the initial recovery.
Introduction of truncated, heat-shock-inducible bar-1 into gld-1 background
To create the LRB406 hsp-16.2pro:: ΔNTbar-1; gld-1(q485) I/hT2 [bli-4(e937) let-? (q782) qIs48] (I; III) strain used in this study, JK3025 gld-1(q485) I/hT2 [bli-4(e937) let-? (q782) qIs48] (I; III) males were allowed to mate with KN53 hsp-16.2pro:: ΔNTbar-1 hermaphrodites. A single hermaphrodite with the hT2 balancer (confirmed phenotypically by expression of GFP in the pharynx) was selected and allowed to self-fertilize. Twenty progenies were then singled and allowed to self-fertilize. A plate-level phenotype (slow growth) was used to identify worms homozygous for the bar-1 overexpression allele. A single hermaphrodite homozygous for bar-1 overexpression and possessing the hT2 balancer was allowed to mate with a JK3025 male, and 20 progenies were singled and allowed to self-fertilize. These progenies were then singled for a second round of self-fertilization, and worms with mutant gld-1(q485) over the hT2 [bli-4(e937) let-? (q782) qIs48] balancer, and homozygous for the hsp-16.2pro:: ΔNTbar-1 allele were identified phenotypically.
Introduction of cwn-1 mutation into gld-1 background
To create the LRB424 cwn-1(ok546) II; gld-1(q485) I/hT2 [bli-4(e937) let-? (q782) qIs48] (I; III) strain used in this study, JK3025 gld-1(q485) I/hT2 [bli-4(e937) let-? (q782) qIs48] (I; III) males were allowed to mate with RB763 cwn-1(ok546) II hermaphrodites. One hermaphrodite expressing the hT2 balancer (confirmed phenotypically) was selected from the progeny and allowed to mate again with JK3025 males. Ten hermaphrodite worms expressing the hT2 balancer were selected from the progeny of this mating event, separated, and allowed to self-fertilize. The presence of the gld-1 mutation was identified by the presence of some offspring which demonstrated the germ-cell differentiation–deficient phenotype. Eight hermaphrodites with intact germ-cell differentiation were selected from each of five of these self-fertilizations that retained the gld-1 mutation. The presence of a homozygous deletion in cwn-1 was identified through PCR amplification of the cwn-1 gene in each of the selected worms, and the presence of the gld-1 mutation was reconfirmed phenotypically.
RNA interference
To prepare RNAi bacteria, E. coli HT115 with plasmids that express double-stranded RNA of various genes were grown on 100 μg/ml carbenicillin and 12.5 μg/ml tetracycline LB plates. Single colonies from these plates were used to inoculate LB starter cultures with 100 μg/ml carbenicillin and 12.5 μg/ml tetracycline. These were transferred into larger cultures containing 50 μg/ml carbenicillin. Cells from large cultures were spun down at 4°C, then resuspended at a concentration of 25 μg/ml in 15% glycerol S-complete medium. The resulting suspensions were separated into aliquots and stored at −80°C. Aliquots were thawed once and seeded onto nematode growth medium plates containing 25 μg/ml carbenicillin and 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Bacteria was grown overnight at ∼22°C before plating worms. RNAi plasmids used include empty vector (pAD12), daf-2 (pAD48), and daf-16 (pAD43). daf-18 RNAi bacteria used is from the Open Biosystems library.
Quantitative image analysis
Strain HS2329 osEx397 [cwn-1p::cwn-1::Venus] was cultured, hypochlorite treated, starved for 8 days, and recovered on empty vector or daf-2 RNAi food as described above. Forty-eight hours after plating on food, worms were washed from plates in S-basal, anesthetized with sodium azide, and transferred to black, clear-bottom 96-well plates for imaging on a Molecular Devices ImageXpress Nano with a 4× Nikon objective. Using the MetaXpress software (Molecular Devices), worms were identified as objects in brightfield to create masks. In the fluorescent channel, masks were used to calculate background pixel intensities across the entire field minus the objects (masked areas). Background intensity was subtracted from pixel intensities for each object, and the background-corrected average pixel intensity was calculated for each object. Objects that intersected the edge of the field were not included, and images were manually curated so that objects that were not single worms were excluded.
Statistical analyses
All statistical tests were performed in R. When performing t-tests, Bartlett's test was first performed to determine if there was a significant difference in variance among conditions that were compared. If the variances were not significantly different, a t-test with pooled variances was performed. Otherwise, a t-test with empirical (unpooled) variance was performed. For quantitative image analysis of CWN-1::VENUS expression, a pairwise linear mixed-effects model was used with conditions as fixed effect and replicate as random effect. All plots were produced with the R package ggplot2 (Wickham 2016).
Results
Early-life starvation and IIS affect Wnt pathway gene expression
We confirmed that worms starved for 8 days as L1-stage larvae (L1 arrest) and recovered in replete conditions (with E. coli as food) develop gonad abnormalities, including proximal germ-cell tumors and uterine masses, as adults (starvation-induced abnormalities; Fig. 1a; Jordan et al. 2019, co-submitted). Reduction of IIS after starvation during development with daf-2/InsR RNAi suppresses starvation-induced abnormalities (Jordan et al. 2019, co-submitted). To identify genes that contribute to development of or suppression of starvation-induced abnormalities, we performed an mRNA sequencing (RNA-seq) experiment to determine the effects of early-life starvation and IIS on adult gene expression (primary results published in Jordan et al., co-submitted). Samples were collected at egg-laying onset as whole worms that had been previously starved for 1 day (control) or 8 (starved) days during L1 arrest and then cultured on empty vector (negative control) or daf-2 (reduced IIS) RNAi food. These results revealed prominent effects on lipid metabolism and membrane biology, as described in our companion manuscript (Jordanet al. co-submitted).
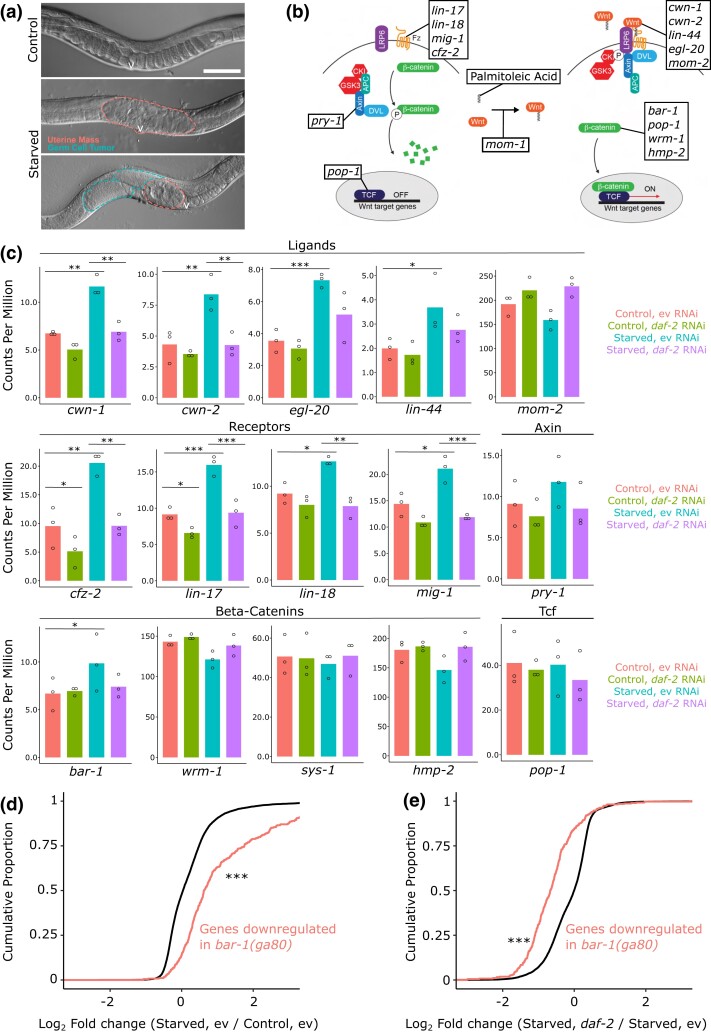
Fig. 1.
Expression of multiple Wnt ligands and receptors is affected by early-life starvation and IIS. a) Representative images of wild-type worms starved 1 day (control) or 8 days (starved) as L1 larvae and recovered for 90 h at 20°C with plentiful E. coli HT115 are presented. Dashed lines indicate starvation-induced abnormalities in the gonadal region, with dashes circumscribing uterine masses and germ-cell tumors. Scale bar represents 50 μm. The letter “v” indicates the vulva. b) Cartoon depicting the canonical Wnt signaling pathway is shown in the absence (left) and presence (right) of a Wnt ligand. Boxes include the genes in C. elegans that encode each component of the pathway. Adapted with permission from WormBook.org (Sawa and Korswagen 2013). c) mRNA counts per million for genes involved in the Wnt signaling pathway is plotted. The exactTest in EdgeR was used for statistics: *P < 0.05; **P < 0.01; ***P < 0.001. d) Cumulative distributions of fold changes in mRNA expression levels for starved vs control worms at egg-laying onset are plotted for genes activated by bar-1 (in red; all detected genes in black). e) Cumulative distributions of fold changes in mRNA expression levels for starved daf-2 RNAi vs starved empty vector worms at egg-laying onset is plotted for genes activated by bar-1. c–e) Samples were collected at egg-laying onset for worms starved for 1 day (control) or 8 days (starved) and recovered on empty vector or daf-2 RNAi food, as indicated in the legend (primary results are published in Jordan et al. co-submitted). d and e) The Kolmogorov–Smirnov test was used for statistics: ***P < 0.001. ev, empty vector.
Given extensive roles that Wnt signaling plays in development and cancer, we wondered if early-life starvation and IIS affect Wnt pathway gene expression. Figure 1b displays components of the canonical Wnt signaling pathway in C. elegans (Sawa and Korswagen 2013). Of the nine C. elegans Wnt ligands and receptors, eight (all but mom-2) had significantly increased relative mRNA expression in adults that had been starved as larvae compared with controls (P < 0.05; Fig. 1c). Notably, daf-2/InsR RNAi decreased expression of all eight of these genes in starved worms back to baseline levels, with the decrease being statistically significant in six cases. Furthermore, no ligand or receptor had significantly different expression between control and starved worms recovered on daf-2 RNAi (Fig. 1c). The ligand-encoding gene mom-2 is the notable exception to these trends, since it did not significantly deviate from control expression levels in any condition. We examined other genes involved in the Wnt signaling pathway, including pry-1/Axin, four β-catenin homologs, and pop-1/Tcf. The only one of these with significant differential expression in any condition was bar-1/β-catenin, which had significantly increased expression in starved worms (Fig. 1c). Consistent with the patterns seen for Wnt ligands and receptors, bar-1 did not have increased expression in starved worms recovered on daf-2 RNAi food. These expression patterns suggest that Wnt signaling is increased following early-life starvation but that it is decreased with reduced IIS during larval development.
Based on the expression patterns of Wnt ligands and receptors, we wondered whether Wnt signaling activity is increased following early-life starvation. To assess Wnt signaling activity, we interrogated expression levels of genes that are downregulated in bar-1/β-catenin loss-of-function mutant worms (Van Der Bent et al. 2015). bar-1 activates expression of these genes, and they were collectively expressed at significantly higher levels in starved worms compared with control worms at egg-laying onset, suggesting increased Wnt signaling activity (Fig. 1d). We also wondered whether the increase in apparent Wnt activity is IIS dependent. The same bar-1 target genes were collectively expressed at significantly lower levels in starved worms fed daf-2 RNAi food during recovery than starved worms fed empty vector RNAi during recovery (Fig. 1e). Together these results support the hypothesis that Wnt signaling activity is increased following early-life starvation and that it is suppressed by reducing IIS in previously starved worms.
Wnt signaling promotes development of starvation-induced abnormalities
We hypothesized that increased Wnt signaling in previously starved worms promotes starvation-induced abnormalities. To test this, we used RNAi and mutations to disrupt Wnt signaling in control and starved worms. daf-2 RNAi significantly reduced starvation-induced abnormalities, as expected (Jordan et al. 2019, co-submitted; Fig 2a). RNAi of Wnt ligands cwn-1, lin-44, and mom-2, and of receptors cfz-2, lin-17, and mig-1 significantly reduced the frequency of starvation-induced abnormalities (Fig. 2a). Notably, RNAi was administered during larval development after starvation, indicating that gene function during recovery rather than L1 starvation itself affects formation of starvation-induced abnormalities. However, suppression of abnormalities was consistently smaller than with daf-2 RNAi. The smaller effect size is consistent with other pathways downstream of IIS promoting starvation-induced abnormalities (Jordanet al. co-submitted) and possibly functional redundancy among Wnt pathway genes.
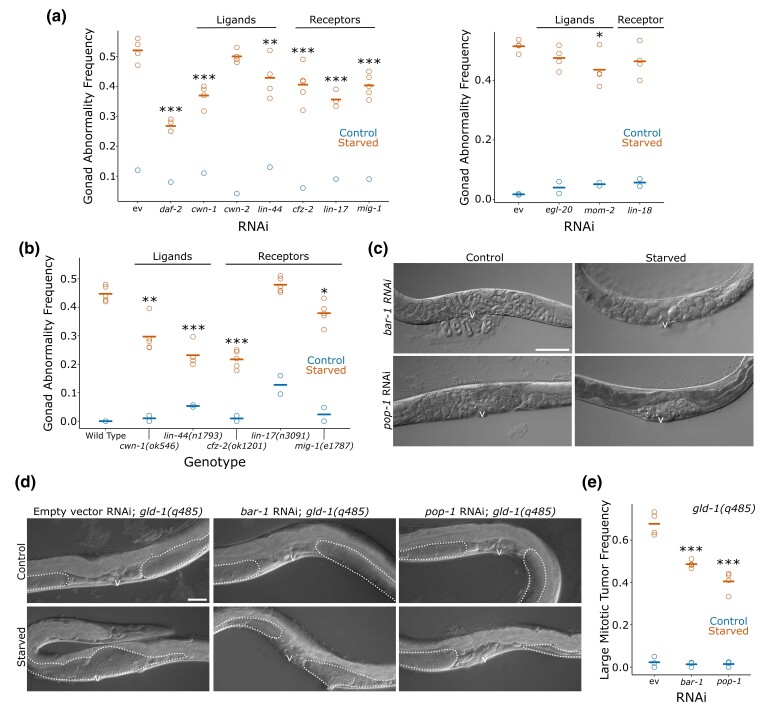
Fig. 2.
Wnt signaling promotes starvation-induced gonad abnormalities. a) Frequency of all gonad abnormalities for worms starved 1 day (control) or 8 days (starved) as L1 larvae and recovered for 72 h on the indicated RNAi food is plotted. There were at least 40 worms per replicate. b) Frequency of all gonad abnormalities for control and starved worms of the indicated genotypes recovered for 72 h. There were at least 35 worms per replicate. c) Representative images of control and starved wild-type worms recovered for 90 h on bar-1 or pop-1 RNAi are presented. Images were taken at 200× total magnification. d) Representative images of control and starved gld-1(q485) mutant worms recovered for 90 h on empty vector, bar-1, or pop-1 RNAi are presented. Visible portions of the gonad are outlined with a dotted line. Images were taken at 400× total magnification. e) Frequency of large mitotic tumors for control and starved gld-1(q485) worms recovered for 90 h on empty vector, bar-1, or pop-1 RNAi are plotted. There were at least 12 worms per replicate; most replicates have >50 worms. a, b, and e) Circles represent biological replicates. Cross bars reflect the mean. *P < 0.05; **P < 0.01; ***P < 0.001; t-tests on means of replicates between empty vector RNAi and the indicated genotype or condition. c and d) Scale bars are 50 μm. The letter “v” indicates the location of the vulva. ev, empty vector.
We assayed mutants affecting the Wnt ligands and receptors that significantly decreased starvation-induced abnormalities with RNAi. Of the five genes examined (cwn-1, lin-44, cfz-2, lin-17, and mig-1), mutation of all but lin-17 suppressed starvation-induced abnormalities (Fig. 2b). These results corroborate our RNAi results. Notably, several phenotypes which resemble starvation-induced abnormalities were observed in control lin-17 mutants, including protruding vulvae and uterine masses, potentially explaining the apparent lack of suppression of starvation-induced abnormalities with the mutant despite suppression with RNAi. The protruding vulva phenotype is consistent with previous characterizations of lin-17, reflecting its role in vulva development (Sawa et al. 1996). Together these results suggest that Wnt signaling promotes starvation-induced abnormalities.
We sought to determine whether global reduction of Wnt signaling could reduce starvation-induced abnormalities more effectively than knockdown of individual ligands and receptors. We chose to target the gene bar-1/β-catenin and the gene pop-1/Tcf because both are important for canonical Wnt signal transduction, and the expression data (Fig. 1c) suggest that bar-1 activity may be increased by early-life starvation. However, RNAi of bar-1 and pop-1 produced confounding phenotypes in a wild-type background, consistent with the role of Wnt signaling in vulva development (Sawa and Korswagen 2013). Control worms exposed to bar-1 or pop-1 RNAi exhibited substantial retention of eggs, and eventually eggs hatching within the parent (bagging; Fig. 2c). Starved worms exposed to bar-1 RNAi during recovery also exhibited substantial bagging, although the phenotype was delayed, consistent with delayed development (Jobson et al. 2015). These phenotypes interfered with reliable identification of starvation-induced abnormalities.
To circumvent the confounding phenotypes produced by bar-1 and pop-1 RNAi in wild-type worms, we used a germ-cell differentiation–deficient gld-1 mutant (Jeong et al. 2011). Following early-life starvation, these worms develop large mitotic germ-cell tumors in the gonad without uterine masses or fertilized embryos (since these phenotypes require differentiation; Fig. 2d; Jordan et al. 2019). Disruption of vulva development therefore does not interfere with scoring starvation-induced gonad abnormalities in this mutant. The frequency of large mitotic tumors in gld-1 mutant worms that had been starved as L1 larvae was significantly decreased by exposure to bar-1 or pop-1 RNAi during recovery (Fig. 2d, e). Because gld-1 mutants are germ-cell differentiation deficient, the observed reduction of large mitotic tumors provides evidence that Wnt signaling promotes starvation-induced abnormalities at least in part by increasing germ-cell proliferation.
Wnt signaling functions downstream of insulin/IGF signaling
Because daf-2/InsR signaling promotes Wnt pathway gene expression (Fig. 1c, e), we hypothesized that reduced IIS suppresses starvation-induced abnormalities by reducing Wnt signaling. To test this, we performed genetic epistasis analysis. We used a strain carrying a heat-shock-inducible, truncated bar-1/β-catenin gene. Truncation of BAR-1 prevents its degradation by the β-catenin destruction complex, causing constitutive activation of Wnt signaling upon heat shock (Jackson et al. 2014). Following heat shock, these worms displayed phenotypes resembling the abnormalities caused by early-life starvation (Fig. 3a). Specifically, the worms developed protruding vulvae and uterine masses. Although daf-2 RNAi during recovery significantly reduced the frequency of starvation-induced abnormalities in wild-type and truncated bar-1 overexpression backgrounds without heat shock, it did not alter the frequency of starvation-induced abnormalities produced by bar-1 overexpression in control worms (Fig. 3b). That daf-2 RNAi fails to reduce starvation-induced abnormalities when Wnt signaling is constitutively active suggests that Wnt signaling acts downstream of IIS to promote development of gonad abnormalities. However, it is possible that the gonad abnormalities resulting from truncated bar-1 overexpression arise through different molecular mechanisms than starvation-induced abnormalities despite similar appearances.
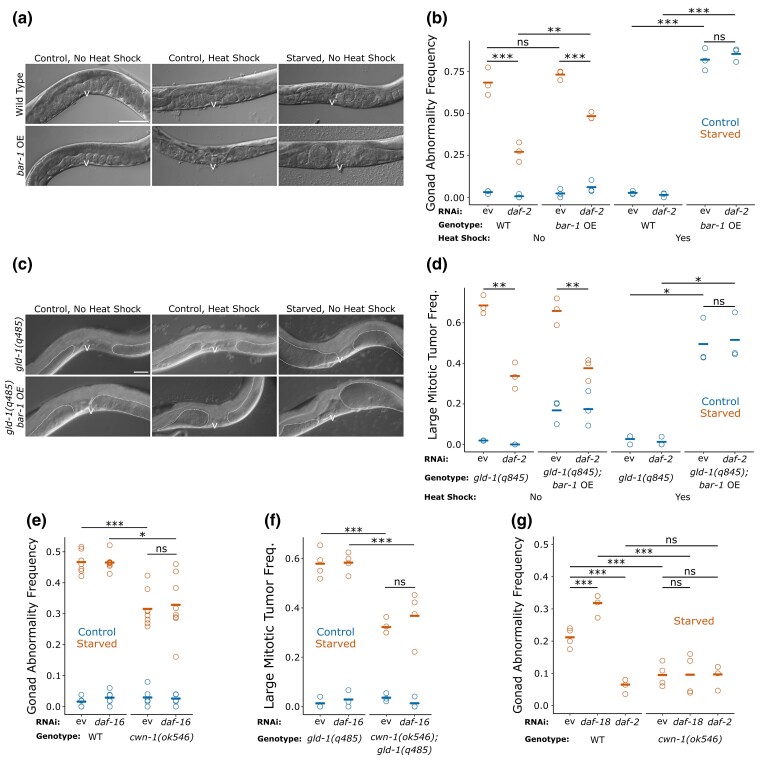
Fig. 3.
Wnt signaling functions downstream of IIS to promote starvation-induced gonad abnormalities. a) Representative images of wild-type and heat-shock-inducible truncated bar-1 overexpression worms (conditional gain-of-function; abbreviated “bar-1 OE”) exposed to the indicated conditions and recovered for 90 h. Images taken at 200× total magnification. b) Frequency of all gonad abnormalities in control and starved worms recovered for 90 h in the indicated conditions. There were at least 37 worms per replicate. c) Representative images of gld-1(q485) mutant and gld-1(q485); bar-1 overexpression worms exposed to the indicated conditions and recovered for 90 h. Images taken at 400× total magnification. d) Frequency of large mitotic tumors for control and starved worms recovered for 90 h in the indicated conditions is plotted. There were at least 36 worms per replicate. e) Frequency of all gonad abnormalities for control and starved wild-type and cwn-1(ok546) mutant worms recovered for 90 h in the indicated conditions is plotted. There were at least 45 worms per replicate. f) Frequency of large mitotic tumors for control and starved gld-1(q485) and gld-1(q485); cwn-1(ok546) worms recovered for 90 h in the indicated conditions is plotted. There were at least 30 worms per replicate. g) Frequency of all gonad abnormalities for starved wild-type and cwn-1(ok546) mutant worms recovered for 72 h in the indicated conditions is plotted. There were at least 28 worms with an average of 45 per replicate. a and c) Scale bars represent 50 μm. The letter “v” indicates the location of the vulva. b, d, e, f, and g) Circles represent biological replicates. Cross bars indicate the mean. *P < 0.05; **P < 0.01; ***P < 0.001; t-test on means of replicates. ev, empty vector; ns, not significant.
We also investigated the effect of truncated bar-1 overexpression in gld-1 mutant worms. This background allowed us to focus on alteration of germ-cell proliferation while avoiding confounding effects of bar-1 on vulva development. Heat-shock-induced overexpression of truncated bar-1 produced large mitotic tumors in control worms (Fig. 3c, d), suggesting that Wnt signaling promotes germ-cell proliferation. Furthermore, daf-2 RNAi during recovery significantly decreased the frequency of large mitotic tumors in starved worms but did not alter the frequency of large mitotic tumors caused by bar-1 overexpression. These results support the conclusion that Wnt signaling acts downstream of IIS to promote excessive germ-cell proliferation as a driver of starvation-induced abnormalities.
We extended our epistasis analysis by simulating increased IIS (with daf-16 RNAi) and reduced Wnt signaling (with mutation of cwn-1) in wild-type and gld-1 mutant backgrounds. We chose the cwn-1 mutant for these experiments because RNAi and mutation of it consistently suppressed starvation-induced abnormalities (Fig. 2a, b). This approach also avoided the confounding phenotypes of bar-1 and pop-1 (Fig. 2c). daf-16 RNAi had no effect on the frequency of gonad abnormalities in wild-type or cwn-1 mutant backgrounds (Fig. 3e). We obtained essentially the same result for large mitotic tumor frequencies when we replicated this experiment in a gld-1 mutant background (Fig. 3f). The lack of an effect of daf-16 RNAi in the cwn-1 mutant, in either a wild-type or gld-1 mutant background, further supports our conclusion that Wnt signaling acts downstream of IIS to promote starvation-induced abnormalities. However, it should be noted that there is no evidence that daf-16 RNAi was effective in this experiment, though we routinely use it in this context (Jordan et al. 2019, co-submitted).
We used daf-18/PTEN RNAi as a complementary way to increase IIS for epistasis analysis with the cwn-1 mutant. daf-18 encodes the phosphatase and tensin homolog PTEN, which dephosphorylates phosphatidylinositol (3,4,5)-trisphosphate to antagonize PI3K signaling, effectively increasing IIS (Ogg and Ruvkun 1998). daf-18 RNAi significantly increased the frequency of starvation-induced gonad abnormalities, supporting the conclusion that elevated IIS promotes these abnormalities (Fig. 3g). Notably, daf-16 RNAi did not increase the frequency of abnormalities (Fig. 3e, f), but PI3K antagonizes SKN-1/Nrf in parallel to DAF-16/FoxO (Tullet et al. 2008), and like daf-16, skn-1 functions downstream of daf-2/InsR to suppress starvation-induced abnormalities (Jordan et al. 2019). daf-2 RNAi and mutation of cwn-1 suppressed the frequency of starvation-induced abnormalities (Fig. 3g), as expected (Figs. 2a, b and 3b, d–f). However, daf-18 RNAi did not increase the frequency of abnormalities in the cwn-1 mutant background, and daf-2 RNAi did not suppress abnormalities in the cwn-1 mutant background (Fig. 3g). Together with the other epistasis results presented, these results support the conclusion that IIS increases Wnt signaling to promote starvation-induced gonad abnormalities.
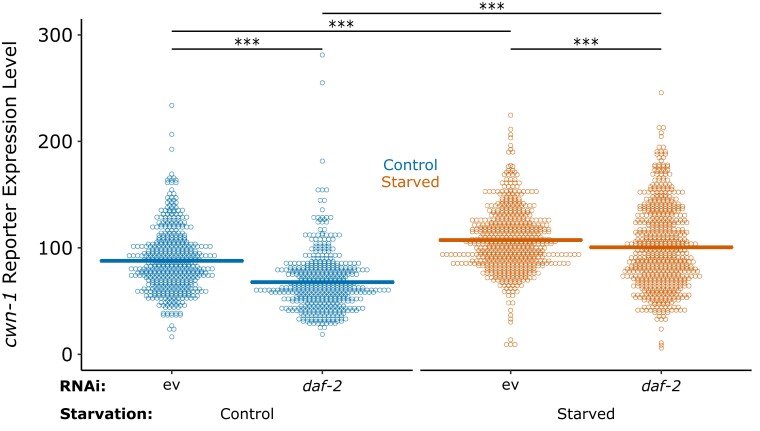
RNA-seq suggests that early-life starvation and IIS promote expression of cwn-1 and other Wnt pathway components, suggesting transcriptional regulation as a mechanism for the epistatic relationship between IIS and cwn-1. To test this hypothesis, we analyzed a cwn-1 translational reporter gene (Yamamoto et al. 2011). CWN-1::VENUS displayed graded expression in the posterior of developing larvae, as reported. Multiple replicates of qualitative analysis of fluorescence levels suggested that CWN-1::VENUS is more abundant in starved than control worms 48 h after recovery from starvation and that daf-2/InsR RNAi decreases expression levels (data not shown). Quantitative image analysis supported these observations, revealing a significant increase in CWN-1::VENUS expression following starvation and a significant decrease with daf-2 RNAi (Fig. 4). These results support the conclusion that IIS promotes cwn-1 expression, accounting for their epistatic relationship.
Fig. 4.
Early-life starvation and insulin/IGF signaling promote cwn-1 reporter-gene expression. Background-corrected mean pixel intensity is plotted for CWN-1::VENUS in control and starved worms recovered for 48 h in the conditions indicated. There is a single data point outside the plotted range for control empty vector and starved daf-2. Each point represents an individual worm. Cross bars indicate the mean. Four biological replicates were performed and pooled for display. ***P < 0.001; pairwise linear mixed-effect model. ev, empty vector.
Wnt signaling functions in somatic tissues to promote gonad abnormalities
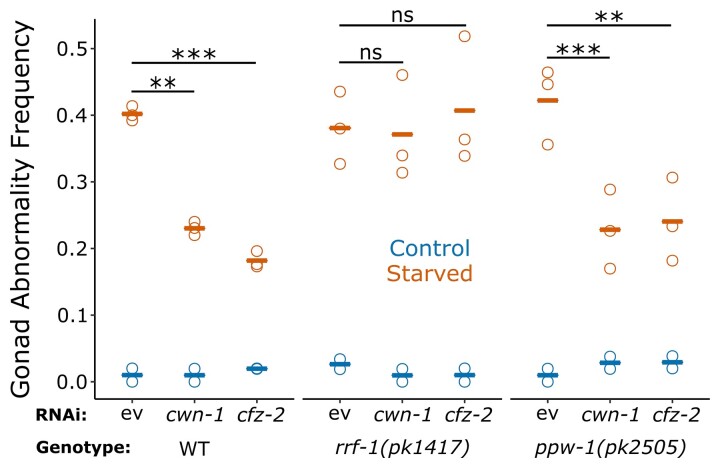
We sought to determine whether Wnt signaling acts cell autonomously to promote starvation-induced abnormalities. To address this, we used RNAi to knockdown expression of Wnt ligand cwn-1 and receptor cfz-2 in wild-type worms as well as somatic RNAi–deficient rrf-1 mutants and germline RNAi–deficient ppw-1 mutants (Tijsterman et al. 2002; Grishok et al. 2005; Kumsta and Hansen 2012). RNAi of cwn-1 and cfz-2 significantly reduced the frequency of starvation-induced abnormalities in both wild-type and ppw-1 mutant worms but had no apparent effect in rrf-1 mutant worms (Fig. 5). The finding that RNAi of a Wnt ligand and receptor in somatic RNAi–deficient worms fails to suppress starvation-induced abnormalities suggests that Wnt signals both originate in and target somatic tissues to promote gonad abnormalities, implying cell-nonautonomous effects on germ-cell proliferation and possibly other developmental processes.
Fig. 5.
cwn-1 and cfz-2 function in somatic tissues to promote starvation-induced gonad abnormalities. Frequency of all gonad abnormalities for control and starved wild-type, rrf-1(pk1417), and ppw-1(pk2505) mutant worms recovered for 90 h in the indicated conditions is plotted. There were at least 50 worms per replicate. Circles represent biological replicates. Cross bars indicate the mean. **P < 0.01; ***P < 0.001; t-test on means of replicates. ev, empty vector; ns, not significant.
Discussion
Our results support a model linking early-life starvation, IIS, and Wnt signaling to development of germline tumors and other gonad abnormalities in adult C. elegans. We show that expression of Wnt pathway genes increases following early-life starvation and that it decreases with reduced daf-2/InsR function. Genetic analysis suggests that Wnt signaling promotes starvation-induced abnormalities, that it does so at least in part by promoting germ-cell proliferation, that it functions downstream of IIS, and that it acts in somatic cells. Together with our companion manuscript (Jordanet al. co-submitted), this work reveals consequences of early-life starvation on gene-regulation and signaling that result in adult pathology.
Transcriptional regulation of Wnt signaling
We show elsewhere that early-life starvation and IIS have overlapping effects on adult gene expression and lipid metabolism (Jordanet al. co-submitted). That is, nearly 2,000 genes are upregulated in adults starved as L1 larvae, and about half of those are downregulated when starved larvae are recovered on daf-2 RNAi food. The fatty-acid synthetase fasn-1/FASN exemplifies this expression profile, it promotes development of starvation-induced abnormalities, and it contributes substantially to the effects of early-life starvation and IIS on adult gene expression (Jordanet al. co-submitted). However, with such widespread effects on gene expression and metabolism, it is likely that early-life starvation and IIS impinge on additional regulatory mechanisms to promote pathological outcomes in adults subjected to early-life starvation.
Results presented here suggest that early-life starvation and IIS also affect Wnt signaling, and that Wnt signaling promotes starvation-induced abnormalities. mRNA expression analysis shows that nearly all the Wnt ligands and receptors as well as bar-1/β-catenin are upregulated in adults starved as L1 larvae but downregulated when such starved worms are recovered on daf-2 RNAi food. This pattern of regulation was confirmed with a cwn-1 reporter gene. These Wnt pathway genes have not been identified as targets of IIS (Tepper et al. 2013), but the magnitude of the effect of daf-2 RNAi on their expression is relatively small in worms that had not been subjected to L1 starvation. This observation highlights the importance of studying worms subjected to early-life starvation and the novelty of our results. We have not identified transcriptional mechanisms affecting Wnt pathway gene expression, but we believe the transcription factor DAF-16/FoxO is involved based on epistasis analysis (here and in our campanion manuscript; Jordanet al. co-submitted). DAF-16 is the major effector of DAF-2/InsR signaling, and daf-16 is required for daf-2 RNAi to suppress starvation-induced abnormalities (Jordan et al. 2019). DAF-16 is thought to function as a transcriptional activator (Schuster et al. 2010), but our data suggest it represses Wnt pathway gene expression. DAF-16 antagonism of the transcription factor PQM-1 contributes to repression of DAF-16 class II targets (Tepper et al. 2013), suggesting pqm-1 could also be involved. In addition, the transcription factor SKN-1/Nrf is also an effector of DAF-2/InsR signaling, and skn-1 is required for daf-2 RNAi to suppress starvation-induced abnormalities (Jordan et al. 2019), suggesting that it too could contribute to transcriptional regulation of Wnt pathway genes.
We provide evidence that Wnt signaling activity is increased following early-life starvation and that Wnt signaling promotes starvation-induced abnormalities. Expression of Wnt-target genes displays the same pattern as Wnt pathway components, with upregulation following early-life starvation and downregulation with reduced IIS, based on RNA-seq. We used RNAi and mutations to show that disruption of two Wnt ligands and three different receptors suppresses starvation-induced abnormalities, as does RNAi of bar-1/β-catenin or pop-1/TCF. Furthermore, constitutive activation of Wnt signaling phenocopies the effects of early-life starvation on gonad development in control worms (without extended starvation). Multiple approaches to epistasis analysis also suggest that Wnt signaling functions downstream of IIS in promoting starvation-induced abnormalities. Furthermore, reporter-gene analysis of cwn-1 show that its expression is promoted by daf-2/InsR. Together these results suggest that early-life starvation and IIS increase Wnt signaling to promote development of germline tumors and other gonad abnormalities.
Somatic regulation of germline proliferation
We used a germ-cell differentiation-defective gld-1 mutant to focus on mitotic germline proliferation as opposed to other developmental mechanisms that could contribute to starvation-induced gonad abnormalities. This mutant background revealed that Wnt signaling promotes excessive germline proliferation following early-life starvation (based on loss-of-function and gain-of-function Wnt signaling mutants). Excessive germline proliferation tentatively explains starvation-induced germline tumors in wild-type worms, and it could also lead to dysregulation of critical signaling interactions between germ cells and the somatic gonad causing additional gonad abnormalities observed (Jordan et al. 2019).
We used tissue-specific RNAi to show that a Wnt ligand- and receptor-encoding gene (cwn-1 and cfz-2, respectively) both function in somatic cells to promote starvation-induced abnormalities. Likewise, mRNA was barely detectable in L2 germ cells but abundant in a variety of somatic cell types for both genes based on single-cell RNA-seq data (Cao et al. 2017). However, it should be noted that we tested only a single ligand and receptor, so we do not know if the same is true for the others. Nonetheless, this result argues against cell-autonomous regulation of germline proliferation by Wnt signaling, instead suggesting that Wnt signaling affects at least one additional signaling pathway from the soma to influence germline proliferation. It is well established that mitotic germline proliferation is controlled by Delta-Notch signaling from the somatic gonad to germ cells, and that perturbation of this signaling can result in formation of germ-cell tumors (Albert Hubbard and Schedl 2019). It is attractive to speculate that Wnt signaling targets the somatic gonad to affect production of Notch ligands. Notably, daf-16/FoxO expression in the epidermis of an otherwise null daf-16 mutant is sufficient for strong suppression of starvation-induced abnormalities, and expression in the intestine and nervous system also suppress starvation-induced abnormalities (Jordan et al. 2019). The somatic gonad was not tested for daf-16 function in these experiments. The epidermis, nervous system, and intestine are also important sites of daf-16 function for starvation resistance and aging (Libina et al. 2003; Zhang et al. 2013; Kaplan et al. 2015). It is unclear if Wnt signaling also functions in a widely distributed anatomical fashion or if signaling in a particular tissue or set of cells is sufficient to drive germline proliferation and formation of gonad abnormalities.
Insulin/IGF and Wnt signaling in other systems
This work resonates with findings in other systems, revealing complexity in the regulation of Wnt signaling. In mice, fasting and refeeding results in transcriptional upregulation of Fasn and Scd1 (Cabrae et al. 2020). We also see increased mRNA expression of fasn-1/FASN and fat-5/SCD1 in well-fed worms following early-life starvation, and disruption of either suppresses starvation-induced abnormalities (Jordanet al. co-submitted). Notably, FASN and SCD1 increase abundance of palmitoleic acid, which is used by PORCN to acylate Wnt and increase signaling activity (Cabrae et al. 2020). These feeding-dependent effects in mice that had been fasted are dependent on insulin and mTOR signaling. Future work on the consequences of early-life starvation in C. elegans should investigate mTOR signaling, but there are nonetheless striking parallels between these two models. Furthermore, there is evidence of cross-talk between insulin and Wnt signaling (Essers et al. 2005; Palsgaard et al. 2012), and that Wnt signaling may increase sensitivity to insulin (Abiola et al. 2009). Additional work is needed to unravel the complex relationship between these critical signaling pathways at the interface of nutrient availability and developmental regulation.
Conclusion
We add to a body of work showing that early-life starvation and IIS promote developmental pathology in well-fed worms, establishing a valuable invertebrate model for developmental origins of adult health and disease (Jobson et al. 2015; Jordan et al. 2019;, co-submitted). Early-life malnutrition causes adult cancer in humans, and this work could lead to predictive diagnostics of disease or therapeutic interventions. Caenorhabditis elegans have remarkably robust starvation responses, and the species thrives despite fluctuations in nutrient availability. Nonetheless, this work sheds light on the limits of developmental homeostasis, revealing mechanisms of dysregulation that result in adult pathology and reduced reproductive success following early-life starvation. Nutrient stress is ubiquitous in nature, and the pathways we implicate are widely conserved among metazoa, suggesting that insights made in worms will translate to other animals.
Acknowledgments
The authors thank the Caenorhabditis Genetics Center (CGC), funded by NIH Office of Research Infrastructure Programs (P40 OD010440), for providing some strains used in the study. The authors also thank WormBase. Rojin Chitrakar helped obtain reagents and maintain worm strains, and Amy Webster helped design genetic crosses. Figure 1a was adapted from wntsignal_fig 1.jpg from “Wnt signaling in C. elegans” by Hitoshi Sawa and Rik Korswagen (Sawa and Korswagen 2013; license: CC BY 2.5).
Contributor Information
Nathan C Shaul, Department of Biology, Duke University, Durham, NC 27708, USA.
James M Jordan, Department of Biology, Duke University, Durham, NC 27708, USA.
Ivan B Falsztyn, Department of Biology, Duke University, Durham, NC 27708, USA.
L Ryan Baugh, Department of Biology, Duke University, Durham, NC 27708, USA; Center for Genomic and Computational Biology, Duke University, Durham, NC 27708, USA.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Funding
The National Institutes of Health funded this work (R01GM117408 to L.R.B.).
Note added in proof
See https://doi.org/10.1093/genetics/iyac172 for a related work by Jordan et al.
Communicating editor: B. Conradt
Literature cited
- Abiola M, Favier M, Christodoulou-Vafeiadou E, Pichard AL, Martelly I, Guillet-Deniau I. Activation of Wnt/β-catenin signaling increases insulin sensitivity through a reciprocal regulation of Wnt10b and SREBP-1c in skeletal muscle cells. PLoS One. 2009;4:e8509. doi: 10.1371/journal.pone.0008509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert Hubbard EJ, Schedl T. Biology of the Caenorhabditis elegans germline stem cell system. Genetics. 2019;213:1145–1188. doi: 10.1534/genetics.119.300238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJP. The origins of the developmental origins theory. J Int Med. 2007;261:412–417. doi: 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- Basu S, Cheriyamundath S, Ben-Ze’Ev A. Cell–cell adhesion: linking Wnt/β-catenin signaling with partial EMT and stemness traits in tumorigenesis. F1000Res. 2018;7:1488. doi: 10.12688/f1000research.15782.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR. To grow or not to grow: nutritional control of development during Caenorhabditis elegans L1 arrest. Genetics. 2013;194:539–555. doi: 10.1534/genetics.113.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrae R, Dubuquoy C, Caüzac M, Morzyglod L, Guilmeau S, Noblet B, Fève B, Postic C, Burnol AF, Moldes M. Insulin activates hepatic Wnt/β-catenin signaling through stearoyl-CoA desaturase 1 and porcupine. Sci Rep. 2020;10:5186. doi: 10.1038/s41598-020-61869-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, et al. . Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017;357:661–667. doi: 10.1126/science.aam8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi AK, Wightman B, Chalfie M. A Transparent Window into Biology: A Primer on Caenorhabditis elegans. WormBook; 2015. doi: 10.1895/wormbook.1.177.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Maloof JN, Simske JS, Kenyon C, Kim SK. The beta-catenin homolog BAR-1 and LET-60 Ras coordinately regulate the Hox gene lin-39 during Caenorhabditis elegans vulval development. Development. 1998;125:3667–3680. doi: 10.1242/dev.125.18.3667. [DOI] [PubMed] [Google Scholar]
- Essers MAG, de Vries-Smits LMM, Barker N, Polderman PE, Burgering BMT, Korswagen HC. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science. 2005;308:1181–1184. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
- Fentiman IS, Allen DS, Ellison GTH. The impact of the occupation of Guernsey 1940-1945 on breast cancer risk factors and incidence. Int J Clin Pract. 2007;61:973–943. doi: 10.1111/j.1742-1241.2007.01288.x. [DOI] [PubMed] [Google Scholar]
- Gillman MW. Developmental origins of health and disease. N Engl J Med. 2005;353:1848–1850. doi: 10.1056/NEJMe058187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrepati L, Krause MW, Chen W, Brodigan TM, Correa-Mendez M, Eisenmann DM. Identification of Wnt pathway target genes regulating the division and differentiation of larval seam cells and vulval precursor cells in Caenorhabditis elegans. G3 (Bethesda). 2015;5:1551–1566. doi: 10.1534/g3.115.017715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Sinskey J, Sharp P. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 2005;19:683–696. doi: 10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibshman JD, Webster AK, Baugh LR. Liquid-culture protocols for synchronous starvation, growth, dauer formation, and dietary restriction of Caenorhabditis elegans. STAR Protoc. 2021;2:100276. doi: 10.1016/j.xpro.2020.100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogeboom D, Essers MAG, Polderman PE, Voets E, Smits LMM, Burgering BM. Interaction of FOXO with β-catenin inhibits β-catenin/T cell factor activity. J Biol Chem. 2008;283:9224–9230. doi: 10.1074/jbc.M706638200. [DOI] [PubMed] [Google Scholar]
- Jackson BM, Abete-Luzi P, Krause MW, Eisenmann DM. Use of an activated beta-catenin to identify Wnt pathway target genes in Caenorhabditis elegans, including a subset of collagen genes expressed in late larval development. G3 (Bethesda). 2014;4:733–747. doi: 10.1534/g3.113.009522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Verheyden JM, Kimble J. Cyclin E and Cdk2 control GLD-1, the mitosis/meiosis decision, and germline stem cells in Caenorhabditis elegans. PLoS Genet. 2011;7:e1001348. doi: 10.1371/journal.pgen.1001348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobson MA, Jordan JM, Sandrof MA, Hibshman JD, Lennox AL, Baugh LR. Transgenerational effects of early life starvation on growth, reproduction, and stress resistance in Caenorhabditis elegans. Genetics. 2015;201:201–212. doi: 10.1534/genetics.115.178699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan JM, Hibshman JD, Webster AK, Kaplan REW, Leinroth A, Guzman R, Maxwell CS, Chitrakar R, Bowman EA, Fry AL, et al. . Insulin/IGF signaling and vitellogenin provisioning mediate intergenerational adaptation to nutrient stress. Curr Biol. 2019;29:2380–2388.e5. doi: 10.1016/j.cub.2019.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan JM, Webster AK, Chen J, Chitrakar R, Baugh LR. Early-life starvation alters lipid metabolism in adults to cause developmental pathology in Caenorhabditis elegans. Genetics. 2022:iyac172. doi: 10.1093/genetics/iyac172 [DOI]
- Joshi PM, Riddle MR, Djabrayan NJV, Rothman JH. Caenorhabditis elegans as a model for stem cell biology. Dev Dyn. 2010;239:1539–1554. doi: 10.1002/dvdy.22296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan REW, Chen Y, Moore BT, Jordan JM, Maxwell CS, Schindler AJ, Baugh LR. dbl-1/TGF-β and daf-12/NHR signaling mediate cell-nonautonomous effects of daf-16/FOXO on starvation-induced developmental arrest. PLoS Genet. 2015;11:e1005731. doi: 10.1371/journal.pgen.1005731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumsta C, Hansen M. C. elegans rrf-1 mutations maintain RNAi efficiency in the soma in addition to the germline. PLoS One. 2012;7:e35428. doi: 10.1371/journal.pone.0035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/S0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- Murphy CT, Hu PJ. Insulin/insulin-like Growth Factor Signaling in C. elegans. WormBook;2013. doi: 10.1895/wormbook.1.164.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA, Desai TJ. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science. 2018;359:1118–1123. doi: 10.1126/science.aam6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S, Ruvkun G. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol Cell. 1998;2:887–893. doi: 10.1016/s1097-2765(00)80303-2. [DOI] [PubMed] [Google Scholar]
- Painter RC, De Rooij SR, Bossuyt PMM, Osmond C, Barker DJP, Bleker OP, Roseboom TJ. A possible link between prenatal exposure to famine and breast cancer: a preliminary study. Am J Human Biol. 2006;18:853–856. doi: 10.1002/ajhb.20564. [DOI] [PubMed] [Google Scholar]
- Palsgaard J, Emanuelli B, Winnay JN, Sumara G, Karsenty G, Kahn CR. Cross-talk between insulin and Wnt signaling in preadipocytes. J Biol Chem. 2012;287:12016–12026. doi: 10.1074/jbc.M111.337048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseboom TJ, Van Der Meulen JHP, Ravelli ACJ, Osmond C, Barker DJP, Bleker OP. Effects of prenatal exposure to the Dutch famine on adult disease in later life: an overview. Mol Cell Endocrinol. 2001;185:93–98. doi: 10.1016/S0303-7207(01)00721-3. [DOI] [PubMed] [Google Scholar]
- Sawa H, Korswagen HC. Wnt Signaling in C. elegans. WormBook; 2013. doi: 10.1895/wormbook.1.7.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa H, Lobel L, Horvitz HR. The Caenorhabditis elegans gene lin-17, which is required for certain asymmetric cell divisions, encodes a putative seven-transmembrane protein similar to the Drosophila frizzled protein. Genes Dev. 1996;10:2189–2197. doi: 10.1101/gad.10.17.2189. [DOI] [PubMed] [Google Scholar]
- Schuster E, McElwee JJ, Tullet JM, Doonan R, Matthijssens F, Reece-Hoyes JS, Hope IA, Vanfleteren JR, Thornton JM, Gems D. DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol Syst Biol. 2010;6:399. doi: 10.1038/msb.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K. The developing world of DOHaD. J Dev Orig Health Dis. 2018;9:266–269. doi: 10.1017/S2040174417000691 [DOI] [PubMed] [Google Scholar]
- Tepper RG, Ashraf J, Kaletsky R, Kleemann G, Murphy CT, Bussemaker HJ. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell. 2013;154:676–690. doi: 10.1016/j.cell.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M, Okihara KL, Thijssen K, Plasterk RHA. PPW-1, a PAZ/PIWI protein required for efficient germline RNAi. Is defective in a natural isolate of C. elegans. Curr Biol. 2002;12:1535–1540. doi: 10.1016/s0960-9822(02)01110-7. [DOI] [PubMed] [Google Scholar]
- Tullet JMA, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bent ML, Sterken MG, Volkers RJ, Riksen JA, Schmid T, Hajnal A, Kammenga JE, Snoek LB. Loss-of-function of β-catenin bar-1 slows development and activates the Wnt pathway in Caenorhabditis elegans. Sci Rep. 2015;4:4926. doi: 10.1038/srep04926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CL, Ho S-M. Developmental reprogramming of cancer susceptibility. Nat Rev Cancer. 2012;12:479–486. doi: 10.1038/nrc3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag; 2016. [Google Scholar]
- Yamamoto Y, Takeshita H, Sawa H. Multiple Wnts redundantly control polarity orientation in Caenorhabditis elegans epithelial stem cells. PLoS Genet. 2011;7:e1002308. doi: 10.1371/journal.pgen.1002308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Judy M, Lee S-J, Kenyon C. Direct and indirect gene regulation by a life-extending FOXO protein in C. elegans: roles for GATA factors and lipid gene regulators. Cell Metab. 2013;17:85–100. doi: 10.1016/j.cmet.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.