Significance
The drug praziquantel has been used for decades to treat infections caused by parasitic flatworms, including schistosomiasis which afflicts over 200 million people worldwide. Most flukes and tapeworms display high sensitivity to praziquantel; however, the molecular basis for the unique sensitivity of some, but not all, parasites to praziquantel is unclear. Here, we comprehensively analyzed sequence variation in the binding pocket of a flatworm ion channel target of praziquantel, known as TRPMPZQ, and define a key residue that underpins the exquisite sensitivity of trematode and certain cestodes to praziquantel. This insight provides impetus for designing new drugs that treat various parasitic flatworm infections where treatments are currently lacking or where improvements over praziquantel could be realized.
Keywords: parasitic flatworm, Ca2+ signaling, TRP channel
Abstract
The drug praziquantel (PZQ) is the key clinical therapy for treating schistosomiasis and other infections caused by parasitic flatworms. A schistosome target for PZQ was recently identified— a transient receptor potential ion channel in the melastatin subfamily (TRPMPZQ)—however, little is known about the properties of TRPMPZQ in other parasitic flatworms. Here, TRPMPZQ orthologs were scrutinized from all currently available parasitic flatworm genomes. TRPMPZQ is present in all parasitic flatworms, and the consensus PZQ binding site was well conserved. Functional profiling of trematode, cestode, and a free-living flatworm TRPMPZQ ortholog revealed differing sensitives (~300-fold) of these TRPMPZQ channels toward PZQ, which matched the varied sensitivities of these different flatworms to PZQ. Three loci of variation were defined across the parasitic flatworm TRPMPZQ pocketome with the identity of an acidic residue in the TRP domain acting as a gatekeeper residue impacting PZQ residency within the TRPMPZQ ligand binding pocket. In trematodes and cyclophyllidean cestodes, which display high sensitivity to PZQ, this TRP domain residue is an aspartic acid which is permissive for potent activation by PZQ. However, the presence of a glutamic acid residue found in other parasitic and free-living flatworm TRPMPZQ was associated with lower sensitivity to PZQ. The definition of these different binding pocket architectures explains why PZQ shows high therapeutic effectiveness against specific fluke and tapeworm infections and will help the development of better tailored therapies toward other parasitic infections of humans, livestock, and fish.
Praziquantel (PZQ), a drug discovered in the 1970s, has been in clinical usage for four decades to treat many different diseases caused by parasitic flatworms (1). Notably, PZQ is the key clinical agent for treating the neglected tropical disease schistosomiasis, which afflicts over 200 million people worldwide (2, 3). PZQ is classified as an essential medication by the World Health Organization and is integral to the current roadmap of interventions that are designed to control schistosomiasis morbidity and eliminate schistosomiasis as a public health problem.
PZQ effects an immediate paralysis of schistosome worms in vitro with concomitant surface damage that facilitates immunological clearance of worms in vivo (1, 4, 5). Despite the long-term clinical use of PZQ as an anthelminthic drug, the flatworm target of PZQ has remained undefined. However, a prime candidate was recently identified in Schistosoma mansoni: a Ca2+-permeable ion channel of the transient receptor potential melastatin family named Schistosoma mansoni TRPMPZQ (Sm.TRPMPZQ, (6)). Heterologous expression of Sm.TRPMPZQ in mammalian cells conferred responsiveness to PZQ, with PZQ elevating cytoplasmic Ca2+ with an observed sensitivity in the submicromolar range and stereoselectivity toward the active enantiomer (R)-PZQ. These features match the known action of PZQ on intact worms (7). Subsequent work validated Sm.TRPMPZQ as a relevant in vivo flatworm target of PZQ, based on more comprehensive pharmacological and genetic analyses of Sm.TRPMPZQ properties (8, 9).
In addition to treating schistosomiasis, PZQ also displays a broad spectrum activity against other parasitic flatworm infections, including diseases caused by other trematodes, monogeneans, and cestodes (1, 5, 10–12). However, PZQ sensitivity of these parasitic flatworms is not identical leading to different clinical dosages required to effectively treat different infections (13–15). PZQ also has effects on free-living flatworms (16–18) but over a much higher concentration range (tens of micromolar) than active against schistosomes and certain tapeworms (hundreds of nanomolar).
Variation in TRPMPZQ binding pocket sequences may account for the varied sensitivities of different flatworms to PZQ. Precedent for this explanation has been established by comparison of a PZQ-sensitive (S. mansoni) and a PZQ-insensitive (Fasciola hepatica) parasitic flatworm. The binding pockets of these TRPMPZQ orthologs harbored a single amino acid difference at a residue (N1388 in S. mansoni, T1270 in F. hepatica) within the first transmembrane helix (S1) of the voltage sensor-like domain (VSLD) of the ion channel (8). This residue was predicted to engage the cyclohexyl carbonyl of PZQ via hydrogen bonding (8). Reciprocal mutations caused either a loss (N1388T in Sm.TRPMPZQ) or gain (T1270N in Fh.TRPMPZQ) of sensitivity to PZQ (8). Natural sequence variation within the TRPMPZQ binding pocket, therefore, underpins the insensitivity of Fh.TRPMPZQ to PZQ, correlating with the loss of PZQ activity against F. hepatica. The refractoriness toward PZQ has necessitated the development of other approaches to combat human and agricultural infections caused by this liver fluke (19, 20).
This example provides impetus for a comprehensive evaluation of TRPMPZQ binding pocket architecture across all parasitic flatworms. This analysis, performed in this study, revealed the following: first, a molecular basis for the uniquely high sensitivity of certain flukes and tapeworms to PZQ, long recognized as a key feature of PZQ action; second, examples where the interaction of PZQ with TRPMPZQ is nonoptimal identifying opportunity to develop new ligands to better treat specific parasitic infections.
Results
PZQ Activation of Schistosome TRPMPZQ Channels.
As PZQ is a broadly used clinical therapy for treating parasitic flatworm infections (1), it is important to define whether other TRPMPZQ orthologs are also sensitive to PZQ. For example, while three schistosome species—S. mansoni, Schistosoma haematobium, and Schistosoma japonicum—are responsible for most human infections worldwide, only Sm.TRPMPZQ has been studied to date (6, 8).
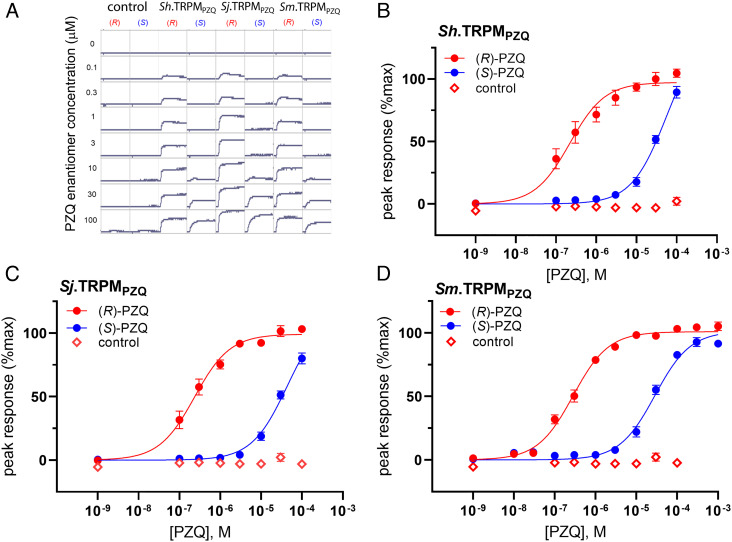
S. haematobium TRPMPZQ was cloned from a cDNA sample prepared from S. haematobium total RNA (see Methods), to yield a full-length sequence for Sh.TRPMPZQ of 2195 amino acids (SI Appendix, Fig. S1). Based on the sequences of Sm.TRPMPZQ and Sh.TRPMPZQ, a full-length sequence for Sj.TRPMPZQ (2179 amino acids) was synthesized from the S. japonicum genomic annotation (SI Appendix, Fig. S1). The three Schistosoma spp. TRPMPZQ orthologs were highly similar in sequence (>80% amino acid identity, SI Appendix, Table S1). The individual schistosome TRPMPZQ channels were transiently expressed in HEK293 cells, and their responsiveness to the PZQ enantiomers, (R)-PZQ and (S)-PZQ, was examined by monitoring fluorescence changes using a cytoplasmic Ca2+-sensitive dye. The addition of (R)-PZQ to cells expressing individual schistosome TRPMPZQ channels caused a rapid increase in cytoplasmic Ca2+, whereas no Ca2+ increase was seen in control cells (Fig. 1A). Activation of each of these channels displayed stereoselectivity for (R)-PZQ over (S)-PZQ (Fig. 1A), consistent with the effects of individual PZQ enantiomers on worms in vitro and in vivo (21). The observed sensitivities to (R)-PZQ were EC50 = 221 ± 85 nM for Sh.TRPMPZQ, EC50 = 234 ± 80 nM for Sj.TRPMPZQ, and EC50 = 281 ± 29 nM for Sm.TRPMPZQ (Fig. 1 B–D).
Fig. 1.
Functional profiling of schistosome TRPMPZQ channels. (A) Examples of real-time fluorescence values from a representative experiment that depict the effects of (R)-PZQ (red) and (S)-PZQ (blue) in untransfected cells (‘control’) as well as HEK293 cells expressing Sh.TRPMPZQ, Sj.TRPMPZQ, and Sm.TRPMPZQ. PZQ enantiomers (concentrations on left hand side) were added after ~20 s of sampling a baseline fluorescence emission. (B–D) Concentration–response relationships for the activation of (B) Sh.TRPMPZQ, (C) Sj.TRPMPZQ, and (D) Sm.TRPMPZQ by PZQ enantiomers, (R)-PZQ (red) and (S)-PZQ (blue). (±)-PZQ was also profiled against untransfected cells (open symbols). Data are expressed as a percentage of the maximum response evoked by (R)-PZQ at Sm.TRPMPZQ and represent the mean ± SEM of n ≥ 3 independent experiments.
(R)-PZQ has been shown to activate Sm.TRPMPZQ via engagement of a binding pocket located at the base of the VSLD of the ion channel (8). This PZQ binding pocket of TRPMPZQ is formed by residues from each of the first four transmembrane spanning helices (S1 to S4), their interconnecting loops, and the juxtamembrane TRP helix. Based on the understanding of how (R)-PZQ engages this binding pocket, point mutants of a key asparagine residue in the first transmembrane helix (S1) were made in Sh.TRPMPZQ (Sh.TRPM[N1314A]PZQ) and Sj.TRPMPZQ (Sj.TRPM[N1299A]PZQ). This S1 asparagine is important for hydrogen bonding to (R)-PZQ in Sm.TRPMPZQ (8). Ligand sensitivity was reduced in these mutants, such that (±)-PZQ (10 μM) caused little activity relative to the wildtype construct (peak response of 7.7 ± 5.9% for Sh.TRPM[N1314A]PZQ, 0.7 ± 1.3% for Sj.TRPM[N1299A]PZQ and 1.9 ± 1.5% for Sm.TRPM[N1388A]PZQ). These data suggest a similar mechanism of PZQ engagement by each of the schistosome TRPMPZQ orthologs.
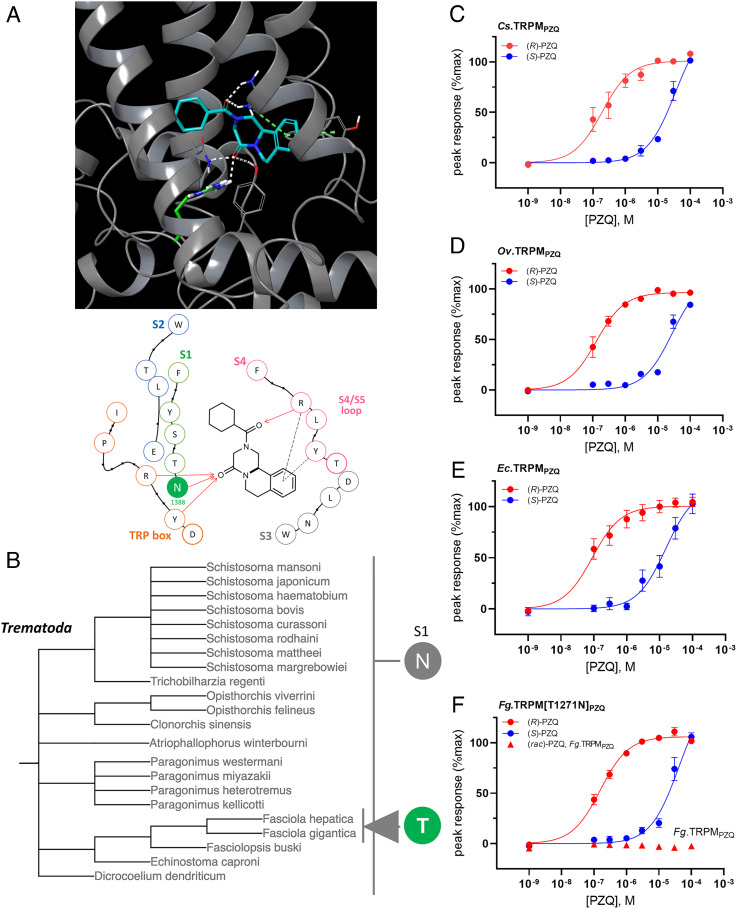
A total of 23 amino acids project within a 5Å radius of the PZQ binding pose (Fig. 2A), providing an approximation of the binding pocket ‘lining’ (8). Using these 23 residues as a barcode that encodes PZQ sensitivity, the identity of amino acids at equivalent positions within TRPMPZQ orthologs from all other schistosome species was scrutinized. Bioinformatic analysis extended to eight other Schistosoma spp. TRPMPZQ sequences revealed the complete conversation of residues lining the PZQ binding pocket across all the Schistosomatidae (SI Appendix, Table S2). This analysis encompassed the three species that were functionally profiled (S. mansoni, S. haematobium, and S. japonicum), four other schistosome species (S. bovis, S. curassoni, S. mattheei, and S. margrebowiei), as well as the avian schistosome Trichobilharzia regenti, which causes cercarial dermatitis (swimmer’s itch) in humans (22). The conservation of the PZQ binding pocket in all schistosome TRPMPZQ channels is consistent with the in vitro sensitivity to PZQ reported in each of these species and the known efficacy of PZQ for treating human, livestock, and wild animal schistosome infections (1, 3, 13, 22, 23).
Fig. 2.
TRPMPZQ binding pocket variation and functionality in trematode flatworms. (A) Top, docking pose of (R)-PZQ (blue) within the VSLD of Sm.TRPMPZQ. Key interactions are shown as follows: hydrogen bond (white) and π–π stacking and cation–π interactions (dashed green line). Residue N1388 (S1) is shown in green. Bottom, interaction map of (R)-PZQ within the transmembrane binding pocket of Sm.TRPMPZQ, showing residues within a 5Å radius of the proposed binding pose, modified from (8). Residues are colored to indicated channel domains (S1 to S4 and TRP domain). The asparagine residue shown in solid color—‘N’ (S1, green)—represents the location of variation within Fasciola spp. TRPMPZQ. Interactions are highlighted as follows (hydrogen binding, arrows; cation–π, long dash; π–π, short dash). (B) Summary of sequence variation across parasitic trematode TRPMPZQ orthologs. The presented classification is reconstructed from the evolutionary pedigree of Lifemap (24) and is not a bootstrapped phylogeny. The Fasciola spp. threonine variant (‘T’) within the S1 helix is highlighted. (C–F) Concentration–response relationships for (R)-PZQ (red) and (S)-PZQ (blue) profiled against (C) Clonorchis sinensis (Cs.TRPMPZQ), (D) Opisthorchis viverrini (Ov.TRPMPZQ), (E) Echinostoma caproni TRPMPZQ (Ec.TRPMPZQ) and (F) Fasciola gigantica TRPMPZQ (Fg.TRPMPZQ, up triangle) and Fg.TRPM[T1271N]PZQ (circles). Data are expressed as mean ± SEM from n = 3 independent transient transfections of HEK293 cells.
PZQ Activation of Other Trematode TRPMPZQ Orthologs.
TRPMPZQ sequence information from other trematodes was then collated (SI Appendix, Table S2). Across all trematode TRPMPZQ sequences, the twenty-three residues lining the PZQ binding pocket were highly conserved (SI Appendix, Table S2), with the exception of Fasciola spp. The conservation of the binding pocket sequence would suggest that, with the exception of Fasciola spp., PZQ sensitivity is retained across all trematodes.
To test this prediction, several trematode TRPMPZQ orthologs were functionally profiled. Each of these trematode TRPMPZQ orthologs exhibited high sensitivity to (R)-PZQ (Fig. 2 C–E). These were TRPMPZQ from Clonorchis sinensis (the ‘oriental liver fluke’, EC50 = 193 ± 110 nM for (R)-PZQ at Cs.TRPMPZQ), Opisthorchis viverrini (the ‘Southeast Asian liver fluke’, EC50 = 129 ± 52 nM for Ov.TRPMPZQ), and Echinostoma caproni (EC50 = 90 ± 31 nM for Ec.TRPMPZQ), Fig. 2 C–E). This is consistent with the known sensitivity of each of these parasites to PZQ in vitro (25) and the efficacy of PZQ in treating clinical diseases caused by these agents (1, 10). Bioinformatic analyses of the remaining trematode TRPMPZQ binding pockets (Paragonimus spp. and Dicrocoelium dendriticum, Fasciolopsis buski) revealed conservation of these binding pocket residues, again consistent with the known sensitivity of these flukes to PZQ (25–31).
The Fasciolidae provided the only example where the sequence of the PZQ binding pocket showed variation from the schistosome consensus (Fig. 2B). Both F. hepatica and F. gigantica harbored a threonine residue in S1 where an asparagine residue is present in all other digeneans. For F. hepatica, the presence of this S1 threonine residue rendered Fh.TRPMPZQ insensitive to PZQ (8), consistent with the lack of efficacy of PZQ against F. hepatica in vitro and in vivo. Here, Fasciola gigantica TRPMPZQ (Fg.TRPMPZQ) was examined. The addition of (R)-PZQ did not increase cytoplasmic Ca2+ in cells expressing wild-type Fg.TRPMPZQ (Fig. 2F). Wild-type Fg.TRPMPZQ was refractory to PZQ, consistent with the known lack of efficacy of PZQ against F. gigantica (32). A mutant of wild-type Fg.TRPMPZQ to mimic the schistosome TRPMPZQ binding pocket consensus (Fg.TRPM[T1271N]PZQ) was generated. In cells expressing Fg.TRPM[T1271N]PZQ, (R)-PZQ evoked a concentration-dependent increase in Ca2+ (EC50 = 155 ± 30 nM).
In summary, analysis of 22 trematode TRPMPZQ orthologs revealed, with the exception of the Fasciola spp., a complete sequence conservation of the PZQ binding pocket (Fig. 2B). Functional profiling of TRPMPZQ orthologs, with the exception of the two Fasciola spp., showed stereoselective activation by (R)-PZQ. These data are consistent with the known sensitivity of trematode parasites to PZQ in vitro and in vivo and TRPMPZQ serving as a relevant in vivo target for PZQ.
PZQ Activation of Other Flatworm TRPMPZQ Orthologs.
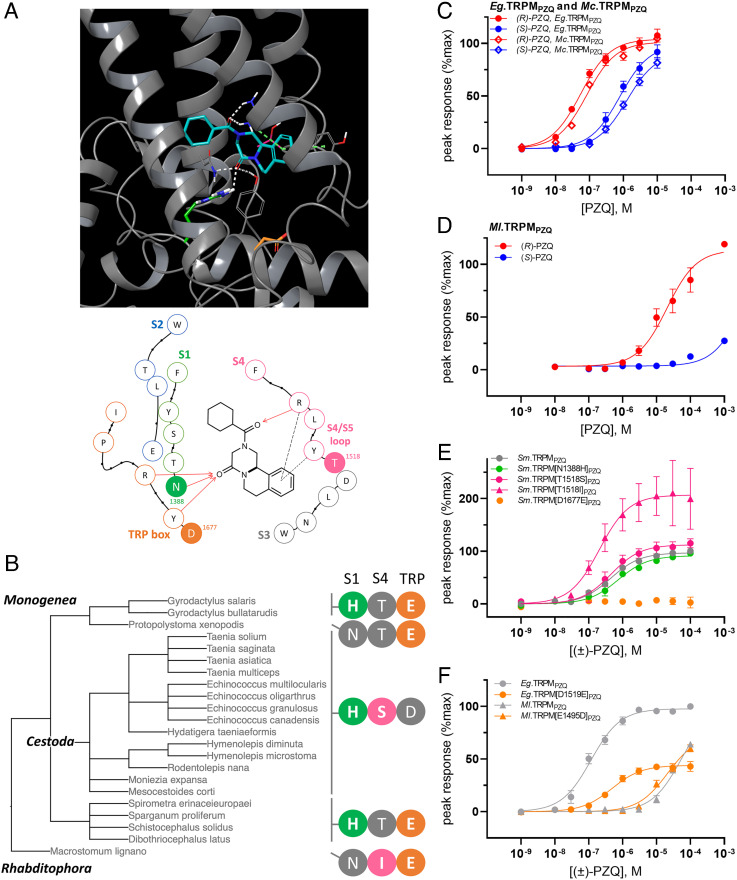
Bioinformatic analysis was extended to a total of 43 parasitic flatworm TRPMPZQ sequences. This analysis encompassed 35 different parasitic flatworm genomes currently aggregated on the WormBase ParaSite portal (33) and 8 examples where TRPMPZQ sequences have been annotated in other genomic datasets (SI Appendix, Table S2). TRPMPZQ orthologs were identifiable in every parasitic flatworm genome analyzed (6). SI Appendix, Table S2 collates the predicted PZQ binding pocket residues across all these TRPMPZQ orthologs, encompassing all available parasitic flatworm TRPMPZQ sequences, three free-living flatworm TRPMPZQ representatives (Macrostomum lignano, Schmidtea mediterranea, and Dugesia japonica) and the human TRPM2 and TRPM8 channels with which TRPMPZQ displays some domain homology (SI Appendix, Table S2). The 23 residues lining the VSLD binding pocket were well conserved across all the parasitic flatworm representatives, with only three positions showing variation from the trematode TRPMPZQ binding pocket consensus (Fig. 3A). The extent of variation at these positions is shown in Fig. 3B and SI Appendix, Table S2.
Fig. 3.
Variation in the binding pocket and functional sensitivity of TRPMPZQ in other parasitic flatworms. (A, Top) docking pose of (R)-PZQ (blue) within the VSLD of Sm.TRPMPZQ. Key interactions are shown as follows: hydrogen bond (white) and π–π stacking and cation–π interactions (dashed green line). Residues N1388 (S1, green), D1677 (TRP helix, orange) and T1518 (S4/S5 loop, pink) are highlighted. (Bottom) Interaction map of (R)-PZQ within the transmembrane binding pocket of Sm.TRPMPZQ as detailed in Fig. 2A. The residues in solid color: ‘N’ (S1, green), ‘T’ (S4/S5 loop, magenta), and ‘D’ (TRP box, orange) highlight the three sites of variation between wild-type TRPMPZQ sequences of parasitic flatworms. (B) Summary of sequence variation in TRPMPZQ orthologs using a classification reconstructed from Lifemap (24). Binding pocket sequence combinations define two cestode groupings (‘HSD’ vs ‘HTE', cyclophyllidean versus pseudophyllidean cestodes), and two groupings of monogeneans (‘HTE’ vs ‘NTE’, monopisthocotylean versus polyopisthocotylean monogeneans). The sequence from a free-living flatworm representative (Macrostomum lignano) is also shown (‘NIE’). (C) Concentration–response curve for activation of Echinococcus granulosus TRPMPZQ (Eg.TRPMPZQ) and Mesocestoides corti TRPMPZQ (Mc.TRPMPZQ) by (R)-PZQ (red) and (S)-PZQ (blue). (D) Concentration–response curve for the activation of Macrostomum lignano TRPMPZQ (Ml.TRPMPZQ) by PZQ enantiomers, (R)-PZQ (red) and (S)-PZQ (blue). (E) Concentration–response relationships for four different point mutants made within the full-length Sm.TRPMPZQ backbone (Sm.TRPM[N1388H]PZQ , Sm.TRPM[T1518S]PZQ ,Sm.TRPM[T1518I]PZQ, and Sm.TRPM[D1677E]PZQ) expressed relative to the maximal response of the wild-type Sm.TRPMPZQ sequence. (F) Effects of mutation of the natural TRP helix acidic residue in wild-type Eg.TRPMPZQ (‘D’ to ‘E’, orange circle) and Ml.TRPMPZQ (‘E’ to ‘G’, orange triangle). Data are expressed as mean ± SEM from n = 3 independent transfections.
The first residue exhibiting variation was the previously characterized S1 residue (Fig. 3 A and B). This S1 binding pocket residue—represented by asparagine residue in all trematodes (with the exception of Fasciola spp.)—was represented by histidine in all cestodes as well as monopisthocotylean monogeneans. The second variant residue was in the S4–S5 loop (Fig. 3 A and B) which was represented by a threonine in Sm.TRPMPZQ (T1518) but as a serine in cyclophyllidean cestodes and an isoleucine in M. lignano. The final position showing variation was within the TRP helix (Fig. 3 A and B) where an aspartic acid residue in Sm.TRPMPZQ (D1677) was represented by a glutamic acid residue in monogeneans, pseudophyllidean cestodes, and free-living flatworms.
Therefore, three unique, natural TRPMPZQ binding pocket configurations were resolved in parasitic flatworms that diverged from the ‘NTD’ configuration seen in trematodes (Fig. 3B). These configurations were ‘HSD’ (variation underlined) in cyclophyllidean cestodes, ‘HTE’ in pseudophyllidean cestodes and Gyrodactylus spp. (monopisthocotylean monogeneans), and ‘NTE’ in Protopolystoma xenopodis (the sole polyopisthocotylean monogenean represented). The binding pocket consensus was less conserved in free-living planarians, with six amino acid differences observed between the Dugesia japonica and Schmidtea mediterranea TRPMPZQ binding pocket with Sm.TRPMPZQ (SI Appendix, Table S2). However, the TRPMPZQ pocket of free-living flatworm Macrostomum lignano exhibited only two amino acid changes from the Sm.TRPMPZQ sequence, which were an isoleucine in the S4 to S5 loop as well as the glutamic acid residue in the TRP domain (‘NIE’ configuration). Human TRPM2 (5 differing residues) and TRPM8 (10 differing residues) exhibited greater divergence.
PZQ sensitivity was then assessed in several of these TRPMPZQ orthologs. These included TRPMPZQ from the cyclophyllidean cestodes Echinococcus granulosus (Eg.TRPMPZQ) and Mesocestoides corti (Mc.TRPMPZQ), as well as TRPMPZQ from a free-living flatworm representative, M. lignano (Ml.TRPMPZQ). Both cestode channels exhibited high sensitivity to (R)-PZQ (EC50 = 55 ± 6 nM for Eg.TRPMPZQ and EC50 = 82 ± 3 nM for Mc.TRPMPZQ, Fig. 3C), consistent with the known activity of PZQ against E. granulosus, M. corti and other cyclophyllidean cestodes (1, 15, 34, 35). Ml.TRPMPZQ was also activated by (R)-PZQ but displayed considerably lower sensitivity (EC50 = 18 ± 0.8 µM, Fig. 3D) but a sensitivity consistent with PZQ action on M. lignano (18). These data evidence a broad range in (R)-PZQ sensitivity (~330-fold) between these flatworm TRPMPZQ orthologs.
Molecular Basis for Varied PZQ Sensitivity.
To investigate the basis for this differential sensitivity, we examined the functional impact of mutants at each of the three identified binding pocket residues that show natural variation. Mutations were generated in Sm.TRPMPZQ (Sm.TRPM[N1388H]PZQ, Sm.TRPM[T1518S]PZQ, Sm.TRPM[T1518I]PZQ, and Sm.TRPM[D1677E]PZQ, Fig. 3E), and each was functionally profiled using the Ca2+ reporter assay. Two mutants—Sm.TRPM[N1388H]PZQ and Sm.TRPM[T1518S]PZQ—exhibited equivalent sensitivity toward PZQ as the wild-type channel (Fig. 3E). One mutant—Sm.TRPM[T1518I]PZQ—displayed higher sensitivity (EC50 = 207 ± 27 nM) than the wild-type channel (EC50 = 442 ± 55 nM, Fig. 3E). However, Sm.TRPM[D1677E]PZQ displayed no responsiveness to (±)-PZQ (Fig. 3E).
With the clear caveat that these point mutations were all made within the S. mansoni TRPMPZQ binding pocket and not in the context of full-length sequences of the different TRPMPZQ orthologs, the results suggest that PZQ sensitivity is retained at naturally occurring S1 (‘H’) and S4 variants (‘S’ and ‘I’). However, the acidic residue substitution in the TRP domain (from aspartic acid ‘D’ to glutamic acid ‘E’) markedly impaired PZQ action. The presence of the glutamic acid residue was also associated with poor sensitivity to PZQ in Ml.TRPMPZQ (Fig. 4D) where this glutamic acid residue occurs naturally (Fig. 3D). The importance of this TRP helix acidic residue was also highlighted by reciprocal mutagenesis. Introduction of a (‘D’ to ‘E’) point mutant within the backbone of the cestode TRPMPZQ channel lowered (±)-PZQ sensitivity by ~4-fold (EC50 for (±)-PZQ = 485 ± 51nM in Eg.TRPM[D1519E]PZQ versus 116 ± 20 nM in wild-type Eg.TRPMPZQ, Fig. 3F). Reciprocally, the introduction of a (‘E’ to ‘D’) point mutant in the Ml.TRPMPZQ channel increased (±)-PZQ sensitivity by ~2.5-fold (EC50 for (±)-PZQ = 21.1 ± 3.1 μM in Ml.TRPM[E1495D]PZQ versus 53.9 ± 0.4 μM in wild type, Fig. 3F).
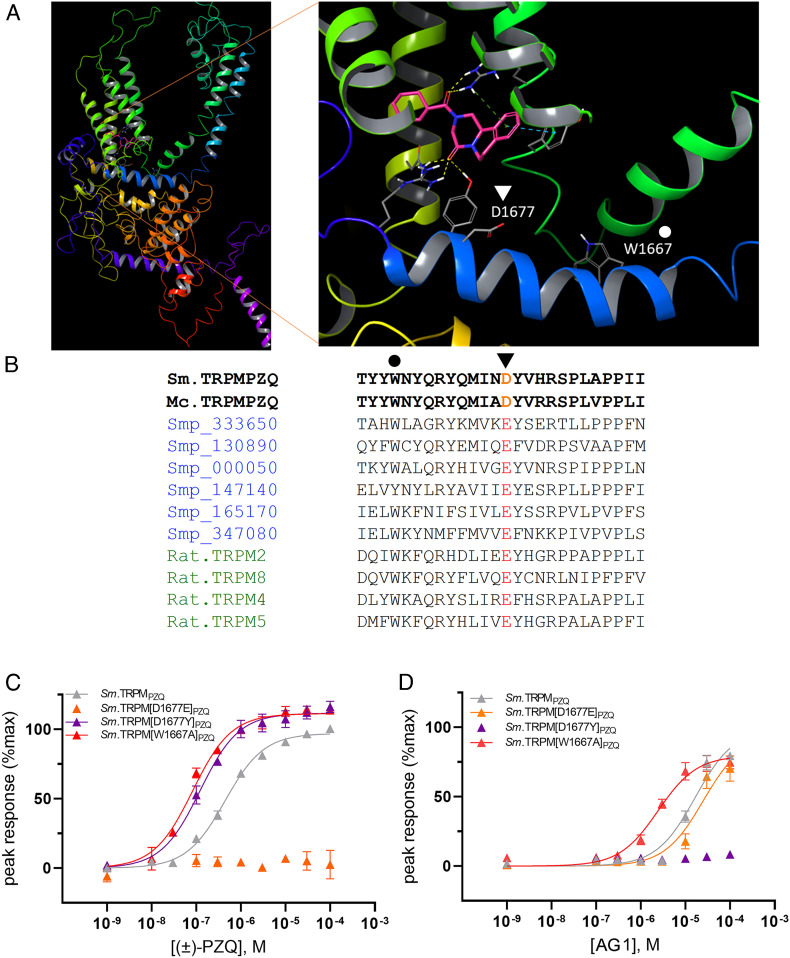
Fig. 4.
Role of an acidic TRP helix residue as a gatekeeper regulating PZQ engagement within the VSLD binding pocket. (A) Sm.TRPMPZQ homology model (from (8)) with enlargement showing the binding pose of (R)-PZQ within the VSLD binding pocket in relation to TRP helix residues D1677 (triangle) and W1667 (circle). (B) Sequence alignment of a trematode TRPMPZQ (Sm.TRPMPZQ) and cyclophyllidean cestode TRPMPZQ (Mc.TRPMPZQ) highlighting the TRP domain aspartic acid residue (D1677, triangle) that is present in parasites that display the highest sensitivity to PZQ. Other predicted schistosome TRPM channels (gene identifiers in blue) and vertebrate Ca2+-sensitive TRPM channels (green) possess a glutamic acid residue at the same position. A residue that regulates TRP channel gating (W1667) interrogated by mutagenesis in (C&D) is also highlighted (circle). (C and D) Differential effects of TRP helix mutants on ligand activation of Sm.TRPMPZQ. Concentration–response curves for (C) (±)-PZQ and (D) AG1 activation of Sm.TRPM[D1677Y]PZQ (purple), Sm.TRPM[D1677E]PZQ (orange), and Sm.TRPM[W1667A]PZQ compared with wild-type Sm.TRPMPZQ (gray).
Collectively, these data highlight the importance of this variant acidic residue (‘D’ or ‘E’) found in the TRP helix as a key determinant of TRPMPZQ sensitivity toward PZQ (Fig. 4A). The aspartic acid variant was unique to trematode and cyclophyllidean cestode TRPMPZQ, and these channels and these parasites exhibit the highest sensitivity to PZQ. The glutamic acid variant, associated with lower sensitivity to PZQ, was naturally present in monogeneans, pseudophyllidean cestodes, and free-living flatworm TRPMPZQ (SI Appendix, Table S2). This glutamic acid residue is also found in the TRP helix of all other schistosome TRPM family members (Fig. 4B), none of which have been reported to show sensitivity to PZQ (6). It is also present in the vertebrate TRPM channels TRPM2 and TRPM8 (which most closely resemble TRPMPZQ (7), Fig. 4B). Human TRPM8 responds to (S)-PZQ only in the micromolar range [EC50 for human TRPM8 to (S)-PZQ = ~20 µM (36, 37)].
How Does This Residue Impact PZQ Sensitivity?.
This variant TRP helix residue (D1677) is situated just beneath the PZQ binding pocket mapped within the VSLD of TRPMPZQ (Fig. 4A). This would suggest that the identity of the acidic residue impacts ligand binding in the pocket. However, given that the TRP domain is important for transducing the binding of ligands into channel activation, an alternative possibility is that this variation impacts channel gating rather than ligand binding. If this were the case, then the ‘D’ to ‘E’ variation would have the same deleterious effect for different ligands. Assays performed with the two known activating ligands of Sm.TRPMPZQ—PZQ and AG1 (38)—revealed that the effects of this residue on gating were not the explanation for the differential sensitivity to PZQ. While Sm.TRPM[D1677E]PZQ ablated sensitivity to PZQ, responses to AG1 were unaffected (Fig. 4 C and D). In contrast, a different mutant at this same position—Sm.TRPM[D1677Y]PZQ—enhanced PZQ sensitivity but ablated AG1 responsiveness (Fig. 4D). Therefore, variation at this position confers differential effects on different ligands, implicating effects on ligand binding. In contrast, mutation of a tryptophan residue (W1667A, Fig. 4A) in the TRP helix that is positioned further away from the binding pocket and known to regulate channel gating (39, 40) caused increased sensitivity to both ligands (Fig. 4 C and D). Therefore, variation at D1677 impacts channel sensitivity to PZQ by altering PZQ engagement within the VSLD binding pocket.
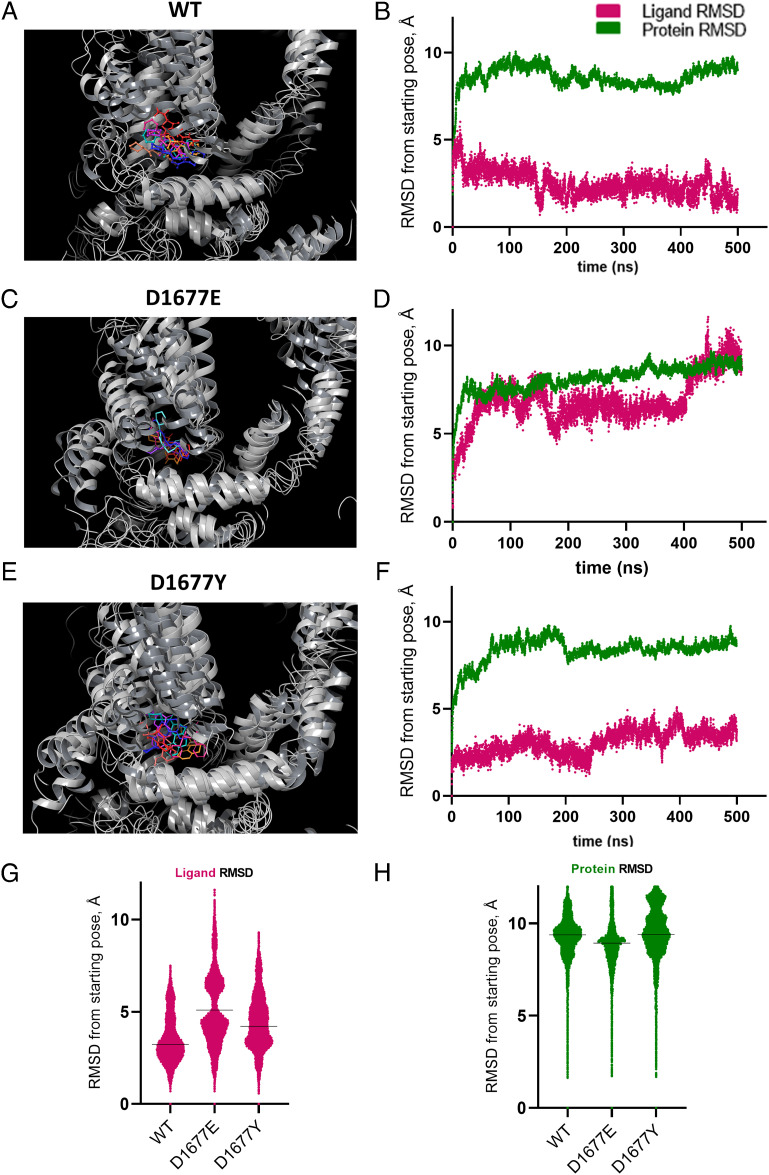
To investigate how amino acid variation at residue 1677 impacted ligand binding, we utilized molecular dynamics (MD) to model residue flexibility and conformational changes at the atomic level. MD permits the appreciation of key ligand–receptor interactions and ligand stability within a binding pocket. For each of the TRP helix variants (wild type, D1677E, and D1677Y), seven independent MD simulations (500 ns duration) of the channel complexed with (R)-PZQ were simulated, compiling 3500 ns of data for each variant (70,000 total frames). Each trajectory was clustered to resolve the most likely binding pose from all those sampled during each run, and these poses were superimposed. For all three variants, (R)-PZQ occupied the VSLD of the channel (Fig. 5 A, C, and E).
Fig. 5.
MD Simulations of (R)-PZQ complexed with Sm.TRPMPZQ TRP helix variants. (A) Superposition of the top pose from each of seven independent 500 ns MD runs of (R)-PZQ in complex with WT Sm.TRPMPZQ. The channel is depicted in gray, (R)-PZQ is colored. (B) Representative RMSD trace of a single 500-ns trajectory (10,000 frames) of (R)-PZQ complexed with wild-type Sm.TRPMPZQ with both ligand RMSD (maroon) and protein RMSD (green) represented. RMSD is measured in reference to the Cα of the channel. (C and D) As in A and B but with Sm.TRPM[D1677E]PZQ. (E and F) As in A and B but with Sm.TRPM[D1677Y]PZQ. (G) Plot of ligand RMSD of all combined 70,000 frames of MD simulation for each TRP helix variant referenced to the Cα of the channel. The black line indicates the median RMSD of the cumulative 3,500 ns of simulation (WT = 2.9 Å, D1677E = 5.1 Å, D1677Y = 4.1 Å). (H) Plot of the protein RMSD of all combined 70,000 frames of MD simulation for each TRP helix variant referenced to the Cα of the channel. The black line indicates the median RMSD of the cumulative 3500 ns of simulation (WT = 9.3 Å, D1677E = 8.9 Å, D1677Y = 9.4 Å).
In wild-type Sm.TRPMPZQ, the (R)-PZQ binding poses clustered together and recapitulated critical ligand–protein interactions (Fig. 5A and ref. 8). For the D1677E variant, the clustered poses showed more variability (Fig. 5C). The top poses were displaced from the wild-type pose, and the interactions important for channel activation were absent (8). Poses from the Sm.TRPM[D1677Y]PZQ simulations clustered together but were deeper into the VSLD pocket (Fig. 5E), suggesting additional attractive interactions between the ligand and channel.
The root-mean-square deviation (RMSD) of the ligand during simulation was obtained from these MD trajectories as a measure of ligand stability in the binding pocket with respect to the modeled protein. Larger RMSD values indicated greater movement of the ligand from the reference binding pose, inversely correlating with the stability of the initial pose. For (R)-PZQ in the WT channel, the ligand was stable throughout these simulations (Fig. 5B): analysis of the RMSD across all 70,000 frames yielded a median RMSD of 2.9 Å (Fig. 5G). This contrasted with MD simulations of Sm.TRPM[D1677E]PZQ, where (R)-PZQ displayed an increased RMSD, and in multiple runs, the ligand diffused away from the binding pocket. Fig. 5D shows the RMSD of the ligand immediately jumped to >5 Å and never returned to the baseline. Analysis of the RMSD of all 70,000 frames from the Sm.TRPM[D1677E]PZQ simulation yielded a median RMSD of 5.1 Å (Fig. 5G). This suggested that (R)-PZQ is less stable within the D1677E variant binding pocket compared to the WT channel. For Sm.TRPM[D1677Y]PZQ, (R)-PZQ stabilized almost immediately in the VSLD pocket and remained in place throughout the simulation (Fig. 5F). Analysis of the RMSD of all 70,000 frames from this simulation revealed a median RMSD of 4.1 Å (Fig. 5G). The RMSD in this variant represented a shift of the ligand into a stabilized pose deeper within the binding pocket than seen with the WT channel. For each of the three variants, protein RMSD stabilized at ~9 Å across each individual run (green tracings, Fig. 5 B, D, F, and H), with analyses of all 70,000 frames suggesting similar protein flexibility for each variant. Overall, these unbiased MD simulations revealed that when starting from the ligand–channel pose containing the proposed critical residue interactions (8), (R)-PZQ remains stable in its binding orientation. The D1677E variant disrupted these interactions with (R)-PZQ exhibiting increased movement. The D1677Y variant captured (R)-PZQ deeper within the binding pocket.
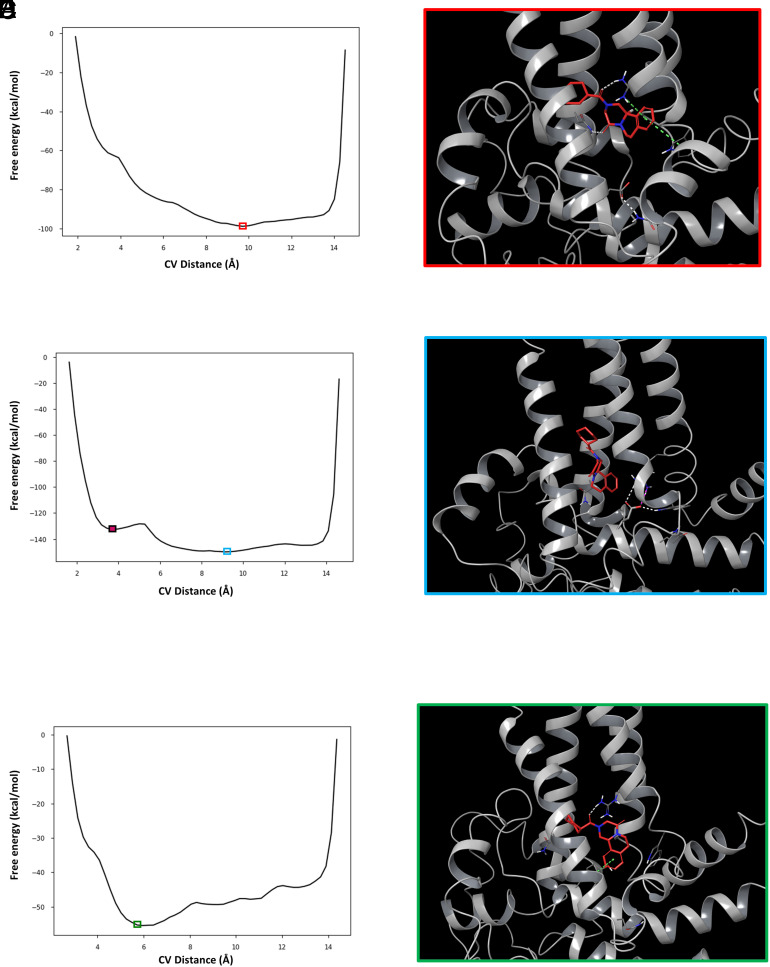
To sample more of the conformational landscape of these ligand–channel complexes, we employed metadynamics as a second modeling approach (41, 42). Metadynamics facilitates extended sampling of ligand–receptor binding conformations, and this methodology has been used to reconstruct the full free-energy landscape of protein-ligand binding (41, 43, 44). With this approach, the lowest energy binding pose and corresponding ligand–channel interactions can be determined along a given collective variable (CV). When the wild-type complex was subjected to metadynamics, employing the distance between the center of mass of (R)-PZQ and the center of mass of D1677 as the CV, the lowest free-energy pose reproduced key interactions between (R)-PZQ and VSLD binding pocket residues (Fig. 6 A and B). (R)-PZQ was anchored in the pocket by hydrogen bonding and cation–π interactions with R1514 (S4) and a hydrogen bond with N1388 (S1). These interactions have previously shown to be important for engaging (R)-PZQ (8). However, D1677 formed a hydrogen bond with Q1673 one turn further along the TRP helix. This interaction pulled the D1677 side chain away from the VSLD binding pocket, precluding any interference with (R)-PZQ association.
Fig. 6.
Metadynamics simulations of (R)-PZQ complexed with Sm.TRPMPZQ TRP helix variants. (A) Representative free-energy plot resulting from metadynamics simulations of (R)-PZQ complexed with wild-type Sm.TRPMPZQ. The x-axis is the CV, set as the distance between the center of mass of (R)-PZQ and the center of mass of D1677 (in Å). The y-axis shows the free energy of that interaction (in kcal/mol). The red box encloses the lowest energy frame from the simulation. (B) The binding pose of (R)-PZQ within Sm.TRPMPZQ from the lowest energy frame from panel A. (R)-PZQ forms hydrogen bonds (dashed white lines) with R1514 and N1388, cation–π interactions with R1514, and π–π stacking interactions with W1451 (dashed green lines). D1677 is pulled away from the pocket via a hydrogen bond with Q1673. (C) Representative free-energy plot resulting from metadynamics simulations of (R)-PZQ complexed with Sm.TRPM[D1677E]PZQ as in A. The solid maroon box encloses the frame representing the relative minima of the simulation (pose shown in SI Appendix, Fig. S2). The blue box encloses the lowest energy frame from the simulation. (D) The binding pose of (R)-PZQ within Sm.TRPM[D1677E]PZQ in the lowest energy frame from the simulation. (R)-PZQ is displaced from any binding interactions and E1677 points into the binding pocket, forming hydrogen bonds (dashed white lines) and a salt bridge with R1514 (dashed magenta line) and a hydrogen bond with W1451. (E) Representative free-energy plot from metadynamics simulations of (R)-PZQ complexed with Sm.TRPM[D1677Y]PZQ as in A. The green box encloses the lowest energy frame from the simulation. (F) The binding pose of (R)-PZQ within Sm.TRPM[D1677Y]PZQ in the lowest energy frame from the simulation. (R)-PZQ binds within the VSLD of the channel, forming a hydrogen bond (dashed white line) with R1514 and π–π stacking (dashed green line) with Y1677.
The lowest free-energy pose for (R)-PZQ in Sm.TRPM[D1677E]PZQ was distinctly different from the lowest free-energy pose of (R)-PZQ in the wild-type channel. Metadynamics revealed two energy minima—a relative minimum (‘metastable state’) and an absolute minimum (Fig. 6C). In the pose associated with the relative minimum, the side chain of the variant glutamate residue projects into the VSLD binding pocket, resulting in (R)-PZQ displacement (SI Appendix, Fig. S2) In the pose associated with the absolute minimum, (R)-PZQ was also completely displaced from any binding pocket interactions, with E1677 projected into the binding pocket (Fig. 6D). The observed lower free energy resulted from new stabilizing hydrogen bond interactions that formed within the pocket: E1677 with R1514 and W1451 (Fig. 6D). These new hydrogen bonding interactions prevent (R)-PZQ interaction with R1514, an interaction critical for (R)-PZQ-evoked channel activation (8). The Sm.TRPM[D1677Y]PZQ complex with (R)-PZQ was also subjected to metadynamics. In the lowest free-energy pose, (R)-PZQ acquired new π–π stacking interactions with the phenol of Y1677 (Fig. 6 E and F) and was anchored in place through the hydrogen bond interaction with R1514. This prediction of a new interaction with PZQ was consistent with the increased potency of PZQ in functional assays (Fig. 4C). A comparison of the (R)-PZQ binding pose in Sm.TRPMPZQ and Sm.TRPM[D1677E]PZQ with the binding pose adopted by TRPM8 ligands within Hs.TRPM8 (45, 46) is shown in SI Appendix, Fig. S3.
Therefore, both MD analyses (Fig. 5) and metadynamics (Fig. 6) demonstrated that the TRP helix aspartate to glutamate variation in Sm.TRPMPZQ displaced (R)-PZQ from productive interactions within the VSLD binding pocket, consistent with the decreased PZQ sensitivity of parasitic flatworm TRPMPZQ channels containing this variation.
Discussion
In this study, we have profiled the PZQ binding pocket of TRPMPZQ orthologs in all currently available parasitic flatworm genomes: TRPMPZQ is present in all parasitic flatworms examined, and the PZQ binding site is well conserved. In trematodes, functional profiling of six TRPMPZQ orthologs demonstrated stereoselective activation by (R)-PZQ in the submicromolar range (EC50 for (R)-PZQ = ~90 to 320 nM, Figs. 1 and 2). These include TRPMPZQ from the three species of schistosomes (S. mansoni, S. haematobium, and S. japonicum) responsible for the majority of infections worldwide, as well as TRPMPZQ from E. caproni, C. sinensis, and O. viverrini. In all these TRPMPZQ orthologs, the key PZQ binding pocket residues are identical (Fig. 2A and SI Appendix, Table S2), and PZQ sensitivity of this TRP channel is consistent with the known PZQ sensitivity of these parasites in vitro and in vivo. We predict the consensus TRPMPZQ pocketome will also be conserved in other trematodes where TRPMPZQ sequence information is currently lacking (1, 25, 31, 47–49), given the known efficacy of PZQ for treating other diseases caused by these parasites. The only exception found was the identification of a single amino acid variant within the TRPMPZQ pocket of Fasciola spp. (F. hepatica and F. gigantica). Fh.TRPMPZQ and Fg.TRPMPZQ were refractory to PZQ, consistent with the lack of PZQ efficacy in treating infections caused by these liver flukes (10). However, both Fasciola spp. TRPMPZQ channels gained PZQ sensitivity when the S1 variant threonine was substituted for an asparagine residue to mimic the trematode consensus sequence (Fig. 2F and ref. 8). It will be of interest to further analyze the Fasciolidae to understand the evolution of this pocket between closely related liver flukes (50) and determine if this S1 residue variation is specific to just the Fasciola genus. Key representatives for analysis would be from the genera Fascioloides (e.g., F. magna), Protofasciola, and Parafasciolopsis.
While Fasciola provide a stark example showing how differential species sensitivity to a TRP channel ligand has impact on disease treatment, the principle that TRP channel sequence variation imparts species-specific ligand sensitivity has been previously recognized. Differences between avian and mammalian TRPV1 sequences underpin the tolerance of birds to capsaicin, an adaptation thought to aid seed dispersal (51, 52). Variation of an S4 residue in tree shrew TRPV1 confers insensitivity to an analog of capsaicin, so that this particular species of tree shrew is indifferent to the consumption of spicy plants (53). This specific variant is predicted to remove a critical hydrogen bonding interaction with the ligand (53), paralleling the effect of sequence variation at a key hydrogen-binding position in the S1 helix of TRPMPZQ underlying the insensitivity of Fasciola spp. TRPMPZQ to PZQ (8). Possibly, this binding pocket variation within Fasciola spp. TRPMPZQ holds significance for (dis)engagement of natural products or metabolites that are uniquely encountered during the Fasciola lifecycle.
PZQ is also effective for combating other parasitic flatworm infections (1, 5, 10–12). PZQ is highly effective against cyclophyllidean cestodes (1, 54–57), with deleterious effects occurring at very low PZQ concentrations in vitro [as low as 320 pM, (55)]. This is consistent with functional analyses of E. granulosus TRPMPZQ (Eg.TRPMPZQ) and M. corti TRPMPZQ (Mc.TRPMPZQ) which both displayed extremely high sensitivity to PZQ. This sensitivity occurred despite differences between these tapeworm (‘HSD’ configuration) and trematode (‘NTD’) TRPMPZQ binding pockets (Fig. 3B). Interestingly, the structure–activity relationships of PZQ analogs against trematodes and cestodes are different (1) such that these variant residues (Fig. 3B) likely contribute to the ability of trematode and cestode TRPMPZQ to accommodate different ligands.
For all other flatworms, the presence of the glutamic acid TRP domain residue (Fig. 4A) is important as this variant confers lower sensitivity to PZQ (supramicromolar rather than submicromolar). This holds clinical relevance as pseudophyllidean cestode species harboring this residue can cause a rare, but potentially serious, disease in humans (58, 59). There is prior evidence that pseudophyllidean tapeworms exhibit lower sensitivity to PZQ (15); for example, larval and adult diphyllobothriids required PZQ concentrations in excess of 320 μM to show lethality (60). This glutamic acid residue is also present in all monogeneans; the effective treatment of which is a priority for the aquaculture industry (12, 61). In polyopisthocotylean monogeneans, this glutamic acid variant occurs as the sole change from the trematode TRPMPZQ binding pocket consensus (‘NTE’ in Protopolystoma xenopodis) compared to monopisthocotylean monogeneans (‘HTE’ configuration in Gyrodactylus spp.) where the more accommodating S1 histidine variant is present. Differential sensitivities of monogenean species have been reported (12, 61), with evidence that polyopisthocotylean species require higher PZQ concentrations to manifest deleterious effects (62, 63). Similarly, the biological efficacy of PZQ against free-living flatworms, which also harbor the glutamic acid TRP domain residue, is in the micromolar range (EC50 = 35 ± 7 μM for PZQ evoking bipolarity in D. japonica (16)). Such lower sensitivity was supported here by functional analysis of M. lignano TRPMPZQ (‘NIE’ configuration) which was activated by micromolar (±)-PZQ concentrations (Fig. 3F). This molecular insight provides pause should this TRP domain variant (E1677 in S. mansoni, E1588 in S. japonicum, and E1605 in S. haematobium), which has evolved naturally in several flatworm lineages, were to be found in schistosome populations, for example, occurring in response to selection pressure evoked by mass drug administrative campaigns. Any decrease in the effectiveness of PZQ as a cheap, effective drug for treating schistosomiasis would be a significant public health challenge.
Mechanistically, the application of MD (Fig. 5) and metadynamics (Fig. 6) to simulate PZQ engagement within the wild-type Sm.TRPMPZQ and Sm.TRPM[D1677E]PZQ binding pocket confirmed the deleterious impact of the glutamic acid variant. The glutamic acid residue (E1677) projected into the VSLD, rather than along the TRP helix as occurred with the wild-type aspartic acid residue (D1677) which was anchored away from the binding pocket via an interaction with Q1673 (Fig. 6). The internal projection of E1677 within the VSLD positioned negative charge in too close proximity to the preferred (R)-PZQ binding pose or removed a key interaction for (R)-PZQ engagement as E1677 formed a hydrogen bond with R1514. These outcomes were represented as discrete energy minima in the metadynamics simulations (Fig. 6). Therefore, the identity of this TRP domain gatekeeper residue provides an answer to why infections with parasites carrying a glutamate at this position (monogeneans, pseudophyllidean cestodes) are clinically less sensitive to treatment with PZQ, while infections with parasites carrying an aspartate at this position (most trematodes and cyclophyllidean cestodes) uniquely display a high sensitivity to PZQ. This provides an opportunity to design better ligands tailored for the TRPMPZQ orthologs displaying lower sensitivity toward PZQ. Different ligands utilize different poses to engage the VSLD binding pocket, such that alternative chemotypes (for example, AG1 (38)) may better tolerate the glutamic acid TRP helix variant that impairs PZQ engagement (Fig. 4D).
In conclusion, the presence of TRPMPZQ in all parasitic flatworms sensitive to PZQ, and the correlation between TRPMPZQ sensitivity to PZQ and worm sensitivity to PZQ provides further support for TRPMPZQ acting as the relevant in vivo target of PZQ. The definition of the TRPMPZQ pocketome revealed the unique presence of an aspartic acid (‘D’) residue within the TRP helix of TRPMPZQ is critical for conferring high sensitivity to PZQ, and this underpins the unique efficacy of PZQ against trematode and cestode infections first observed over four decades ago (1).
Materials and Methods
Materials and Reagents.
(±)-PZQ was purchased from Sigma, and the individual enantiomers (R)-PZQ and (S)-PZQ were resolved following published protocols (64). HEK-293 cell lines were sourced from ATCC (CRL-1573) and found to be negative for mycoplasma contamination by monthly scheduled testing (LookOut® Mycoplasma PCR Detection Kit, Sigma). All cell culture reagents were from Invitrogen. Total RNA from adult male and female S. haematobium (Egyptian Strain, NR-31801) was provided by the NIAID Schistosomiasis Resource Center for distribution through BEI Resources, NIAID, NIH. Biological samples of different parasites were generously provided from various sources: F. hepatica (Paul McCusker, Maule lab, Queen’s University, Belfast), Gyrodactylus spp. (Eric Leis, La Crosse Fish Health Center, US Fish and Wildlife Service), M. corti (Arun Chauhan, Mishra lab, University of North Dakota), H. diminuta (Elise Nanista, Rozario lab, University of Georgia), and M. lignano (Lisa Glazenburg, Berezikov lab, University of Groningen).
Bioinformatic Analyses.
Genome sequences were accessed either through WormBase ParaSite v16 (33) or individual project data repositories deposited on NCBI. TRPMPZQ candidate sequences were identified by BLAST searches, and then genome sequence information was manually curated. For the calculation of TRPMPZQ amino acid identity, sequences were aligned using MAFFT (v6.864) and aligned sequences analyzed using the Ident and Sim interface using standard groups for amino acid similarity (65). The full-length sequence of S. haematobium TRPMPZQ (Sh.TRPMPZQ) was cloned from S. haematobium total RNA using RT-PCR and 5'-RACE. 5′-RACE was performed using the SMARTer RACE 5′3′ Kit (Clontech). Briefly, total RNA (0.5 to 1 µg) was reverse-transcribed using a SMARTer II A Oligonucleotide (42 °C for 1.5 h) to obtain first-strand cDNA. 5′RACE RT-PCR was performed using first-strand cDNA, Sh.TRPMPZQ gene-specific primers, and a universal primer mix using the following touchdown PCR conditions: 5 cycles (94 °C for 30 s and 70 °C for 3 min), 5 cycles (94 °C for 30 s, 68 °C for 30 s, and 72 °C for 3 min), and then, the mixture was amplified for 30 cycles (94 °C for 30 s; 63 °C for 30 s and 72 °C for 3 min). The RACE PCR products were cloned into pRACE vector using In-Fusion Cloning system (Clontech) and sequenced (Retrogen, Inc).
FLIPR Ca 2+ Assay.
The Fluorescence Imaging Plate Reader (FLIPR) Ca2+ reporter assay was performed in black-walled, clear-bottomed 384-well plates coated with poly-D-lysine (Greiner Bio-One, Germany). Briefly, nontransfected or transfected HEK293 cells were seeded (20,000 cells/well) in DMEM growth media containing 10% FBS. After 24 h, the medium was removed and replaced with 20 µL of Fluo-4 NW dye loading solution (Molecular Devices), previously reconstituted in assay buffer (Hanks' balanced salt solution with Ca2+, Mg2+, 20 mM HEPES, and 2.5 mM probenecid). Cells were incubated for 30 min at 37 °C (5% CO2) followed by an additional 30-min incubation at room temperature. Drug dilutions were prepared in an assay buffer, but without probenecid and fluorescent dye, in 384-well plates (Greiner Bio-One). Using a FLIPRTETRA (Molecular Devices), basal fluorescence (filter settings λex = 470 to 495 nm, λem = 515 to 575 nm) from each well was monitored for 20 s, then 5 μL of drug or vehicle solution was added (25 μL total volume) and the signal was recorded over 250 s. Changes in fluorescence were represented as relative fluorescence units after subtracting the average basal fluorescence (average basal fluorescence over 20 s) from the recorded values. Concentration–response analysis was performed using sigmoidal curve fitting functions in Prism using data from n ≥ 3 independent transfections, with n ≥ 3 technical replicates per assay.
Computational Procedures.
Generation of the Sm.TRPMPZQ homology model has been previously described (8). Sm.TRPM[D1677E]PZQ and Sm.TRPM[D1677Y]PZQ homology models were created by making the desired mutant of the Sm.TRPMPZQ wild-type homology model in the Schrodinger Computational Suite (v2022-1) followed by minimization of the protein using the Protein Preparation Wizard in the OPLS4 force field at pH = 7.4 (66). To prepare ligands for modeling, (R)-PZQ was drawn in ChemDraw Professional (v21.0.0), imported into the Maestro GUI (v. 13.1), and prepared using the LigPrep tool with default settings in the OPLS4 force field at pH = 7.4. The output structure was used for subsequent modeling.
MD simulations.
Unbiased MD were performed using Desmond within the Schrodinger Computational Suite (67). For Sm.TRPMPZQ, the protein–ligand complex was inserted into a POPC membrane bilayer using the ‘System Builder’, aligning the membrane coordinates to PDB 6NR3 in the OPM database. Solvation was treated explicitly using the SPC water model with 0.15 M NaCl, charges were neutralized by adding Na+ or Cl− ions when necessary, and the membrane was generated in the OPLS4 force field. The system was minimized to a protein RMSD <0.3 Å, and the minimized system was used as the starting pose for MD simulations. Seven independent runs of 500 ns were completed. The simulations were run in the NPγT ensemble using both the Langevin thermostat (300 K) and semiisotropic barostat (1.01325 bar). The system was relaxed before simulation and gradually brought to temperature with decreasing constraints as per the default series of Desmond simulations. Each simulation began from a random seed, the velocities were randomized, and frames were recorded at an interval of 50 ps which allowed for the collection of 10,000 frames in each simulation. Sm.TRPM[D1677E]PZQ and Sm.TRPM[D1677Y]PZQ were prepared, and simulations were executed in an identical manner. Each of the seven runs for each channel variant was independently clustered using the ‘Trajectory Clustering Tool’ within the Maestro GUI. The poses of the most populated cluster from each run were then superimposed as depicted in Fig. 5. Ligand and protein RMSD—the average change in displacement of selected atoms for a particular MD frame with respect to a reference frame—were calculated from MD simulations with reference to the Cα of the protein in the input structure using the Maestro GUI of the Schrodinger Computational Suite. Images were plotted, and median calculations were performed, using GraphPad Prism (v9.3.1).
Metadynamics.
The minimized, solvated systems for metadynamics simulations were generated as described for the MD Simulations and then run using Desmond (67). The CV was set as the distance between the center of mass of (R)-PZQ and the center of mass of the side chain of residue 1677 (D1677 for WT Sm.TRPMPZQ, E1677 for Sm.TRPM[D1677E]PZQ, and Y1677 for Sm.TRPM[D1677Y]PZQ). The Gaussian width and height were set at 0.025 Å and 0.015 kcal/mol, respectively, and deposited at 0.09 ps intervals, and a necessary wall was placed at 15 Å to keep the ligand in the VSLD of the channel. Simulations were run in the NPγT ensemble using the Langevin thermostat (300 K) and Langevin barostat (1.01325 bar). Independent simulations of 50 ns were run for each channel variant, starting from a random seed number and randomizing atom velocities for each simulation. Using the ‘metadynamics analysis’ tool in Maestro, free energy of the system (ΔG) was plotted versus the CV (Å) to produce a free-energy diagram corresponding to every frame in the metadynamics simulation, as presented in Fig. 6.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This research in the Marchant lab was supported by the National Institutes of Health (R01-AI145871 to J.S.M.). N.J.M. was supported from T32 GM080202 (Medical Scientist Training Program). D.J.S. acknowledges the support from the NIH (T32 HL134643) and the MCW Cardiovascular Center’s A.O. Smith Fellowship Scholars Program. We acknowledge the computational resources and technical support provided by the Research Computing Center at the Medical College of Wisconsin.
Author contributions
C.M.R., D.J.S., S.-K.P., and J.S.M. designed research; C.M.R., D.J.S., and S.-K.P. performed research; N.J.M. contributed new reagents/analytic tools; C.M.R., D.J.S., and S.-K.P. analyzed data; and C.M.R., D.J.S., S.-K.P., and J.S.M. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Andrews P., Thomas H., Pohlke R., Seubert J., Praziquantel. Med. Res. Rev. 3, 147–200 (1983). [DOI] [PubMed] [Google Scholar]
- 2.Colley D. G., Bustinduy A. L., Secor W. E., King C. H., Human schistosomiasis. Lancet 383, 2253–2264 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McManus D. P., et al. , Schistosomiasis. Nat. Rev. Dis. Primers 4, 13 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Mehlhorn H., et al. , In vivo and in vitro experiments on the effects of praziquantel on Schistosoma mansoni. A light and electron microscopic study. Arzneimittelforschung. 31, 544–554 (1981). [PubMed] [Google Scholar]
- 5.Gonnert R., Andrews P., Praziquantel, a new board-spectrum antischistosomal agent. Z. Parasitenkd. 52, 129–150 (1977). [DOI] [PubMed] [Google Scholar]
- 6.Park S. K., et al. , The anthelmintic drug praziquantel activates a schistosome transient receptor potential channel. J. Biol. Chem. 294, 18873–18880 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park S. K., Marchant J. S., The journey to discovering a flatworm target of praziquantel: A long TRP. Trends Parasitol. 36, 182–194 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park S. K., et al. , Mechanism of praziquantel action at a parasitic flatworm ion channel. Sci. Transl. Med. 13, eabj5832 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Clec’h W., et al. , Genetic analysis of praziquantel response in schistosome parasites implicates a transient receptor potential channel. Sci. Transl. Med. 13, eabj9114 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chai J. Y., Praziquantel treatment in trematode and cestode infections: An update. Infect. Chemother. 45, 32–43 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas H., Gonnert R., The efficacy of praziquantel against cestodes in animals. Z. Parasitenkd. 52, 117–127 (1977). [DOI] [PubMed] [Google Scholar]
- 12.Bader C., Starling D. E., Jones D. E., Brewer M. T., Use of praziquantel to control platyhelminth parasites of fish. J. Vet. Pharmacol. Ther. 42, 139–153 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Mikes L., et al. , In vitro stimulation of penetration gland emptying by Trichobilharzia szidati and T. regenti (Schistosomatidae) cercariae. Quantitative collection and partial characterization of the products. Parasitol. Res. 96, 230–241 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Hirazawa N., Akiyama K., Umeda N., Differences in sensitivity to the anthelmintic praziquantel by the skin-parasitic monogeneans Benedenia seriolae and Neobenedenia girellae. Aquaculture 404, 59–64 (2013). [Google Scholar]
- 15.Gemmell M. A., Johnstone P. D., Cestodes. Antibiot. Chemother. 1971, 54–114 (1981). [DOI] [PubMed] [Google Scholar]
- 16.Nogi T., Zhang D., Chan J. D., Marchant J. S., A novel biological activity of praziquantel requiring voltage-operated Ca2+ channel beta subunits: subversion of flatworm regenerative polarity. PLoS Negl. Trop. Dis. 3, e464 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang D., Chan J. D., Nogi T., Marchant J. S., Opposing roles of voltage-gated Ca2+ channels in neuronal control of stem cell differentiation in vivo. J. Neurosci. 31, 15983–15995 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horan G. H., “The microturbellarian Macrostomum ligano as a model for the study of parasitic worms”, PhD thesis, Queen’s University, Belfast, United Kingdom; (2014). [Google Scholar]
- 19.Cwiklinski K., O’Neill S. M., Donnelly S., Dalton J. P., A prospective view of animal and human Fasciolosis. Parasite Immunol. 38, 558–568 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McManus D. P., Recent progress in the development of liver fluke and blood fluke vaccines. Vaccines (Basel) 8, 553 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meister I., et al. , Activity of praziquantel enantiomers and main metabolites against Schistosoma mansoni. Antimicrob. Agents Chemother. 58, 5466–5472 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horak P., et al. , Avian schistosomes and outbreaks of cercarial dermatitis. Clin. Microbiol. Rev. 28, 165–190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monrad J., et al. , Treatment efficacy and regulatory host responses in chronic experimental Schistosoma bovis infections in goats. Parasitology 133, 151–158 (2006). [DOI] [PubMed] [Google Scholar]
- 24.de Vienne D. M., Lifemap: Exploring the entire tree of life. PLoS Biol 14, e2001624 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehlhorn H., et al. , Ultrastructural investigations on the effects of praziquantel on human trematodes from Asia: Clonorchis sinensis, Metagonimus yokogawai, Opisthorchis viverrini, Paragonimus westermani and Schistosoma japonicum. Arzneimittelforschung 33, 91–98 (1983). [PubMed] [Google Scholar]
- 26.Lee S. H., et al. , In vitro effect of praziquantel on Paragonimus westermani by light and scanning electron microscopic observation. Kisaengchunghak Chapchi. 25, 24–36 (1987). [DOI] [PubMed] [Google Scholar]
- 27.Becker B., et al. , Light and electron microscopic studies on the effect of praziquantel on Schistosoma mansoni, Dicrocoelium dendriticum, and Fasciola hepatica (Trematoda) in vitro. Z. Parasitenkd. 63, 113–128 (1980). [DOI] [PubMed] [Google Scholar]
- 28.Dadak A. M., Wieser C., Joachim A., Franz S., Efficacy and safety of oral praziquantel against Dicrocoelium dendriticum in llamas. Vet. Parasitol. 197, 122–125 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Johnson R. J., et al. , Paragonimiasis: diagnosis and the use of praziquantel in treatment. Rev. Infect. Dis. 7, 200–206 (1985). [DOI] [PubMed] [Google Scholar]
- 30.Harinasuta T., Bunnag D., Radomyos P., Efficacy of praziquantel on fasciolopsiasis. Arzneimittelforschung. 34, 1214–1215 (1984). [PubMed] [Google Scholar]
- 31.Taraschewski H., et al. , Effects of praziquantel on human intestinal flukes (Fasciolopsis buski and Heterophyes heterophyes). Zentralbl. Bakteriol. Mikrobiol. Hyg. A 262, 542–550 (1986). [DOI] [PubMed] [Google Scholar]
- 32.Arafa W. M., Shokeir K. M., Khateib A. M., Comparing an in vivo egg reduction test and in vitro egg hatching assay for different anthelmintics against Fasciola species, in cattle. Vet. Parasitol. 214, 152–158 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Howe K. L., et al. , WormBase ParaSite - a comprehensive resource for helminth genomics. Mol. Biochem. Parasitol. 215, 2–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris D. L., Richards K. S., Chinnery J. B., Protoscolicidal effect of praziquantel–in-vitro and electron microscopical studies on Echinococcus granulosus. J. Antimicrob. Chemother. 18, 687–691 (1986). [DOI] [PubMed] [Google Scholar]
- 35.Markoski M. M., et al. , Praziquantel and albendazole damaging action on in vitro developing Mesocestoides corti (Platyhelminthes: Cestoda). Parasitol. Int. 55, 51–61 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Gunaratne G. S., Yahya N. A., Dosa P. I., Marchant J. S., Activation of host transient receptor potential (TRP) channels by praziquantel stereoisomers. PLoS Negl. Trop. Dis. 12, e0006420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Babes R. M., Selescu T., Domocos D., Babes A., The anthelminthic drug praziquantel is a selective agonist of the sensory transient receptor potential melastatin type 8 channel. Toxicol. Appl. Pharmacol. 336, 55–65 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Chulkov E. G., et al. , Identification of novel modulators of a schistosome transient receptor potential channel targeted by praziquantel. PLoS Negl. Trop. Dis. 15, e0009898 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Y., et al. , Architecture of the TRPM2 channel and its activation mechanism by ADP-ribose and calcium. Nature 562, 145–149 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Teng J., Loukin S. H., Anishkin A., Kung C., A competing hydrophobic tug on L596 to the membrane core unlatches S4–S5 linker elbow from TRP helix and allows TRPV4 channel to open. Proc. Natl. Acad. Sci. U.S.A. 113, 11847–11852 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laio A., Parrinello M., Escaping free-energy minima. Proc. Natl. Acad. Sci. U.S.A. 99, 12562–12566 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leone V., Marinelli F., Carloni P., Parrinello M., Targeting biomolecular flexibility with metadynamics. Curr. Opin. Struct. Biol. 20, 148–154 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Incerti M., et al. , Metadynamics for perspective drug design: Computationally driven synthesis of new protein-protein interaction inhibitors targeting the EphA2 receptor. J. Med. Chem. 60, 787–796 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Gervasio F. L., Laio A., Parrinello M., Flexible docking in solution using metadynamics. J. Am. Chem. Soc. 127, 2600–2607 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Yin Y., et al. , Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel. Science 363, eaav9334 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin Y., et al. , Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP2. Science 378, eadd1268 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chai J. Y., Shin E. H., Lee S. H., Rim H. J., Foodborne intestinal flukes in Southeast Asia. Korean J. Parasitol. 47, S69–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee S. H., In vitro effects of praziquantel on Fibricola seoulensis. Seoul J. Med. 26, 41–51 (1985). [Google Scholar]
- 49.Foreyt W. J., Gorham J. R., Evaluation of praziquantel against induced Nanophyetus salmincola infections in coyotes and dogs. Am. J. Vet. Res. 49, 563–565 (1988). [PubMed] [Google Scholar]
- 50.Choi Y. J., et al. , Adaptive radiation of the flukes of the family fasciolidae inferred from genome-wide comparisons of key species. Mol. Biol. Evol. 37, 84–99 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jordt S. E., Julius D., Molecular basis for species-specific sensitivity to "hot" chili peppers. Cell 108, 421–430 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Chu Y., Cohen B. E., Chuang H. H., A single TRPV1 amino acid controls species sensitivity to capsaicin. Sci. Rep. 10, 8038 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han Y., et al. , Molecular mechanism of the tree shrew’s insensitivity to spiciness. PLoS Biol 16, e2004921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Becker B., Mehlhorn H., Andrews P., Thomas H., Ultrastructural investigations on the effect of praziquantel on the tegument of five species of cestodes. Z. Parasitenkd. 64, 257–269 (1981). [DOI] [PubMed] [Google Scholar]
- 55.Andrews P., Thomas H., The effect of praziquantel on Hymenolepis diminuta in vitro. Tropenmed. Parasitol. 30, 391–400 (1979). [PubMed] [Google Scholar]
- 56.Thomas H., Gonnert R., Pohlke R., Seubert J., "Experimental and clinical studies with a new compound against tapeworms" in Paper Read at the 2nd European Multi-Colloquy of Parasitology Trogir, Yugoslavia, 1975), pp. 1–4. [Google Scholar]
- 57.Thomas H., Gonnert R., The efficacy of praziquantel against cestodes in cats, dogs and sheep. Res. Vet. Sci. 24, 20–25 (1978). [PubMed] [Google Scholar]
- 58.Liu Q., et al. , Human sparganosis, a neglected food borne zoonosis. Lancet Infect Dis 15, 1226–1235 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Chen Y., Chen X., Kang H., Case report: Moving tumor-like foci behind refractory epilepsy-cerebral sparganosis successfully treated by surgery after failure of praziquantel treatment. Front Neurol 13, 838849 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bylund G., Bang B., Wikgren K., Tests with a new compound (Praziquantel) against Diphyllobothrium latum. J. Helminthol. 51, 115–119 (1977). [DOI] [PubMed] [Google Scholar]
- 61.Norbury, Praziquantel use in aquaculture–Current status and emerging issues. Int. J. Parasitol. Drugs Drug Resist. 18, 87–102 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sitja-Bobadilla A., de Felipe M. C., Alvarez-Pellitero P., In vivo and in vitro treatments against Sparicotyle chrysophrii (Monogenea: Microcotylidae) parasitizing the gills of gilthead sea bream (Sparus aurata L.). Aquaculture 261, 856–864 (2006). [Google Scholar]
- 63.Schmahl G., Mehlhorn H., Treatment of fish parasites. 1. Praziquantel effective against Monogenea (Dactylogyrus vastator, Dactylogyrus extensus, Diplozoon paradoxum). Z. Parasitenkd. 71, 727–737 (1985). [DOI] [PubMed] [Google Scholar]
- 64.Woelfle M., et al. , Resolution of praziquantel. PLoS Negl. Trop. Dis. 5, e1260 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stothard P., The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28, 1102–1104 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Sastry G. M., et al. , Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 27, 221–234 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, 2021. Maestro-Desmond Interoperability Tools, Schrödinger,New York, NY: 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.