Significance
ABC transporters constitute a major class of multidrug resistance proteins, e.g., in cancer cells. Membrane lipids have been long known to influence The newly solved structures and functional dynamics of these proteins. Here, using cryo-EM studies of ABCG2, a prominent member of the ABC family, in its ligand-bound form and in its native turnover state, combined with extended molecular simulations, we show that lipids as well as the ligand size can impact the drug-binding mode of ABC transporters. Not only do we demonstrate that phospholipids from the cellular membrane can penetrate into the protein and directly interact with bound drugs, we also provide evidence for differential behavior of cholesterol which is not transported by ABCG2 but ironically needed for its proper transport function.
Keywords: ABC transporters, cryo-EM, molecular dynamics, lipids, membrane proteins
Abstract
ABCG2 is an ATP-binding cassette (ABC) transporter that extrudes a wide range of xenobiotics and drugs from the cell and contributes to multidrug resistance in cancer cells. Following our recent structural characterization of topotecan-bound ABCG2, here, we present cryo-EM structures of ABCG2 under turnover conditions in complex with a special modulator and slow substrate, tariquidar, in nanodiscs. The structures reveal that similar to topotecan, tariquidar induces two distinct ABCG2 conformations under turnover conditions (turnover-1 and turnover-2). μs-scale molecular dynamics simulations of drug-bound and apo ABCG2 in native-like lipid bilayers, in both topotecan- and tariquidar-bound states, characterize the ligand size as a major determinant of its binding stability. The simulations highlight direct lipid-drug interactions for the smaller topotecan, which exhibits a highly dynamic binding mode. In contrast, the larger tariquidar occupies most of the available volume in the binding pocket, thus leaving little space for lipids to enter the cavity and interact with it. Similarly, when simulating ABCG2 in the apo inward-open state, we also observe spontaneous penetration of phospholipids into the binding cavity. The captured phospholipid diffusion pathway into ABCG2 offers a putative general path to recruit any hydrophobic/amphiphilic substrates directly from the membrane. Our simulations also reveal that ABCG2 rejects cholesterol as a substrate, which is omnipresent in plasma membranes that contain ABCG2. At the same time, cholesterol is found to prohibit the penetration of phospholipids into ABCG2. These molecular findings have direct functional ramifications on ABCG2’s function as a transporter.
ABCG2 is a human ATP-binding cassette (ABC) transporter expressed in a variety of tissues and with broad substrate specificity. It exports a wide range of endobiotics and xenobiotics from the cell (1–3) and, thereby, plays a major role in the traffic and distribution of diverse compounds between different compartments of the human body. The ability of ABCG2 to extrude a wide spectrum of substrates is exploited by some cancer cells, which overexpress the transporter and thereby actively pump out various chemotherapeutic agents, a phenomenon generally referred to as multidrug resistance (MDR) (4–6).
ABCG2 functions as a homodimer, and its transport activity is mediated by large-scale conformational changes between the inward-facing (IF) and outward-facing (OF) states, in which the substrate-binding site is alternatively exposed to the cytoplasmic and extracellular sides of the membrane (7, 8). Structurally, ABCG2 is composed of two transmembrane domains (TMDs), which form the substrate-binding cavity and carry on the actual transport process, as well as two highly conserved nucleotide-binding domains (NBDs), which act as the engine for the transporter and drive the conformational changes required for active transport by binding to and hydrolyzing ATP (9–12).
High-resolution structures of ABCG2 have been resolved by cryoelectron microscopy (cryo-EM), shedding light on the protein’s functional conformations arising during the transport cycle (13–17). The IF conformation reveals a slit-like binding cavity, acting as the binding site for both substrates and inhibitors (13, 14). The binding cavity accommodates flat, polycyclic, hydrophobic small molecules that stack in between two prominent phenylalanine side chains (5, 18, 19). Available structures of ABCG2 suggest that while substrates bind as a single copy, the size of the inhibitors might affect their binding stoichiometry; relatively small inhibitors (e.g., MZ29) bind in pairs, whereas larger inhibitors (e.g., MB136) occupy the binding pocket as a single copy (8, 14, 15, 17). Variable binding stoichiometry has also been observed for a few ligands of other ABCG transporters (20, 21).
All the aforementioned ABCG2 structures have been captured via trapping a particular conformation by maneuvers such as the removal of the substrates or ATP (13), addition of inhibitors (14), mutation of the catalytic glutamate to glutamine (15), or complexing with inhibitory Fab fragments (13, 14). Recently, structures of ABCG2 have been resolved under turnover conditions (in the presence of ATP, ADP, Mg2+, and substrates, without trapping), and in the presence of bound endogenous or exogenous substrates, estrone-3-sulfate (E1S), and topotecan, respectively (8). The topotecan-bound structures exhibited two distinct conformations, termed turnover-1 (TO1) and turnover-2 (TO2) (8). These structures may represent the most unperturbed states of the transporter. While both represent IF-like states of ABCG2, TO1 demonstrates a higher separation of the NBDs and a wider substrate binding cavity in comparison to TO2. They are considered to be intermediates of the transport cycle in which first TO1 and then TO2 form during the transition to the OF state where the binding cavity opens to the extracellular side to complete the substrate translocation process (8). Interestingly, even though the residues interacting with topotecan in both TO1 and TO2 show well-defined densities, topotecan’s densities are less clear and show fewer features (8). However, the structures of ABCG2 under turnover conditions have been resolved only for the abovementioned two substrates, leaving the question as to whether similar conformations can exist in the presence of other ligands.
Transport in ABCG2 has been shown to be modulated by the lipid environment (22–26). ABCG2’s transport function is known to be potentiated by the presence of cholesterol (22–24), a fact that aligns with the presence of the protein in the cholesterol-rich regions of the plasma membrane (25, 26). However, unlike its counterparts in the ABCG family, e.g., the homodimeric ABCG1 (20, 27, 28) and the heterodimeric ABCG5/G8 (20, 29–31), cholesterol is not a substrate for ABCG2. Moreover, tightly bound annular lipids have been captured in the nanodisc-reconstituted cryo-EM structures of ABCG2 in different states (8, 13, 14, 17). These boundary lipids are seen to insert into the clefts between the TMD helices of the protein, either bound to the hydrophobic surface of the protein or near the TMD gate of the substrate binding site (8, 13, 14, 17). Moreover, biochemical and optical microscopy studies have revealed that the lipid environment significantly affects ABCG2’s transport activity (23, 24, 26). However, these structural and biochemical studies are limited either by their static description or low spatial resolution to provide a microscopic view of lipid dynamics in the vicinity of the protein.
Molecular dynamics (MD) simulations have been used extensively to shed light on the conformational dynamics of ABC transporters (32–35) as well as relevant lipid-protein interactions at an atomic scale (36, 37). All-atom (AA) MD simulations of P-glycoprotein (Pgp, ABCB1), another MDR ABC transporter, in IF-apo and IF ATP-bound conformations revealed partial entrance of phospholipids from the cytoplasmic leaflet into the substrate-binding cavity of the transporter (34). Based on this observation, the study suggested a putative pathway for drug recruitment by Pgp directly from the lipid bilayer (34). In addition to the IF conformations, lipid penetration to the substrate-binding pocket of Pgp from the extracellular side has also been reported in MD simulations of its modeled OF conformation (38, 39). In addition to AA simulations, coarse-grained MD simulations of Pgp in a complex membrane bilayer have also captured multiple instances of lipid penetration into the ligand-binding cavity (40). Additionally, μs-long AA simulations of a bacterial ABC exporter, Sav1866, in different membranes revealed how the protein’s conformational dynamics can be dependent on the lipid composition of the embedding membrane (41).
In this study, we exploited the power of cryo-EM and μs-scale AA simulations to further investigate ligand-binding properties of ABCG2 and its interplay with lipids. We present the cryo-EM structures of tariquidar-bound ABCG2 under turnover conditions, TO1 and TO2, revealing a unique binding mode for this special modulator and slow substrate. Simulations of ABCG2 in complex with the ligands capture the ligand size as a major determinant of the binding mode. More stable binding of tariquidar (larger ligand) correlates with the close overlap of the EM density and the chemical model for this ligand, while the smaller size of topotecan and the resulting higher fluctuation within the binding pocket might substantiate the low correlation between the EM density and the structural model in this case. Furthermore, for topotecan-bound ABCG2, we observe phospholipid penetration into the binding pocket and their direct interaction. The observed lipid penetration pathway from the cytoplasmic leaflet, which is also observed in the IF-apo conformation, might represent a putative hydrophobic pathway for drug recruitment in ABCG2.
Results and Discussion
Structures of Tariquidar-bound ABCG2 Under Turnover Conditions.
Unlike bona fide substrates such as E1S, and distinct from inhibitors such as Ko143 or MB-136, tariquidar has aspects of both a substrate and an inhibitor of ABCG2. At low concentrations, it acts as an ABCG2 substrate, while at higher concentrations (> 1 μM), it inhibits the transport of other substrates competitively (80, 81). We measured and compared the ATPase activity of nanodisc-reconstituted ABCG2 alone or in the presence of different ligands (SI Appendix, Fig. S4 A and C). As reported previously, the ATPase rate of nanodisc-reconstituted ABCG2 is highest in the presence of E1S, and it is reduced in the presence of topotecan or tariquidar (17, 80). The presence of even 5 μM tariquidar is sufficient to reduce the ATPase activity to 40% of that measured with 50 μM topotecan. This is even lower than our previously observed ATPase rate reduction, where 0.5 μM tariquidar was able to reduce the ATPase rate to 60% of that observed with 50 μM topotecan (17). To ensure a high degree of tariquidar occupancy in ABCG2 nanodisc samples used for structure determination, we incubated them with a tariquidar concentration of 20 μM under turnover conditions (5 mM ATP, 0.5 mM ADP, and 5 mM MgCl2) before preparing cryo-EM grids. From a single turnover data set, we determined two high-resolution cryo-EM structures (Fig. 1 A and B) of tariquidar-bound ABCG2, each including a single copy of tariquidar bound between the TMDs and two ATP bound to the NBDs.
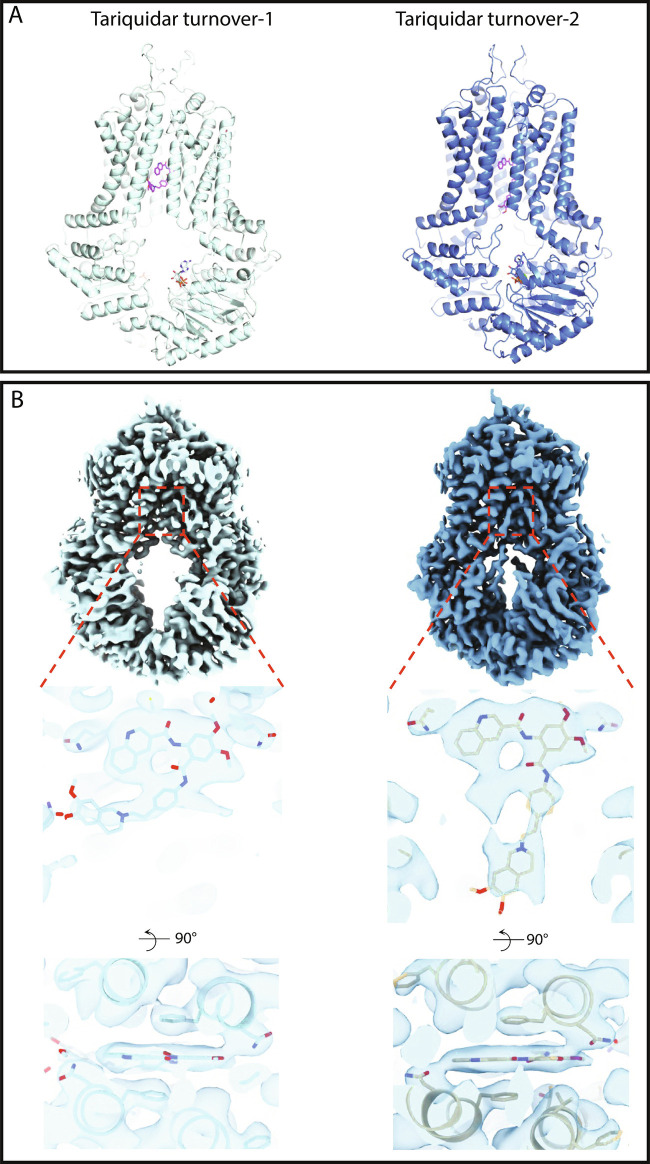
Fig. 1.
Cryo-EM structures of nanodisc-reconstituted, tariquidar-bound ABCG2. (A) Models of tariquidar-bound ABCG2 in TO1 (Left) and TO2 (Right) states shown in cartoon representation, determined at 3.1 and 3.2 Å, respectively. The bound tariquidar and ATP molecules are drawn as stick models. (B) Cryo-EM maps of tariquidar TO1 (Left) and TO2 (Right). Top, Overall cryo-EM map. Middle, Close-up view of tariquidar viewed from within the membrane. Bottom, Close-up view of tariquidar viewed from the cytoplasm.
Similar to the previously reported turnover structures of ABCG2 with topotecan (8), tariquidar-bound ABCG2 exhibits two distinct turnover states: tariquidar turnover-1 features a wider gap between the NBDs (Fig. 1) and constitutes the main class of the dataset, encompassing 74% of the well-ordered particles. The second structure featured semiclosed NBDs and was termed tariquidar turnover-2 (Fig. 1). To simplify the name of the 4 turnover structures, hereby we name them “drug” TO1/2. Tariquidar TO2 is a minor class, including 26% of the well-ordered particles. Both structures were refined in C1 symmetry but displayed evidence of averaging effects in the region of the bound ligand (Fig. 1) due to the pseudosymmetry.
Both structures revealed well-defined densities for two nucleotides bound in the nucleotide-binding sites. There is space only for a single tariquidar molecule in cavity 1 (Fig. 1 A and B). We interpreted the density in the nucleotide-binding site as ATP and Mg2+ in analogy to the previously determined structures of TO2 states bound with E1S or topotecan (8). The density covering bound tariquidar, along the two-fold symmetry axis of the ABCG2 homodimer, captures two poses related by 180° rotation, resulting from the averaging of particles that differ only in the pose of the bound ligand (SI Appendix, Fig. S5 A and B). As observed previously for turnover states of E1S and topotecan (8), the central cavity between the ABCG2 monomers is narrower in TO2. While bound tariquidar is sandwiched between the phenyl rings of the two opposing F439 residues from the ABCG2 monomers in both TO1 and TO2 structures, its conformation is notably different: in the TO1 state, tariquidar adopts a C-shaped conformation, whereas in TO2, it adopts an L-shaped form. The C-shaped conformation in TO1 is similar to that observed in a previously reported, inward-open and nonturnover structure of tariquidar-bound ABCG2 (SI Appendix, Fig. S6 and S7) (17). In TO1, tariquidar is best fitted with its 3-quinolinyl moiety stacking against the phenyl rings of the two F439 residues (SI Appendix, Fig. S7 A and B). The L-shaped conformation adopted by tariquidar in TO2 is probably a consequence of the reduced size of the substrate-binding cavity. The tariquidar density in the drug-binding pocket of TO2 extends along the two-fold symmetry axis of ABCG2 and is greatly affected by the pseudosymmetry of the protein, with the density of L-shaped tariquidar showing more symmetrized features than the C-shaped tariquidar in TO1. As a result, in TO2, only a single copy of tariquidar can be fitted inside the density in either of the two orientations related to each other by a rotation of ∼180° (SI Appendix, Fig. S5). Our structures demonstrate how the conformation of tariquidar might adapt as the transporter changes its shape.
Comparison of Tariquidar and Topotecan Turnover Structures of ABCG2.
The tariquidar TO1 and topotecan TO1 structures of ABCG2 are similar, with an overall RSMD of 0.56 Å. Similarly, the tariquidar TO2 state is nearly indistinguishable from topotecan TO2, with an overall RSMD of only 0.35 Å. Due to improved local resolution, we also observe the density for the bound Mg2+ in the tariquidar TO2 structure, which was not observed before in the topotecan TO2 structure (SI Appendix, Fig. S8).
Within the substrate-binding pocket, however, there are differences between the structures. The EM density for topotecan under turnover conditions is less well defined compared to that of tariquidar, suggesting more flexibility for the bound topotecan (SI Appendix, Fig. S5 A and B). In contrast, the EM density features for bound tariquidar are better defined both in TO1 and TO2 structures. This may reflect the higher binding affinity to ABCG2 or result from the larger size and mass of tariquidar, limiting its flexibility in the binding pocket.
Intriguingly, both topotecan and tariquidar exhibit orientational changes between TO1 and TO2 structures. While topotecan shows only a relative rotation between the two structures, tariquidar also changes its conformation from a C-shaped conformer in the TO1 structure to an L-shaped conformer in the TO2 one. The observed conformational change appears necessary for the molecule to fit the narrowed substrate-binding cavity because a simply rotated form of C-shaped tariquidar would not fit into the binding cavity of the TO2 structure. The distal end of L-shaped tariquidar reaches as far as the cytoplasmic entrance of the substrate-binding cavity. This feature likely explains why tariquidar is slowly transported by ABCG2 since in order to fully close the cytoplasmic side of the binding cavity, tariquidar would have to move further along the translocation pathway. Notably, tariquidar is the largest compound reported to date as a substrate for ABCG2.
Size-Dependent Differential Binding of Ligands to ABCG2.
The dynamics of tariquidar and topotecan in the binding pocket of the TO1 and TO2 states were investigated by μs-scale all-atom molecular dynamics (MD) simulations of membrane-embedded ABCG2 in the presence of either substrate. The stability of the ligand in its binding pose was quantified by RMSD calculations during the course of 2-μs simulations (duplicate in each case). RMSD of tariquidar in TO1 and TO2 in both simulated replicates remains, on average, at ∼2 Å, indicating a stable binding (Fig. 2A and SI Appendix, Fig. S9A and Movies S1 and S2), although transient fluctuations are observed in TO1, during which the substrate-binding pose becomes more similar to that in TO2. Transition of tariquidar binding from a TO1 pose to the TO2 one is aligned with the proposed transition cycle of ABCG2 in which TO1 precedes TO2 (8). In contrast to tariquidar, topotecan fluctuates wildly in both TO1 and TO2 states, with RMSD values ranging 2 to 12 Å (Fig. 2B and SI Appendix, Fig. S9B and Movie S3 and S4). Moreover, the overall stability/symmetry of different domains in ABCG2 show relatively stable conformations (SI Appendix, Figs. S10–S13). Volume calculation reveals that free space in the binding pocket for topotecan-bound TO1 and TO2 states are 20 and 46% larger compared to the tariquidar-bound structures, respectively (Fig. 2 C and D and SI Appendix, Table S3). The additional free volume in the topotecan-bound structures allows for the observed rocking motion of the substrate and sampling different binding poses within the binding pocket (Fig. 2 C and D and SI Appendix, Table S3).
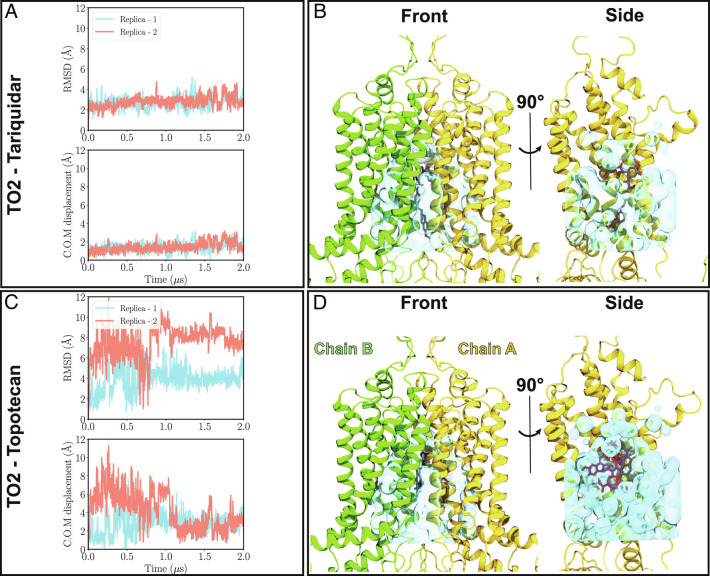
Fig. 2.
Binding stability of tariquidar and topotecan to ABCG2. (A) and (B) Top: Heavy-atom RMSD values for tariquidar and topotecan with respect to their initial positions in the cryo-EM model of the TO2 structure are shown for both simulation replicas (pink and blue traces). While tariquidar is stable in the binding pocket throughout the simulation (RMSD ≈2 Å), topotecan fluctuates extensively, with RMSD values ranging 2 to 12 Å. Bottom, Minimal center of mass distance of tariquidar or topotecan with respect to their two symmetry-related binding poses in the cryo-EM model. (C) and (D) show front and side protein views highlighting the empty volume (shown in cyan) within the binding cavity of ABCG2 for tariquidar and topotecan cryo-EM structures, respectively. Cryo-EM-modeled ligand poses are shown in gray. In the side views, one of the protein chains (chain B) is removed for an unobstructed view of the internal cavity. The larger size of tariquidar leaves less room for fluctuation and therefore a more stable binding mode. In contrast, topotecan’s smaller size allows for more freedom and larger fluctuations of the ligand. Representative topotecan poses arising during the simulations are shown in red in the side view.
ABCG2 can accommodate either one or two small-molecule ligands in its substrate binding pocket (8, 14, 15, 17). While the stoichiometry of binding for inhibitors varies depending on the ligand’s size, all substrates bind as a single copy based on the available structures of ABCG2 (8, 14, 15, 17). While substrate binding stoichiometry seems to be size-independent, our simulation results suggest that substrate stability can be largely affected by its size, which in turn determines the available sampling space within the binding pocket. As observed in the simulations, in the case of topotecan and tariquidar, the larger substrate (tariquidar) is more confined in the binding pocket due to its size and thus experiences less positional and conformational flexibility. The observed fluidity of topotecan binding might substantiate the relatively smaller overlap between the cryo-EM electron density and the modeled ligand structure (8) (SI Appendix, Fig. S5 A and B). On the other hand, the higher degree of overlap between tariquidar’s density and its structure in the cryo-EM model (SI Appendix, Fig. S5 A and B) is in line with its observed stable binding in both turnover states and in all simulation replicas (Fig. 3).
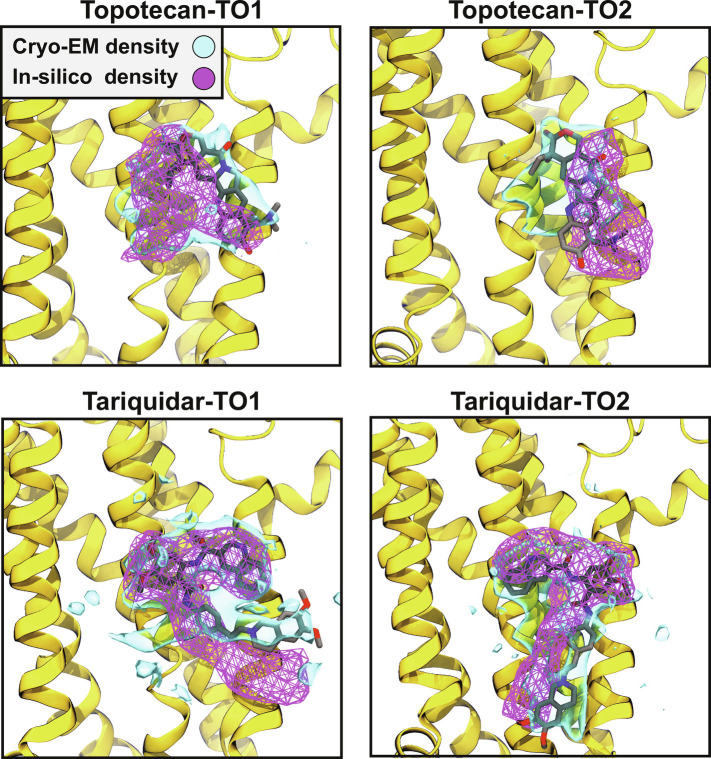
Fig. 3.
Comparison of in silico and cryo-EM obtained densities of the ligands. Cryo-EM and MD obtained densities (accumulated for both replicas of each system during 2 μs of production runs) of the ligands are shown for both topotecan and tariquidar in TO1 and TO2 conformations, demonstrating a good degree of overlap. The cryo-EM densities are shown at isovalue 0.2, whereas the in-silico densities are represented at an isovalue of 0.5. Only one of the monomers is shown to render a more visible binding pocket. The snapshots are viewed from the side. The cryo-EM and in silico densities are represented in solid blue surfaces and magenta meshes, respectively. The cryo-EM modeled ligand in each case is also represented.
Lipid Penetration into the Ligand-Binding Cavity in Topotecan-Bound Structure.
As described, the primary reason for topotecan’s dynamic binding observed in our simulations and the imperfect match between its cryo-EM model and density appears to be its relatively smaller size compared to ABCG2’s substrate binding pocket. However, the simulations hinted at an additional factor that might impact topotecan binding. Inspecting the topotecan-bound simulations, we observed multiple events of phospholipid penetration into ABCG2’s binding pocket through the TMD gate in both TO1 and TO2 states. It is worth noting that the lipid penetration into the lumen is accompanied by an increase in the distance between the two TMDs in topotecan-bound TO2, as the domains move to accommodate the phospholipids (SI Appendix, Fig. S11). Individual lipids penetrating the binding pocket were then classified, and their proximity to the bound ligand was quantified (lipid phosphorus distance to topotecan’s titratable nitrogen; SI Appendix, Fig. S3).
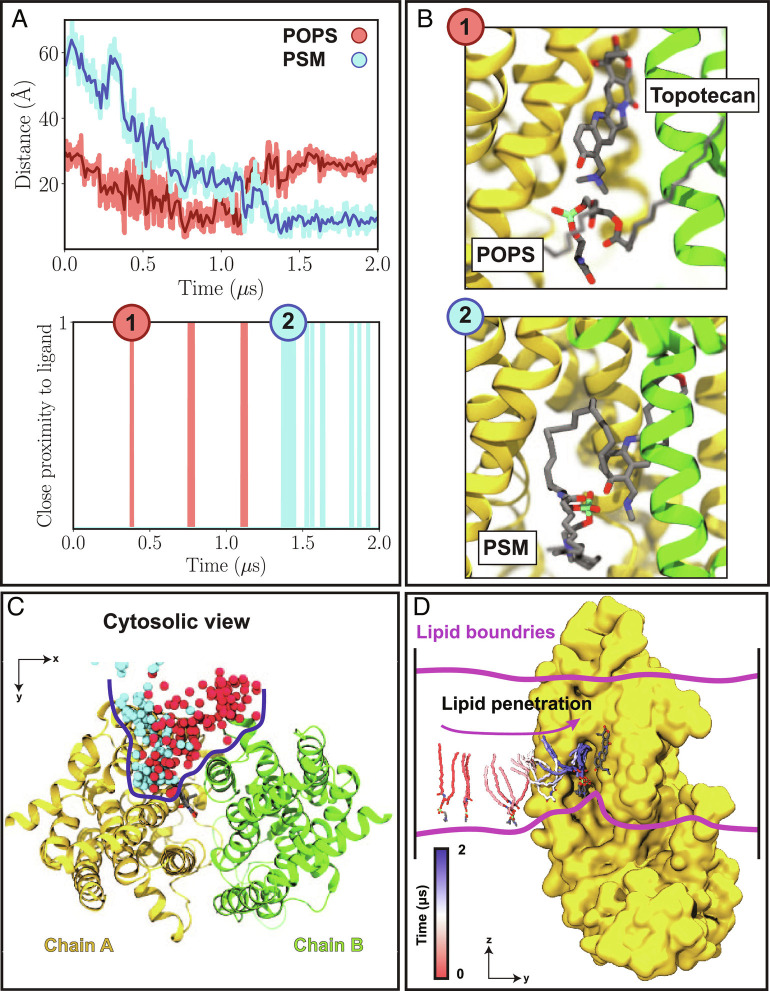
As an example, in the first simulation replica of topotecan-bound TO2, we captured two different lipids, a POPS and later a PSM, that penetrated the binding pocket at different times to the degree that they established direct contacts with the bound topotecan. In the first lipid-penetrating event, a POPS lipid enters the binding pocket and remains in that area during the t≈ 0.4–1.1 μs segment of the simulation (pink bars in Fig. 4A). Departure of this lipid from the TMD cavity allowed for a second lipid (a PSM in this case) to gain access to the binding pocket and to establish direct contacts with the substrate at multiple instances during the remainder of the simulation time (blue bars in Fig. 4A). Therefore, we demonstrate direct interactions of lipids with ligands in a membrane protein. Two representative snapshots of ABCG2-penetrating phospholipids shown in Fig. 4B exemplify the direct nature of interactions between the lipids and the ligand. Tracking the phosphorus atoms of these two lipids during the simulation shows that both lipids originated from the cytoplasmic leaflet of the bilayer (Fig. 4 C and D). The phospholipid entry into the binding cavity starts with the lipid headgroup followed by one of the acyl chains (Fig. 4D). A similar behavior was observed for the second TO2 simulation replica as well as for both topotecan-bound TO1 simulation replicas (SI Appendix, Figs. S14–S16 A–D). Overall, during the course of each simulation (2 μs), 2 to 4 lipids were observed to visit the binding pocket. These lumen-penetrating lipids included POPE, POPC, or even POPI, suggesting that topotecan-phospholipid interaction is not limited to a specific lipid type. Furthermore, the binding pocket seems to only accommodate only a single phospholipid at any given time.
Fig. 4.
Direct lipid–ligand interaction in ABCG2. (A) Top, Distance time series between the phosphorus atom of lipids that penetrate the binding pocket and the titratable nitrogen atom of the bound topotecan. The distances for the first (POPS) and second (PSM) visiting lipids are shown in red and cyan, respectively. Bottom, Instances of lipid–topotecan proximity (P-N distances < 5.0 Å) during the simulations. (B) Two representative snapshots (1 to 2, corresponding to time stamps in (A)) highlighting close lipid–topotecan interactions (1: POPS; 2: PSM). The two pseudosymmetrically related halves of ABCG2 (chains A and B) are shown in yellow and green, respectively. (C) Cytosolic view of lipid penetration into the binding pocket of ABCG2 observed during the 2 μs trajectory, with the phosphorus atoms of the first and second visiting lipids in red and cyan, respectively. Lipid penetration range into the protein is highlighted with a blue line, and the ligand is drawn in licorice to highlight its proximity to lipids. (D) Snapshots describing the trajectory of the PSM lipid that penetrated into the binding pocket shown from the side. Lipid tails are colored according to the time in the trajectory, with red and blue representing the beginning and end of the simulation, respectively. Chain B is removed for a better view of the luminal binding pocket.
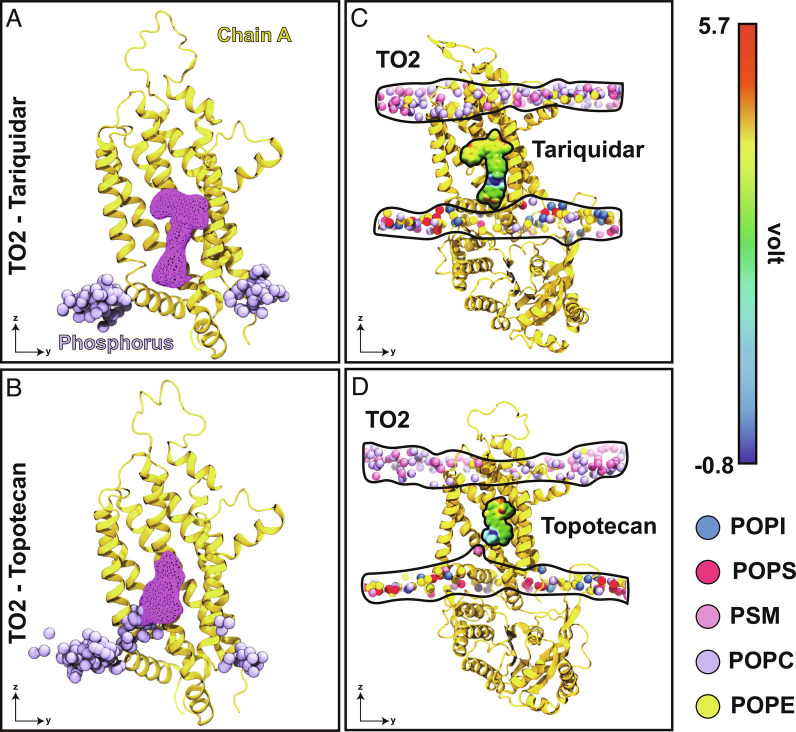
In contrast to topotecan, phospholipid penetration into the substrate binding pocket was not observed in tariquidar-bound simulations (Fig. 5 A and B and SI Appendix, Fig. S9 A and B). Given its larger size, tariquidar is observed to occupy the binding pocket more fully, not only achieving a more stable binding but also leaving less space for penetration of other molecular species such as lipids (Fig. 5 A and C). Furthermore, in contrast to topotecan (Fig. 5D), the positive moiety (amino group) of tariquidar is rather buried in the molecule and unavailable for direct charge–charge interactions with the lipids (Fig. 5C and SI Appendix, Fig. S9C), which likely contributes to the interaction between them. Consistent observations were made in all simulation replicas and for both TO1 and TO2 states, that is, a more stable, less fluctuating pose for tariquidar and direct lipid-ligand interaction for topotecan (SI Appendix, Fig. S9 A–D). Direct, dynamic topotecan-lipid interactions captured in our simulations may further contribute to the relatively large fluctuation of topotecan.
Fig. 5.
Ligand fluctuation and differential lipid penetration into the protein for topotecan- and tariquidar-bound structures. (A) Superimposed snapshots of the phosphorus atoms (purple) that come closer than 20 Å to tariquidar (collected from both TO2 simulated replicas with 125 ns intervals). Spatial density representing the ligand occupancy obtained from the aggregate of two simulation replicas is shown as a purple mesh. (B) Similar data from topotecan-bound simulations. Topotecan shows much larger fluctuations and is observed to directly interact with lipids. (C) and (D) Charge–charge interactions between phospholipids and substrates in the TO2 state for tariquidar and topotecan, respectively. The electrostatic potential map for each ligand highlights the positive center of each molecule (blue). Topotecan’s positive charge is much more accessible for direct interactions/contacts with lipids, whereas tariquidar’s positive charge is more buried and less accessible. Phosphorus atoms of different lipids are shown as spheres and colored according to the legend. To provide a clear view into the binding cavity, only chain A is shown (yellow).
The interconnection between lipids and the drug has been implicated in transport properties of other ABC transporters. In two studies employing fluorescence microscopy, it was shown that lipid A transport via MsbA can be affected by the presence of different drugs (82, 83). In the case of ABCG2, biochemical and optical microscopy studies have shown that the lipid environment can significantly affect its transport activity (23, 24, 26). However, to the best of our knowledge, there has been no study reporting direct lipid-substrate interactions in ABC transporters.
Additional simulations performed on ABCG2 in the IF-apo conformation showed that phospholipids can also enter its empty binding cavity (SI Appendix, Fig. S17 A–D and Movie S5). This is in line with previous MD simulations that have shown phospholipid entrance into the empty binding site of another ABC transporter, Pgp (34, 36, 39, 40). Furthermore, experimental studies have established the capability of ABCG2 and Pgp in directly recruiting their substrates directly from their surrounding lipid bilayer (84–90). We propose that the pathway for the lipid entrance into the ABCG2 cavity captured in our simulations presents a putative general pathway for any molecular species in the membrane to partition into the transporter protein.
Cholesterol Blocking Phospholipids from Penetrating ABCG2.
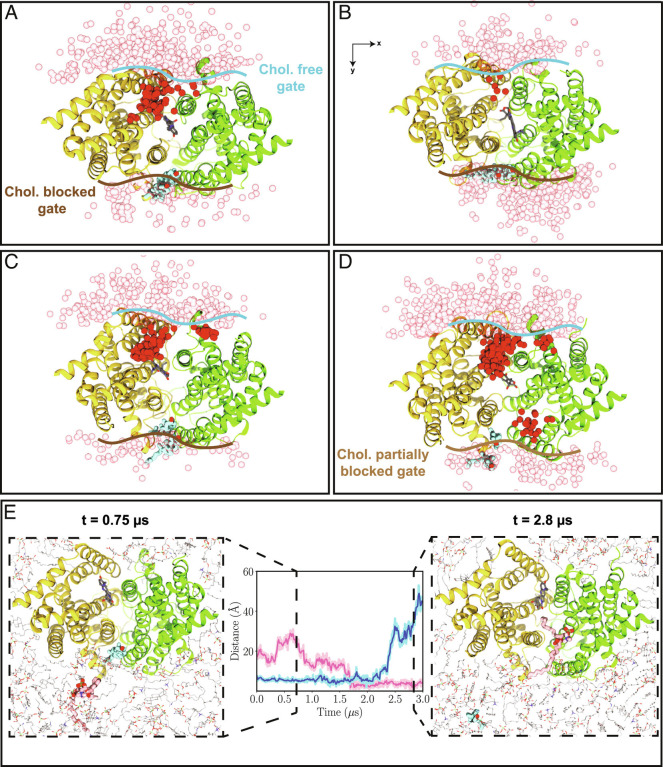
Given the homodimeric structure of ABCG2, two symmetrically related portals within the transmembrane region of ABCG2 separate its ligand-binding cavity from the surrounding membrane. During our simulations, however, we observed a curious asymmetry in the engagement of these two openings in phospholipid penetration into the ABCG2 lumen (Fig. 6). While one of the portals was observed to be heavily engaged in exchange of lipids between the membrane and the lumen, a substantially lower level of lipid traffic was observed for the other portal. Upon further examination, we discovered that the inactive portal was physically blocked by a pair of cholesterol molecules (Fig. 6) in all the simulation systems, which due to their bulky structure, when compared to phospholipid tails, could not further penetrate the luminal cavity. As shown in Fig. 6, in all the cases, the presence of cholesterol in the portal area diminished the penetration of phospholipids into the ABCG2 lumen. Consistent with such a phospholipid-blocking role for cholesterol, its departure from the portal region, observed after extending one of the simulations to 3 μs, resulted in an immediate phospholipid occupancy of the lumen through that portal (Fig. 6E). While at this point we can only speculate on the functional ramifications of this behavior of cholesterol, we propose that due to preventing phospholipids from penetrating and wedging the protein, cholesterol may keep the protein mechanism free of interference and thereby accelerate the transition between its functional states as needed for active transport.
Fig. 6.
Differential lipid penetration into the binding pocket through cholesterol-free and cholesterol-blocked portals. (A–D) Phospholipid penetration into the lumen of ABCG2 from the cholesterol-free portal and its blockade in the cholesterol-blocked portal in TO1 (A and B) and TO2 (C and D) replicas. The boundaries of cholesterol-free, -blocked, and -partially blocked gates are shown with blue, dark-brown, and light-brown lines, respectively. Snapshots of the phosphorus atoms within 20 Å of the ligand are shown from the cytoplasmic view in red for phospholipids inside the lumen, and in pink for those outside the lumen. Ligand is shown in gray, whereas cholesterol molecules occupying the gate (gate-keepers) are represented in cyan. Protein monomers are illustrated in yellow and green. (E) Opening of the portal and phospholipid penetration into the lumen upon departure of the gate-keeper cholesterol for the system shown in (D). Center of mass distances of the gate-keeper cholesterol (cyan) and the penetrating POPC (pink) with respect to the center of mass of the gating helices is shown in the center panel. Two snapshots corresponding to the departure of the gate-keeper cholesterol are shown in the left (t = 0.75 μs) and right (t = 2.8 μs) panels. All the snapshots are shown from the cytosolic view.
Concluding Remarks
ABCG2 is a human ABC transporter contributing to MDR in cancer cells while participating in the general chemoimmunity defense system. Understating the mechanism of transport in ABCG2 and various factors regulating and/or impacting the process has been the main goal of numerous molecular studies over the past two decades. In this study, we employed cryo-EM to resolve the structure of ABCG2 under turnover conditions bound to tariquidar, a special modulator, and a slow substrate of ABCG2. Our results reveal that, similar to topotecan, tariquidar induces two main conformational states in ABCG2 under turnover conditions, namely TO1 and TO2. To further study the dynamics of ABCG2 and its bound ligands, we utilized MD simulations to investigate and compare the topotecan- and tariquidar-bound ABCG2 in the TO1 and TO2 states. In a sharp contrast to tariquidar binding, our simulations demonstrate a high degree of flexibility in topotecan’s binding mode in both TO1 and TO2 states. Topotecan’s dynamic binding to ABCG2 captured in the simulations substantiates the less well-defined density of topotecan compared to that of its interacting residues in the cryo-EM maps. Tariquidar, on the other hand, being larger than topotecan, occupies the binding pocket more fully and, by establishing more interactions, remains more stably bound throughout the simulations. This behavior is also reflected in the well-defined densities of tariquidar captured in our cryo-EM maps.
Besides the fact that the size and the level of drug-protein interactions can directly control the binding mode of different molecules to ABCG2, our MD simulations reveal active participation of phospholipids in this process. In our topotecan-bound simulations, we capture different types of phospholipids entering ABCG2’s binding pocket and directly interacting with topotecan in both TO1 and TO2 states. This direct ligand–lipid interaction further modulates topotecan’s binding mode. In contrast, in the case of tariquidar, phospholipids do not enter the binding pocket as the drug fills the cavity more fully and leaves less space for the penetration of lipids. Additional simulations show that phospholipids can also enter ABCG2’s binding pocket in the apo-IF conformation. As many compounds interacting with ABCG2 contain relatively large hydrophobic moieties, it is not hard to imagine that they first partition into the membrane and from there enter the binding cavity of the transporter. Hence, the mechanism and pathway of lipids penetrating into ABCG2’s binding cavity captured in our simulations provide a putative general pathway for small-molecule compounds to enter ABCG2 in the IF state before being exported to the other side of the membrane.
While ABCG2 does not appear to impose any selectivity for or against a particular type of phospholipids to enter its binding cavity, no penetration of cholesterol molecules into the lumen was observed in any of our simulated systems. Upon reaching the entry portal, cholesterol molecules appear to even block the portal and prohibit phospholipids from entering the lumen. This aspect can be of two functional ramifications. First, while ABCG2 is localized in cholesterol-rich regions of the plasma membrane, unlike other ABCG transporters, e.g., ABCG1 and ABCG5/G8, cholesterol is not a substrate for ABCG2. One can speculate that the entrance of cholesterol into ABCG2’s lumen might have an inhibitory effect on the transporter. The observed discrimination against cholesterol therefore might be physiologically beneficial for ABCG2’s transport by keeping the substrate binding pocket free from cholesterol, which is omnipresent in the environment. On the other hand, the transport function of ABCG2 depends on the presence of cholesterol. Our simulations not only show exclusion of cholesterol but also provide a hint at how cholesterol may potentiate ABCG2’s transport by preventing phospholipids from entering the protein’s lumen and interfering with the conformational changes required for transport.
Materials and Methods
Expression and Purification of ABCG2.
Full-length FLAG-ABCG2 from Homo sapiens was expressed in HEK293-EBNA (Thermo Fisher Scientific) cells by transient transfection (13). Cells were cultured at 37 °C under 8% CO2 and harvested 48 to 60 h after transfection. The collected cells were resuspended in buffer A (40 mM HEPES, pH 7.5, and 150 mM NaCl) with 10% glycerol, 1 mM PMSF (phenylmethylsulfonyl fluoride), 2 μg/mL DNaseI (Roche), and protease inhibitor cocktail (Sigma) and lysed using a Dounce homogenizer. Cell lysates were further solubilized with 1% DDM and 0.1% (w/v) CHS (cholesteryl hemisuccinate, Anatrace). After solubilization, cell lysates were spun at 100,000 g, and the supernatant was incubated with anti-Flag M2 affinity agarose gel (Sigma). Proteins were eluted by flag peptide and concentrated for gel-filtration purification with the Superdex 200 Increase 10/300 column (GE Healthcare) in buffer A with 0.026% DDM and 0.0026% CHS (w/v). Fresh peaks were collected for further experiments.
Reconstitution of ABCG2-Nanodisc.
Membrane scaffold protein (MSP) 1D1 was expressed in E. coli strain BL21(DE3) and purified as previously described in ref. (42). A mixture of brain polar lipid (Avanti Polar Lipids) and CHS at a 4:1 ratio (w/w) was diluted to 10 mM with a buffer containing 20 mM HEPEs and 150 mM NaCl, pH 7.5. Lipids were solubilized with a 3x molar excess of sodium cholate using a sonic bath. Solubilized lipids were further incubated with detergent-purified ABCG2 and MSP1D1 at a molar ratio of 100:5:0.2 (lipid:MSP:ABCG2). Detergents were then removed by the addition of activated Bio-Beads (Bio-Rad) at 4 °C overnight. Bio-Beads were then removed by Econo-Pac Columns (Bio-Rad), and the flow-through was spun at 100,000g for 30 min (13). The supernatant was collected and concentrated before loading on a Superdex 200 Increase column equilibrated with buffer A. The peak fraction of ABCG2 nanodisc was collected for cryo-EM sample preparation, and purity was checked by SDS-PAGE.
ATPase Assay.
The ATPase assay was done at 37 °C in the presence of 2 mM ATP and 10 mM MgCl2. When indicated, 50 μM E1S, 50 μM topotecan, or 5 μM tariquidar was added. The concentration of inorganic phosphate released by the hydrolysis of ATP was measured by tracking absorbance at 850 nm following the classical molybdate method (43). The ATPase rate was determined using linear regression in GraphPad Prism v8 and v9.
Preparation of cryo-EM Samples.
Quantifoil carbon grids (300 meshes, R 1.2/1.3 copper) were glow discharged immediately before use. Nanodisc-reconstituted ABCG2 at 1 mg/mL was incubated with 5 mM ATP, 5 mM MgCl2, 0.5 mM ADP, and 20 μM tariquidar at room temperature for 10 min. Then, 3.5 μL tariquidar turnover sample was applied on grids using Vitrobot IV (FEI), with the environmental chamber set at 100% humidity and 4 °C. Grids were blotted for 2.5 s with blot force 1 and flash frozen in a mixture of liquid ethane and propane.
Cryo-EM Data Acquisition.
The Cryo-EM dataset was collected with a 300-keV Krios G3 Cryo-transmission electron microscope (Thermo Fisher Scientific) equipped with a Gatan BioQuantum 1967 filter and a Gatan K3 camera. Images were recorded with 3 exposures per hole using EPU 2 in the superresolution mode with a 20-eV slit width of the energy filter and at a nominal magnification of 130,000 x, resulting in a calibrated superresolution pixel size of 0.33 Å (physical pixel size 0.66 Å). Defocus was set to vary from −0.6 to −2.2 μm. The image was dose-fractionated to 40 frames with a total dose of 50 e−/Å2. The total exposure time was 1.49 s, and 21,211 movies were collected. Superresolution movies were down-sampled twice and motion-corrected by Fourier cropping, drift-correction, and dose-weighting with MotionCor2 (44). Cryo-EM data collection statistics in this study are presented in SI Appendix, Table S1.
Cryo-EM Image Processing.
The cryo-EM image processing pipeline is shown in SI Appendix, Fig. S1I. The drift-corrected, dose-weighted 2D micrographs were imported in cryoSPARC v2.15 (45). Gctf was used to measure the contrast transfer function (CTF) parameters (46). A total of 20,741 2D micrographs were further selected by manual inspection. To generate 2D templates for Auto-pick, 394 particles were manually picked from six selected micrographs. Based on the template generated from manual picking, 2,939,835 particles were picked and extracted from 20,741 micrographs. After 5 rounds of 2D classifications, 1,513,184 particles belonging to “good” 2D classes were selected. Topotecan TO1 and TO2 structures were used as initial reference models for heterogeneous refinement. Selected good particles after 2D classifications were down-sampled 3 times and subjected to 14 rounds of heterogeneous refinement. In each round, 3D classes with clear secondary structural features were selected and used for further data processing (SI Appendix, Fig. S1I). A total of 426,371 particles from a good 3D class were selected and used for homogeneous refinement, global CTF refinement, local CTF refinement, and nonuniform refinement. The resulting map was in the TO1 state at 3 Å overall resolution. To separate different conformational states of ABCG2, the particles underwent further heterogeneous refinement without binning. A well-defined 3D class with 211,906 particles in the TO1 state was selected and used for further homogeneous refinement and nonuniform refinement. The nonuniform refinement led to a final map at 3.1 Å named tariquidar TO1 state with an automatically determined B factor of −117 Å2. Another 3D class with 208,541 particles was in the TO2 state. To further purify this 3D class, one more round of heterogeneous refinement was performed without binning particles. After the final heterogeneous refinement for TO2 state class, a 3D class in TO1 state with 85,013 and a 3D class without clear features were discarded (SI Appendix, Fig. S1I). The 3D class in the TO2 state with 122,001 particles was selected. Further, global CTF refinement, local CTF refinement, and homogeneous refinement of the 3D class resulted in a 3.2-Å final map named tariquidar TO2, with an automatically determined B factor of −114.9 Å2. Both final maps were refined in C1 symmetry and were directly used for structure modeling. Local resolution maps were generated using cryoSPARC v2.15 (45).
Cryo-EM Model Building and Refinement.
Model building was performed in Coot 0.9 (47). ABCG2 topotecan TO1 (PDB ID: 7OJH) and topotecan TO2 (PDB ID: 7OJI) were docked into two tariquidar turnover maps and used as the reference for manual rebuilding, respectively (8). The coordinate of tariquidar was from the ABCG2-tariquidar-Fabs structure (PDB ID: 7NEQ) (17). The restraint of tariquidar was generated in eLBOW of Phenix (48). Tariquidar turnover structures were refined against their final maps in the real-space refinement of Phenix (49). Reciprocal-space refinement of the B factors and minimization global refinements were applied along with the Ramachandran plot, standard geometry, rotamer, Cα, Cβ, noncrystallographic symmetry (NCS), and secondary structure restraints in the final refinement. The quality of the final model was assessed by MolProbity (50). Refinement statistics are provided in SI Appendix, Table S1.
Molecular Dynamics Simulations.
Simulation setup.
The following structures were used to prepare the simulation systems: 1) topotecan-bound ABCG2 TO1 and TO2 states (PDB IDs: 7OJH and 7OJI (8)), 2) tariquidar-bound ABCG2 TO1 and TO2 states (resolved in this study), and, 3) IF-apo ABCG2 (PDB ID: 6ETI (14), after removing the bound inhibitors and antibody fragments). These ABCG2 structures were subjected to the following steps. Unresolved side chains and hydrogen atoms were added to the structures employing the https://www.ks.uiuc.edu/Research/vmd/plugins/psfgen/ plugin of VMD (51). The experimental structural models include two identical monomers, each missing disordered regions that were modeled with Modeller (52), with the exception of residues 302 to 327 in topotecan-bound TO1, 302 to 325 in topotecan-bound TO2, 302 to 326 in tariquidar-bound TO1, and 305 to 325 in tariquidar-bound TO2 (SI Appendix, Fig. S2A). In these cases, neutral N-terminal and C-terminal caps were added with PSFGEN to the beginning and end of each polypeptide segment, respectively. The coordinates of the ligand in ligand-bound structures were obtained from the cryo-EM modeled poses in the binding pocket (SI Appendix, Fig. S2A). The protonation states of titratable residues were estimated with PROPKA (53, 54). The protein models were then internally hydrated with DOWSER (55, 56).
The initial lipid bilayer used to embed the proteins was constructed with CHARMM-GUI (57, 58). The orientation of the protein in the bilayer was obtained from the OPM (Orientations of Proteins in Membranes) database (59). The structures were then inserted into a heterogeneous lipid bilayer, followed by removing steric clashes with lipid molecules. The bilayer was composed of palmitoyl-oleoyl-phosphatidyl-choline (POPC), palmitoyl-oleoyl-phosphatidyl-ethanolamine (POPE), palmitoyl-oleoyl-phosphatidyl-serine (POPS), palmitoyl-oleoyl-phosphatidyl-inositol (POPI), palmitoyl-oleoyl-phosphatidic-acid (POPA), palmitoyl-sphingomyelin (PSM), and cholesterol (CHL) at molar ratios of 39:6:0:0:0:21:34 and 17:25:11:8:1:9:29 in the extracellular and cytoplasmic leaflets, respectively (SI Appendix, Fig. S2B). The protein–membrane complex was then solvated with a 150 mM NaCl solution in VMD (system size: ∼359,000 atoms). To avoid any bias from the initial lipid configuration and to improve sampling, for each structure, two independent lipid bilayers (with randomly placed initial lipids) were generated using the Membrane Mixer plugin (MMP) in VMD (60).
Simulation conditions.
All systems were simulated following this protocol: 1) 10,000 steps of minimization, followed by 5 ns of equilibration, during which positional harmonic restraints (k = 10 kcal mol−1 Å−2) were applied to the protein’s heavy atoms of the cryo-EM model as well as the heavy atoms of the ligand (for ligand-bound systems) in the binding pocket. Furthermore, the z position (normal to the membrane plane) of the phosphorus atoms of phospholipids and oxygen atoms of cholesterols were harmonically restrained in this step (k = 5 kcal mol−1 Å−2); 2) 10 ns of equilibration while only the protein backbone and the heavy atoms of the ligand (for ligand-bound systems) were restrained (k = 10 kcal mol−1 Å−2); 3) 40 ns of equilibration with restraints applied only to the protein backbone heavy atoms (k = 10 kcal mol−1 Å−2); 4) 5 ns of equilibration during which all the restraints were gradually removed by stepwise-decreasing the force constant from 10 kcal mol−1 Å−2 to 0; 5) 100 ns unrestrained equilibrium simulations; 6) 1 and 2 μs of production runs for IF-apo and ligand-bound systems, respectively, without any restraints (SI Appendix, Table S2). Steps 1 to 5 were for all systems were performed in NAMD2 (61, 62). The production runs in the case of the IF-apo systems were conducted with NAMD2 and the aforementioned parameters. Production runs for the ligand-bound systems were performed on Anton 2 (63).
All simulations were performed employing the fully atomistic CHARMM36m (64) and CHARMM36 (65) force fields for the protein and lipids, respectively. The TIP3P model was employed for water (66). Topotecan and tariquidar were parameterized employing the https://cgenff.paramchem.org (67, 68), followed by further optimization of the parameters with the Force Field Toolkit (ffTK) plugin (69) of VMD.
In NAMD simulations, a 12 Å cutoff was employed for short-range, nonbonded interactions, with switching starting at 10 Å. Long-range electrostatic interactions were calculated using the particle mesh Ewald (PME) algorithm (70) with a grid density of 1 Å−1 and a PME interpolation order of 6. Bonds involving hydrogen atoms were constrained with the SHAKE algorithm (71). Temperature was maintained at 310 K using a Langevin thermostat with a damping coefficient of 1.0 ps−1. Pressure was maintained at 1 atm by the Nosé–Hoover Langevin piston barostat with period and decay of 100 and 50 fs, respectively (72, 73). All systems were simulated in a flexible cell allowing the dimensions of the periodic cell to change independently while keeping the aspect ratio in the xy plane (membrane plane) constant. The simulation timestep was set to 2 fs, and the Lennard–Jones and PME forces were updated at one and two timesteps, respectively.
For Anton simulations, 1 bar pressure and 310 K temperature were maintained by the Martyna–Tuckerman–Klein Nosé–Hoover chain coupling scheme (74), as implemented using a multigrator scheme (75). All the bonds to hydrogen atoms were constrained using the M-SHAKE algorithm (76), and a 2.5-fs timestep was employed. The long-range electrostatic interactions were calculated using the fast Fourier transform (FFT) method on Anton 2 (63).
Analysis.
Phospholipids entering the ABCG2 lumen during the simulations were primarily identified by visually monitoring the position of their phosphorus atoms. Individual lipid-penetration events were further quantified for some of these lipids. For the case of ligand-bound structures, the distance between the phosphorus atom of such lumen-penetrating lipids and the titratable nitrogen atom of the bound ligand (SI Appendix, Fig. S3) was used to classify the lipids. Lipids with a (P–N) distance less than 5 Å were considered ligand-proximal and used for further analysis.
A different set of geometrical parameters were used to analyze the effect of cholesterol on lipid penetration into the lumen. First, all lipid phosphorus atoms within a radius of 20 Å of the bound topotecan were selected to indicate lipids that are physically close to the binding cavity. Then, boundaries of the lumen were defined by the center of mass of the Cα atoms of the first and fifth transmembrane helices (residues 391 to 413 and 534 to 552, respectively) from each protomer. Any phospholipids entering the region within the boundaries was identified as a lumen-penetrating lipid.
To assess the stability of the ligands, RMSD of topotecan and tariquidar in each simulation system was calculated after the transmembrane helices of the protein were aligned to the cryo-EM model. Electrostatic potential maps of the ligands were calculated with Gaussian (77) and visualized with GaussView (78). Molecular images and general analysis were done with VMD (51).
The volume of the binding cavity was calculated using POVME 2.0 (79). The inclusion region was chosen to be bound to the transmembrane domain and extended from V450 to the leucine gate (L554 and L555) marking the cytoplasmic and extracellular boundaries of the binding site, respectively. Grid spacing was set to 0.5 Å, with a distance cutoff of 1.09 Å.
Supplementary Material
Appendix 01 (PDF)
MD trajectory of the tariquidar-bound TO2 state during a 2-μs simulation. ABCG2 is shown in cartoon representation with monomers colored in green and yellow, respectively, and the bound ligands are shown with sticks.
MD trajectory of the tariquidar-bound TO1 state during a 2-μs simulation. ABCG2 is shown in cartoon representation with monomers colored in green and yellow, respectively, and the bound ligands are shown with sticks.
MD trajectory of the topotecan-bound TO2 state during a 2-μs simulation. ABCG2 is shown in cartoon representation with monomers colored in green and yellow, respectively, and the bound ligands are shown with sticks. Two phospholipids, a POPS (red) and a PSM (cyan), that penetrate the binding pocket are shown.
MD trajectory of the topotecan-bound TO1 state during a 2-μs simulation. ABCG2 is shown in cartoon representation with monomers colored in green and yellow, respectively, and the bound ligands are shown with sticks. Two POPE lipids that penetrate the binding pocket during the simulation are shown in red and cyan, respectively.
MD trajectory of ABCG2 in IF-Apo conformation during a 1-μs simulation. ABCG2 is shown in cartoon representation with monomers colored in green and yellow, respectively. Two phospholipids that penetrate the binding pocket during the simulation, a POPS (red) and POPE (cyan), are shown.
Acknowledgments
This study was supported by the NIH through the grants P41-GM104601 and R01-GM123455, and by the Swiss NSF through the National Centre of Competence in Research (NCCR) TransCure, the Scientific Center for Optical and Electron Microscopy (ScopeM) at ETH Zürich for data collection support. Simulations were performed using allocations on Anton at Pittsburgh Supercomputing Center (award MCB100017P), NSF Supercomputing Centers (XSEDE grant number MCA06N060), and the Blue Waters Petascale Computing Facility of National Center for Supercomputing Applications at University of Illinois at Urbana-Champaign, which is supported by the NSF (awards OCI-0725070 and ACI-1238993) and the state of Illinois.
Author contributions
A.R., Q.Y., S.D.-G., P.-C.W., J.K., K.P.L., and E.T. designed research; A.R., Q.Y., S.D.-G., and J.K. performed research; A.R., Q.Y., and S.D.-G. analyzed data; and A.R., Q.Y., S.D.-G., P.-C.W., K.P.L., and E.T. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Kaspar P. Locher, Email: locher@mol.biol.ethz.ch.
Emad Tajkhorshid, Email: emad@illinois.edu.
Data, Materials, and Software Availability
The molecular dynamics trajectories and topology files corresponding to both replicas of each simulated system as well as movies for each system are deposited at https://doi.org/10.5281/zenodo.6621735. The tariquidar turnover-1 EM map and model were deposited to EMDB under accession code EMD-16069 and PDB 8BHT, and tariquidar turnover-2 EM map and model were deposited to EMDB under accession code EMD-16075 and PDB 8BI0, respectively.
Supporting Information
References
- 1.Austin Doyle L., et al. , A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 95, 15665–15670 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miyake K., et al. , Molecular cloning of cDNAs which are highly overexpressed in mitoxantrone-resistant cells: Demonstration of homology to ABC transport genes. Cancer Res. 59, 8–13 (1999). [PubMed] [Google Scholar]
- 3.Allikmets R., Schriml L. M., Hutchinson A., Romano-Spica V., Dean M., A human placenta-specific ATP-binding cassette gene (ABCP) on chromosome 4q22 that is involved in multidrug resistance. Cancer Res. 58, 5337–5339 (1998). [PubMed] [Google Scholar]
- 4.Gottesman M. M., Fojo T., Bates S. E., Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat. Rev. Cancer 2, 48–58 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Imai Y., et al. , Breast cancer resistance protein exports sulfated estrogens but not free estrogens. Mol. Pharmacol. 64, 610–618 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Sarkadi B., Homolya L., Szakács G., Váradi A., Human multidrug resistance ABCB and ABCG transporters: Participation in a chemoimmunity defense system. Physiol. Rev. 86, 1179–1236 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Jardetzky O., Simple allosteric model for membrane pumps. Nature 211, 969–970 (1966). [DOI] [PubMed] [Google Scholar]
- 8.Yu Q., et al. , Structures of ABCG2 under turnover conditions reveal a key step in the drug transport mechanism. Nat. Commun. 12, 4376 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Locher K. P., Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Schneider E., Hunke S.. ATP-binding-cassette (ABC) transport systems: Functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 22, 1–20 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Saurin W., Hofnung M., Dassa E., Getting in or out: Early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J. Mol. Evol. 48, 22–41 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Barry Holland I., Blight M. A., ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J. Mol. Biol. 293, 381–399 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Taylor N. M. I., et al. , Structure of the human multidrug transporter ABCG2. Nature 546, 504–509 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Jackson S. M., et al. , Structural basis of small-molecule inhibition of human multidrug transporter ABCG2. Nat. Struct. Mol. Biol. 25, 333–340 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Manolaridis I., et al. , Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states. Nature 563, 426–430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orlando B. J., Liao M., ABCG2 transports anticancer drugs via a closed-to-open switch. Nat. Commun. 11, 2264 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kowal J., et al. , Structural basis of drug recognition by the multidrug transporter ABCG2. J. Mol. Biol. 433, 166980 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Bakhsheshian J., et al. , Overlapping substrate and inhibitor specificity of human and murine ABCG2. Drug Metab. Dispo. 41, 1805–1812 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gose T., et al. , ABCG2 requires a single aromatic amino acid to “clamp’’ substrates and inhibitors into the binding pocket. FASEB J. 34, 4890–4903 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Y., et al. , Molecular basis of cholesterol efflux via ABCG subfamily transporters. Proc. Natl. Acad. Sci. U.S.A. 118, e2110483118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skarda L., Kowal J., Locher K. P., Structure of the human cholesterol transporter ABCG1. J. Mol. Biol. 433, 167218 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Telbisz A., Hegedus C., Varadi A., Sarkadi B., Ozvegy-Laczka C., Regulation of the function of the human ABCG2 multidrug transporter by cholesterol and bile acids: Effects of mutations in potential substrate and steroid binding sites. Drug Metab. Dispo. 42, 575–585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Telbisz Á., et al. , Membrane cholesterol selectively modulates the activity of the human ABCG2 multidrug transporter. Biochim. Biophys. Acta Biomembr. 1768, 2698–2713 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Pal A., et al. , Cholesterol potentiates ABCG2 activity in a heterologous expression system: Improved in vitro model to study function of human ABCG2. J. Pharmacol. Exp. Ther. 321, 1085–1094 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Szilagyi J. T., Vetrano A. M., Laskin J. D., Aleksunes L. M., Localization of the placental BCRP/ABCG2 transporter to lipid rafts: Role for cholesterol in mediating efflux activity. Placenta 55, 29–36 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Storch C. H., Ehehalt R., Haefeli W. E., Weiss J., Localization of the human breast cancer resistance protein (BCRP/ABCG2) in lipid rafts/caveolae and modulation of its activity by cholesterol in vitro. J. Pharmacol. Exp. Ther. 323, 257–267 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Kennedy M. A., et al. , ABCG1 has a critical role in mediating cholesterol efflux to HDL and preventing cellular lipid accumulation. Cell Metab. 1, 121–131 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Wang N., Lan D., Chen W., Matsuura F., Tall A. R., ATP-binding cassette transporters G1 and G4 mediate cellular cholesterol efflux to high-density lipoproteins. Proc. Natl. Acad. Sci. U.S.A. 101, 9774–9779 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berge K. E., et al. , Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science 290, 1771–1775 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Yu L., et al. , Disruption of ABCG5 and ABCG8 in mice reveals their crucial role in biliary cholesterol secretion. Proc. Natl. Acad. Sci. U.S.A. 99, 16237–16242 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu L., et al. , Expression of ABCG5 and ABCG8 is required for regulation of biliary cholesterol secretion. J. Biol. Chem. 280, 8742–8747 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Nagy T., et al. , The transport pathway in the ABCG2 protein and its regulation revealed by molecular dynamics simulations. Cell. Mol. Life Sci. 78, 2329–2339 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prieß M., Göddeke H., Groenhof G., Schäfer L. V., Molecular mechanism of ATP hydrolysis in an ABC transporter. ACS Cent. Sci. 4, 1334–1343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chao Wen P., Verhalen B., Wilkens S., Mchaourab H. S., Tajkhorshid E., On the origin of large flexibility of P-glycoprotein in the inward-facing state. J. Biol. Chem. 288, 19211–19220 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Francois St-Pierre J., Bunker A., oog T., Karttunen M., Mousseau N., Molecular dynamics simulations of the bacterial ABC transporter SAV1866 in the closed form. J. Phys. Chem. B 116, 2934–2942 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Kapoor K., Thangapandian S., Tajkhorshid E., Extended-ensemble docking to probe evolution of ligand binding sites in proteins undergoing large-scale structural changes for their function. bioRxiv (2021). [DOI] [PMC free article] [PubMed]
- 37.Rasouli A., Kapoor K., Chao Wen P., Tajkhorshid E., Evidence for direct membrane interaction and cholesterol sensing of A-loop in ABC transporter ABCG2. Biophys. J. 118, 527a (2020). [Google Scholar]
- 38.Verhalen B., et al. , Energy transduction and alternating access of the mammalian ABC transporter P-glycoprotein. Nature 543, 738–741 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayne C. G., et al. , The cellular membrane as a mediator for small molecule interaction with membrane proteins. Biochim. Biophys. Acta Biomembr. 1858, 2290–2304 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barreto-Ojeda E., Corradi V., Xu Gu R.-, Peter Tieleman D., Coarse-grained molecular dynamics simulations reveal lipid access pathways in P-glycoprotein. J. Gen. Physiol. 150, 417–429 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Immadisetty K., Hettige J., Moradi M., Lipid-dependent alternating access mechanism of a bacterial multidrug ABC exporter. ACS Cent. Sci. 5, 43–56 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ritchie T. K., et al. , Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Meth. Enzym. 464, 211–231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chifflet S., Torriglia A., Chiesa R., Tolosa S., A method for the determination of inorganic phosphate in the presence of labile organic phosphate and high concentrations of protein: Application to lens ATPases. Addict. Biol. 168, 1–4 (1988). [DOI] [PubMed] [Google Scholar]
- 44.Li X., et al. , Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Wei Zhang Y.-, Turk B. E., Rudnick G., Control of serotonin transporter phosphorylation by conformational state. Proc. Natl. Acad. Sci. U.S.A. 113, E2776–E2783 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Cryst. D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moriarty N. W., Grosse-Kunstleve R. W., Adams P. D., Electronic ligand builder and optimization workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Cryst. D 65, 1074–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Afonine P. V., et al. , Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Cryst. D 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chun Cheng H.-, Ulane C. M., Burke R. E., Clinical progression in Parkinson disease and the neurobiology of axons. Ann. Neurol. 67, 715–725 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- 52.Eswar N., et al. , Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinf., 5–6 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olsson M. H. M., Søndergaard C. R., Rostkowski M., Jensen J. H., PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Søndergaard C. R., Olsson M. H. M., Rostkowski M., Jensen J. H., Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 7, 2284–2295 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Zhang L., Hermans J., Hydrophilicity of cavities in proteins. Proteins: Struct. Func. Gen. 24, 433–438 (1996). [DOI] [PubMed] [Google Scholar]
- 56.Gumbart J., Trabuco L. G., Schreiner E., Villa E., Schulten K., Regulation of the protein-conducting channel by a bound ribosome. Structure 17, 1453–1464 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Jo S., Lim J. B., Klauda J. B., Im W., CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys. J. 97, 50–58 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lomize M. A., Lomize A. L., Pogozheva I. D., Mosberg H. I., OPM: Orientations of proteins in membranes database. Bioinformatics 22, 623–625 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Dehghani-Ghahnaviyeh S., Kapoor K., Tajkhorshid E., Conformational changes in the nucleotide-binding domains of P-glycoprotein induced by ATP hydrolysis. FEBS Lett. 595, 735–749 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Phillips J. C., et al. , Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Phillips J. C., et al. , Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 153, 044130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shaw D. E., et al. , “Anton 2: Raising the bar for performance and programmability in a special-purpose molecular dynamics supercomputer” in Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis (IEEE Press, 2014), pp. 41–53. [Google Scholar]
- 64.Huang J., et al. , CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klauda J. B., et al. , Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 67.Vanommeslaeghe K., et al. , CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vanommeslaeghe K., Prabhu Raman E., A. D. MacKerell, Jr.. Automation of the CHARMM General Force Field (CGenFF) II: Assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 52, 3155–3168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mayne C. G., Saam J., Schulten K., Tajkhorshid E., Gumbart J. C., Rapid parameterization of small molecules using the Force Field Toolkit. J. Comput. Chem. 34, 2757–2770 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Darden T., York D., Pedersen L., Particle mesh Ewald: an N-log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993). [Google Scholar]
- 71.Ryckaert J.-P., Ciccotti G., Berendsen H. J. C., Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comp. Phys. 23, 327–341 (1977). [Google Scholar]
- 72.Martyna G., Tobias D. J., Klein M. L., Constant pressure molecular dynamics algorithms. J. Chem. Phys. 101, 4177–4189 (1994). [Google Scholar]
- 73.Feller S. E., Zhang Y., Pastor R. W., Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 103, 4613–4621 (1995). [Google Scholar]
- 74.Martyna G. J., Tobias D. J., Klein M. L., Constant pressure molecular dynamics algorithms. J. Chem. Phys. 101, 4177–4189 (1994). [Google Scholar]
- 75.Lippert R. A., et al. , Accurate and efficient integration for molecular dynamics simulations at constant temperature and pressure. J. Chem. Phys. 139, 164106 (2013). [DOI] [PubMed] [Google Scholar]
- 76.Kräutler V., Van Gunsteren W. F., Hünenberger P. H., A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 22, 501–508 (2001). [Google Scholar]
- 77.Frisch M. J., et al. , Gaussian 16 (Gaussian Inc., Wallingford CT, 2016). [Google Scholar]
- 78.Dennington R., Keith T. A., Millam J. M., Gaussview Version 6 (Semichem Inc., Shawnee Mission KS, 2019). [Google Scholar]
- 79.Durrant J. D., Amaro R. E., LipidWrapper: An algorithm for generating large-scale membrane models of arbitrary geometry. PLOSCP 10, e1003720 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kannan P., et al. , The “specific’’ P-glycoprotein inhibitor tariquidar is also a substrate and an inhibitor for breast cancer resistance protein (BCRP/ABCG2). ACS Chem. Neurosci. 2, 82–89 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kuhnle M., et al. , Potent and selective inhibitors of breast cancer resistance protein (ABCG2) derived from the P-glycoprotein (ABCB1) modulator tariquidar. J. Med. Chem. 52, 1190–1197 (2009). [DOI] [PubMed] [Google Scholar]
- 82.Woebking B., et al. , Drug-lipid A interactions on the Escherichia coli ABC transporter MsbA. J. Bacteriol. 187, 6363–6369 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Siarheyeva A., Sharom F. J., The ABC transporter MsbA interacts with lipid A and amphipathic drugs at different sites. Biochem. J. 419, 317–328 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Homolya L., Orbán T., Csanády L., Sarkadi B., Mitoxantrone is expelled by the ABCG2 multidrug transporter directly from the plasma membrane. Biochim. Biophys. Acta Biomembr. 1808, 154–163 (2011). [DOI] [PubMed] [Google Scholar]
- 85.Raviv Y., Pollard H. B., Bruggemann E. P., Pastan I., Gottesman M. M., Photosensitized labeling of a functional multidrug transporter in living drug-resistant tumor cells. J. Biol. Chem. 265, 3975–3980 (1990). [PubMed] [Google Scholar]
- 86.Higgins C. F., Gottesman M. M., Is the multidrug transporter a flippase? Trends Biochem. Sci. 17, 18–21 (1992). [DOI] [PubMed] [Google Scholar]
- 87.Homolya L., et al. , Fluorescent cellular indicators are extruded by the multidrug resistance protein. J. Biol. Chem. 268, 21493–21496 (1993). [PubMed] [Google Scholar]
- 88.Bolhuis H., et al. , Multidrug resistance in Lactococcus lactis: Evidence for ATP-dependent drug extrusion from the inner leaflet of the cytoplasmic membrane. EMBO J. 15, 4239–4245 (1996). [PMC free article] [PubMed] [Google Scholar]
- 89.Shapiro A. B., Ling V., Extraction of Hoechst 33342 from the cytoplasmic leaflet of the plasma membrane by P-glycoprotein. Eur. J. Biochem. 250, 122–129 (1997). [DOI] [PubMed] [Google Scholar]
- 90.Shapiro A. B., Corder A. B., Ling V., P-glycoprotein-mediated Hoechst 33342 transport out of the lipid bilayer. Eur. J. Biochem. 250, 115–121 (1997). [DOI] [PubMed] [Google Scholar]
- 91.Durrant J. D., Votapka L., Sørensen J., Amaro R. E., POVME 2.0: An enhanced tool for determining pocket shape and volume characteristics. J. Chem. Theory Comput. 10, 5047–5056 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
MD trajectory of the tariquidar-bound TO2 state during a 2-μs simulation. ABCG2 is shown in cartoon representation with monomers colored in green and yellow, respectively, and the bound ligands are shown with sticks.
MD trajectory of the tariquidar-bound TO1 state during a 2-μs simulation. ABCG2 is shown in cartoon representation with monomers colored in green and yellow, respectively, and the bound ligands are shown with sticks.
MD trajectory of the topotecan-bound TO2 state during a 2-μs simulation. ABCG2 is shown in cartoon representation with monomers colored in green and yellow, respectively, and the bound ligands are shown with sticks. Two phospholipids, a POPS (red) and a PSM (cyan), that penetrate the binding pocket are shown.
MD trajectory of the topotecan-bound TO1 state during a 2-μs simulation. ABCG2 is shown in cartoon representation with monomers colored in green and yellow, respectively, and the bound ligands are shown with sticks. Two POPE lipids that penetrate the binding pocket during the simulation are shown in red and cyan, respectively.
MD trajectory of ABCG2 in IF-Apo conformation during a 1-μs simulation. ABCG2 is shown in cartoon representation with monomers colored in green and yellow, respectively. Two phospholipids that penetrate the binding pocket during the simulation, a POPS (red) and POPE (cyan), are shown.
Data Availability Statement
The molecular dynamics trajectories and topology files corresponding to both replicas of each simulated system as well as movies for each system are deposited at https://doi.org/10.5281/zenodo.6621735. The tariquidar turnover-1 EM map and model were deposited to EMDB under accession code EMD-16069 and PDB 8BHT, and tariquidar turnover-2 EM map and model were deposited to EMDB under accession code EMD-16075 and PDB 8BI0, respectively.