Abstract
Purpose:
PSMA overexpression has been associated with aggressive prostate cancer (PCa). However, PSMA PET imaging has revealed highly variable changes in PSMA expression in response to ADT treatment ranging from increases to moderate decreases. To better understand these PSMA responses and potential relationship to progressive PCa, the PET imaging agent, [18F]DCFPyL, was used to assess changes in PSMA expression in response to ADT using genomically characterized LuCaP patient-derived xenograft mouse models (LuCaP-PDXs) which were found to be sensitive to ADT (LuCaP73 and LuCaP136;CS) or resistant (LuCaP167;CR).
Methods:
[18F]DCFPyL (2-(3-{1-carboxy-5-[(6-[18F]fluoro-pyridine-3-carbonyl)-amino]-pentyl}-ureido)-pentanedioic acid) was used to assess PSMA in vitro (saturation assays) in LuCaP tumor membrane homogenates and in vivo (imaging/biodistribution) in LuCaP-PDXs. Control and ADT-treated LuCaPs were imaged before ADT (0 days) and 2-, 7-, 14-, and 21-days post-ADT from which tumor:muscle ratios (T:Ms) were determined and concurrently tumor volumes were measured (caliper). After the 21-day imaging, biodistributions and histologic/genomic (PSMA, AR) analysis were done.
Results:
[18F]DCFPyL exhibited high affinity for PSMA and distinguished different levels of PSMA in LuCaP tumors. Post-ADT CS LuCaP73 and LuCaP136 tumor volumes significantly decreased at day 7 or 14 respectively vs controls, whereas the CR LuCaP167 tumor volumes were minimally changed. [18F]DCFPyL imaging T:Ms were increased 3–5-fold in treated LuCaP73 tumors vs controls, while treated LuCaP136 T:Ms remained unchanged which was confirmed by day 21 biodistribution results. For treated LuCaP167, T:Ms were decreased (~ 45 %) vs controls but due to low T:M values (<2) may not be indicative of PSMA level changes. LuCaP73 tumor PSMA histologic/genomic results were comparable to imaging/biodistribution results, whereas the results for other tumor types varied.
Conclusion:
Tumor responses to ADT varied from sensitive to resistant among these LuCaP PDXs, while only the high PSMA expressing LuCaP model exhibited an increase in PSMA levels in response to ADT. These models may be useful in understanding the clinical relevance of PSMA PET responses to ADT and potentially the relationship to disease progression as it may relate to the genomic signature.
Keywords: PSMA, Androgen deprivation therapy, ADT therapy, PET imaging, Prostate cancer, PDX, LuCaP
Introduction
Worldwide, prostate cancer (PCa) is one of the leading causes of morbidity and mortality in men [1]. Most intermediate and high risk primary and metastatic prostate cancers overexpress prostate specific membrane antigen (PSMA), a cell surface glycoprotein (~ 100 kDa) which exhibits carboxypeptidase and folate hydrolase enzymatic activities [2]. While PSMA is expressed at relatively low levels in the normal prostate, its high expression in prostate cancer has led to interest in PSMA as both a diagnostic and therapeutic biomarker [3–5]. Several small-molecule inhibitors of PSMA have been developed as PET imaging agents which have shown superior specificity and sensitivity in identifying PCa lesions compared to conventional imaging modalities [6–11].
Prostate cancer is driven by the androgen receptor (AR), and androgen deprivation therapy (ADT) is commonly employed as a first-line therapy in advanced PCa [12]. ADT encompasses a family of drugs that act by lowering serum testosterone levels or by directly blocking binding of androgens to AR. The luteinizing hormone-releasing hormone (LHRH) antagonist, degarelix, represents one of several centrally acting anti-androgen agents [13, 14]. LHRH antagonists indirectly lower testosterone levels and downregulate the AR function which is required for the growth and survival of most PCa [15]. PSMA overexpression has been associated with androgen dependent disease as well as independent disease and increased malignancy; however, some studies have revealed differential PSMA expression levels in response to ADT [16–19]. Although the relevance of this relationship has clinical importance, the relationship between ADT and changes in PSMA expression levels are poorly understood. In part, this is due to lack of preclinical PCa models which exhibit the genetic diversity observed in PCa patients.
Preclinical studies reported thus far using a few conventional human prostate cancer cell lines such as LNCaP do not represent the genetic diversity of PCa found across the patient population [18, 20]. With the recent development of patient-derived prostate cancer, xenograft mouse models (PDX; LuCaP) in which the genomic and phenotypic characteristics of the tumor type have been well characterized a better understanding of the relationship between the tumor’s genomics and PSMA responses to ADT may be possible [21, 22]. Here, three LuCaP PDX mouse models with varying ADT sensitivities (castrate sensitive, CS; castrate resistant, CR) were treated with degarelix and imaged with the PSMA PET imaging agent, [18F]DCFPyL, to ascertain changes in PSMA expression levels over the treatment time course [21]. In addition, at the end of the study, [18F]DCFPyL biodistributions were done from which tumor samples were obtained for histological and genomic analysis to ascertain changes in expression levels of PSMA and AR protein and genes, respectively, in response to ADT.
Methods
Radiosynthesis of [18F]DCFPyL
2-(3-{1-carboxy-5-[(6-[18F]fluoro-pyridine-3-carbonyl)-amino]-pentyl}-ureido)-pentanedioic acid, [18F]DCFPyL, was synthesized according to a literature procedure [23]. The isolated radiochemical yields ranged from 32 to 43 % with a radiochemical purity > 98 %, and molar activity from 1200 to 2600 Ci/mmol (44–96 GBq/μmol).
Xenograft and Drug Treatment Models
Xenograft Models
PC3 Wt (wild type, PSMA negative) and PC3(+) (prostate cancer cell line transfected with human PSMA) were provided by Dr. Hisataka Kobayashi [24]. PC3(+) and PC3 Wt cell suspensions [2 × 106 cells; PBS:Matrigel (70:30)] from in vitro cell culture were subcutaneously implanted (right shoulder) into athymic male mice (FOX/nu, Envigo, 5 weeks old) for use as a positive and negative control respectively for in vitro and in vivo studies. For the PDX models, athymic nude mice (male; FOX/nu, Envigo, 5 weeks old) were subcutaneously implanted (right shoulder) with LuCaP73, LuCaP167, or LuCaP136 tumor cell suspensions [2 × 106 cells; PBS:Matrigel (70:30)] prepared from fresh tumors excised from respective mouse xenografts [21]. When tumors reached the appropriate size (> 100 mg), mice were enrolled in imaging studies or euthanized and tumors excised and fast-frozen for in vitro saturation binding assays to determine PSMA levels (Bmax) for each tumor type and Kd of [18F]DCFPyL (Supplementary information: Materials and methods). All animal studies were performed in accordance with NIH Guidelines for the Care and Use of Laboratory Animals using IACUC-approved protocols.
Drug Treatment
Tumor-bearing mice (tumors >100 mg) were randomly divided (based on tumor volume) into control and ADT (Firmagon (degarelix), Ferring Pharmaceuticals) treated groups. Treated mice were dosed with degarelix (25 mg/Kg) subcutaneously at baseline (following imaging) and day 14 [25, 26].
In vivo PET Imaging Studies
Prior to imaging, all mice were anesthetized using isoflurane/O2 (1.5 % to 3 % v/v) and imaged (BioPET, Bioscan) at 1 h post [18F]DCFPyL injection (100–150 μCi; iv). Whole body images were acquired (2 bed positions; 5 min each) and reconstructed using three-dimensional ordered-subset expectation maximums (3D-OSEM) [27]. Mice in both the groups were imaged before initiation of ADT treatment (baseline) and at days 2, 7, 14, and 21 following treatment. Tumors were measured weekly (concurrent with PET imaging) by caliper, and the volume was calculated using the ellipsoid formula.
PET images were analyzed (ASIPro software, Siemens) by drawing regions of interest (ROI’s) for LuCaP73, LuCaP136, and LuCaP167 which encompassed the whole tumor volume. Additionally, ROIs were drawn for the thigh muscle, from which tumor:muscle PSMA PET uptake ratios (T:M) were calculated for each mouse. Statistical significance (P < 0.05) between the control and treated groups was determined using the student t-test.
Biodistributions, Tumor Histology, and Tumor RT-PCR Analysis
On the last day of imaging, all mice were euthanized (CO2 asphyxiation) 1 h post [18F]DCFPyL injection, and necropsy was performed to obtain blood and tissues from each animal. Radioactivity associated with each sample was determined by gamma counting (counts per minute, CPM). For each tissue, the percentage injected dose per gram normalized to a mouse body weight of 30 g (%ID/gnormalized to 30 g mouse), from which tumor:blood (T:B) and tissue:muscle (T:M) were calculated as follows:
| (1) |
| (2) |
| (3) |
Additionally, excised tumors were either fast-frozen or formalin fixed for H&E and immunohistochemistry (IHC) staining to determine PSMA, AR, and CD31 levels (Supplementary information: Materials and methods). The fast-frozen samples from these tumors were used to perform RT-PCR to determine mRNA levels for FOLH1, AR, KLK3, and FKBP5 (Supplementary information: Materials and methods).
Results
In vitro Binding Studies
[18F]DCFPyL exhibited high affinity binding (Kd = 0.83 ± 0.04 nM) to PSMA using PC3(+) tumor membrane preparations and high specific binding (85–98 %; Fig. 1). PC3(+) tumor membrane preparations [Bmax = 13.95 ± 1.60 femtomoles/μg of protein (fmoles/μg)] had the highest PSMA levels compared to the PDX tumor membrane preparations (SI Table 1, Table 1), which were ~ 7–1400-fold higher than the LuCaP models [Bmax = LuCaP73 (1.87 ± 0.53 fmoles/μg), LuCaP136 (0.038 ± 0.0017 fmoles/μg), LuCaP167 ( 0.013 ± 0.0054 fmoles/μg)].
Fig. 1.
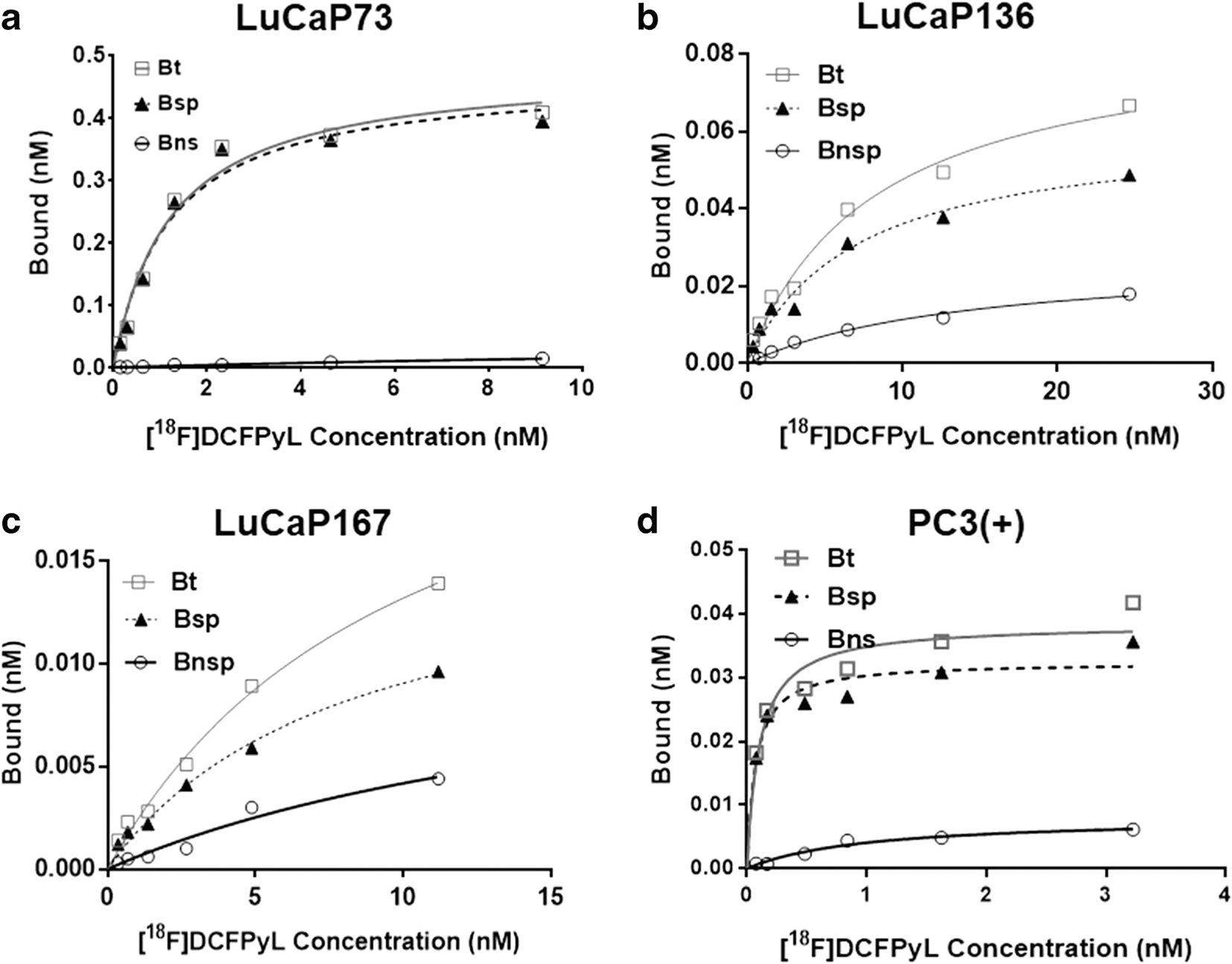
Representative plots of in vitro saturation binding studies using tumor membrane preparations from LuCaP [LuCaP73 (a), LuCaP136 (b), and LuCaP167 (c) or PC3(+) [PC3 cells transfected with human PSMA(d)] xenografts. For each plot: Bt = bound total; Bnsp = bound non-specific; Bsp = bound specific (Bt-Bnsp=Bsp), n= 2 or 3.
Table 1.
LuCaP tumor summary of the origin, histology, microvessel density and protein/mRNA levels of PSMA, AR, KLK3, and FKBP5
| Characteristics of prostate cancer PDXs (LuCaP) | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Tumor models | LuCaP73 | LuCaP136 | LuCaP167 | |||
|
| ||||||
| Tumor origin | Primary prostate cancer | Cells from ascites | Liver metastasis | |||
| Histology | Adenocarcinoma | Adenocarcinoma | Adenocarcinoma | |||
| PSMA (moles/μg)1 | 1.87 ± 0.53 | 0.38 ± 0.0017 | 0.01 ± 1.60 | |||
| Study groups | Control | ADT Tx | Control | ADT Tx | Control | ADT Tx |
| PSMA H-score2 | 28.15 ± 11.59 | 111.04 ± 12.14 | N/D | N/D | N/D | N/D |
| AR H-score2 | 227.93 ± 5.31 | 230.22 ± 21.69 | 263.20 ± 8.60 | 231.00 ± 5.51 | 297.23 ±1.01 | 291.60 ± 1.97 |
| FOLH1 mRNA3 | 8.19 ± 1.27 | 23.78 ± 0.40 | 0.12 ± 0.07 | 0.38 ± 0.029 | 0.08 ± 0.051 | 0.086 ± 0.034 |
| AR mRNA3 | 1.15 ± 0.26 | 2.99 ± 0.39 | 1.06 ± 0.51 | 1.22 ± 0.28 | 29.09 ± 6.75 | 351.28 ±01.33 |
| KLK3 mRNA3 | 50.49 ± 12.13 | 53.85 ± 12.19 | 1.73 ± 0.53 | 0.36 ± 0.18 | 86.93 ± 13.72 | 117.30 ± 20.60 |
| FKBP5 mRNA3 | 9.16 ± 3.11 | 2.22 ± 0.78 | 0.069 ± 0.01 | 0.045 ± 0.02 | 10.73 ± 2.44 | 14.58 ± 3.45 |
| Tumor microvessel density (no. of vessel/μm2)2 | 8.63 × 10−5 | 22.3 × 10−5 | 1.65 × 10−5 | 9.61 × 10−5 | 17.64 × 10−5 | 21.10 × 10−5 |
Notes:
PSMA (femtomoles/μg) was determined in vitro from tumor membrane preparations.
H-score represents the protein levels determined from IHC staining of tumor samples. Tumor microvessel density was determined by CD31 staining of tumor samples.
mRNA levels of FOLH1 (PSMA), AR, KLK3, and FKBP5 were determined by genetic analysis of tumor samples. KLK3: Kallikrein-related peptidase 3 gene; FKBP5: FK506-binding protein 51 gene; FOLH1: folate hydrolase I gene; AR: androgen receptor gene; N/D not determined.
Tumor Volume Responses to ADT Treatment in LuCaP PDXs
LuCaP PDXs had variable tumor volume responses to ADT treatment compared to controls (Figs. 2a, 3a, and 4a). LuCaP73 and LuCaP136 control tumor volumes increased 1.5- to 1.8-fold and 2- to 8-fold, respectively, over the time course, whereas treated tumor volumes decreased for LuCaP73 (15–223 %) and LuCaP136 (20–114 %). LuCaP73 and LuCaP136 PDXs were sensitive (CS) to ADT treatment; however, LuCaP73 exhibited greater sensitivity than LuCaP136 (Figs. 2a and 3a). ADT-treated LuCaP73 tumor volumes showed a significant decline on days 7, 14, and 21 compared to control tumor volumes at the same times (P < 0.05; Fig. 2a), whereas LuCaP136 tumor volumes showed significant decreases at days 14 and 21 compared to controls (P < 0.05; Fig. 3a). LuCaP167, characterized as castrate resistant (CR), exhibited slower tumor growth in both control (1.7- to 2.5-fold) and treated groups (1.3- to 2.0-fold) compared to the CS LuCaPs and demonstrated no significant differences in tumor volumes between the treatment and control tumors throughout the study time course (Fig. 4a).
Fig. 2.
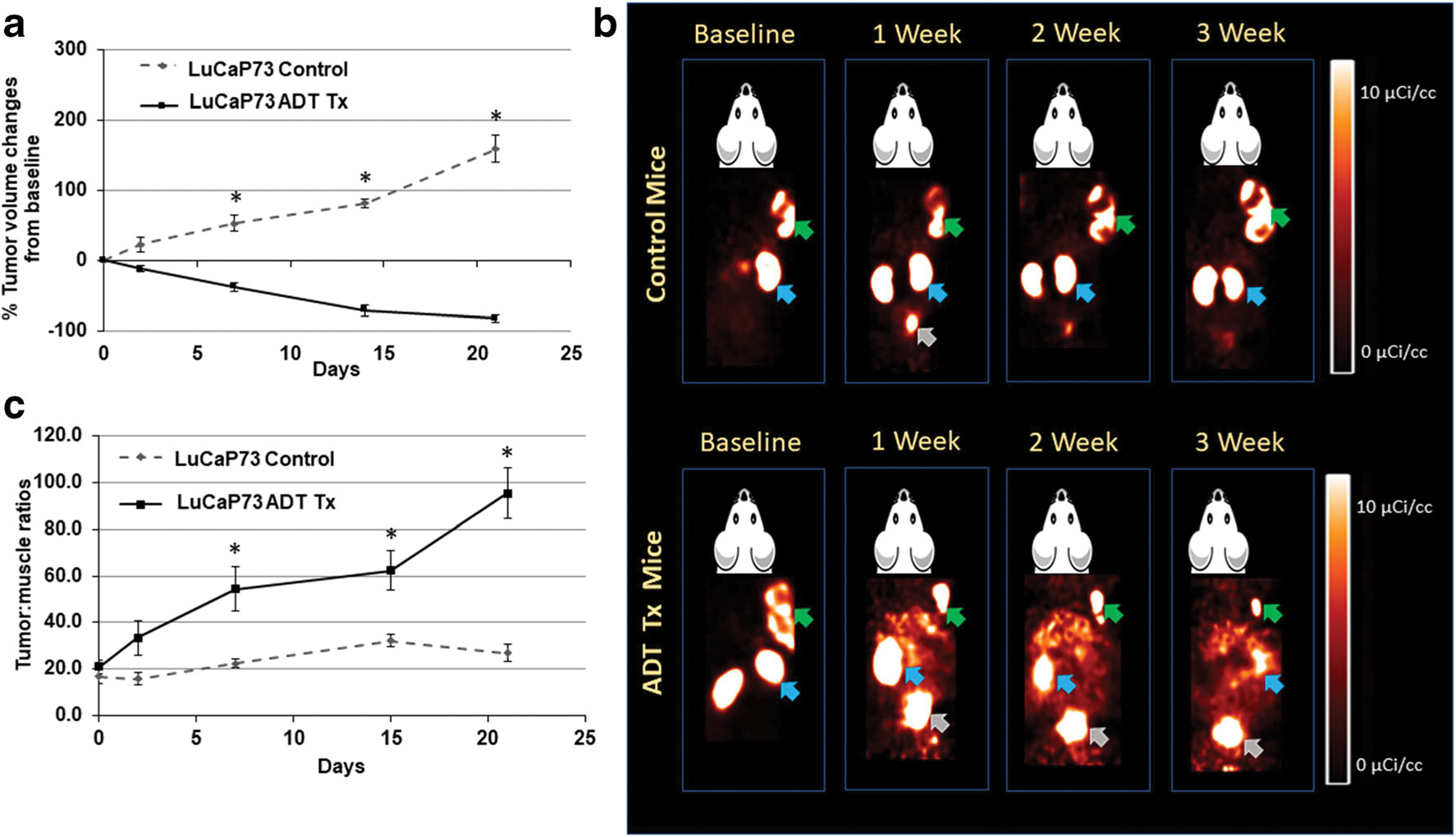
a Comparison of % changes in tumor volumes from baseline in control and ADT-treated LuCaP73 tumor-bearing mice at baseline (0), 2, 7, 14, and 21 days (mean ± SE, n = 5–7 mice; *significant difference between groups, P < 0.05). b Representative [18F]DCFPyL PET images (60 min post-injection) in LuCaP73 control and ADT-treated tumor-bearing mouse over the course of therapy. Green, blue, and grey arrows indicate tumor, kidneys, and bladder respectively. c LuCaP73 tumor:muscle ratios calculated from PET image analysis in control and treated groups (mean tumor:muscle ratios ± SE, n = 5–7 mice; *significant difference between groups, P < 0.05).
Fig. 3.
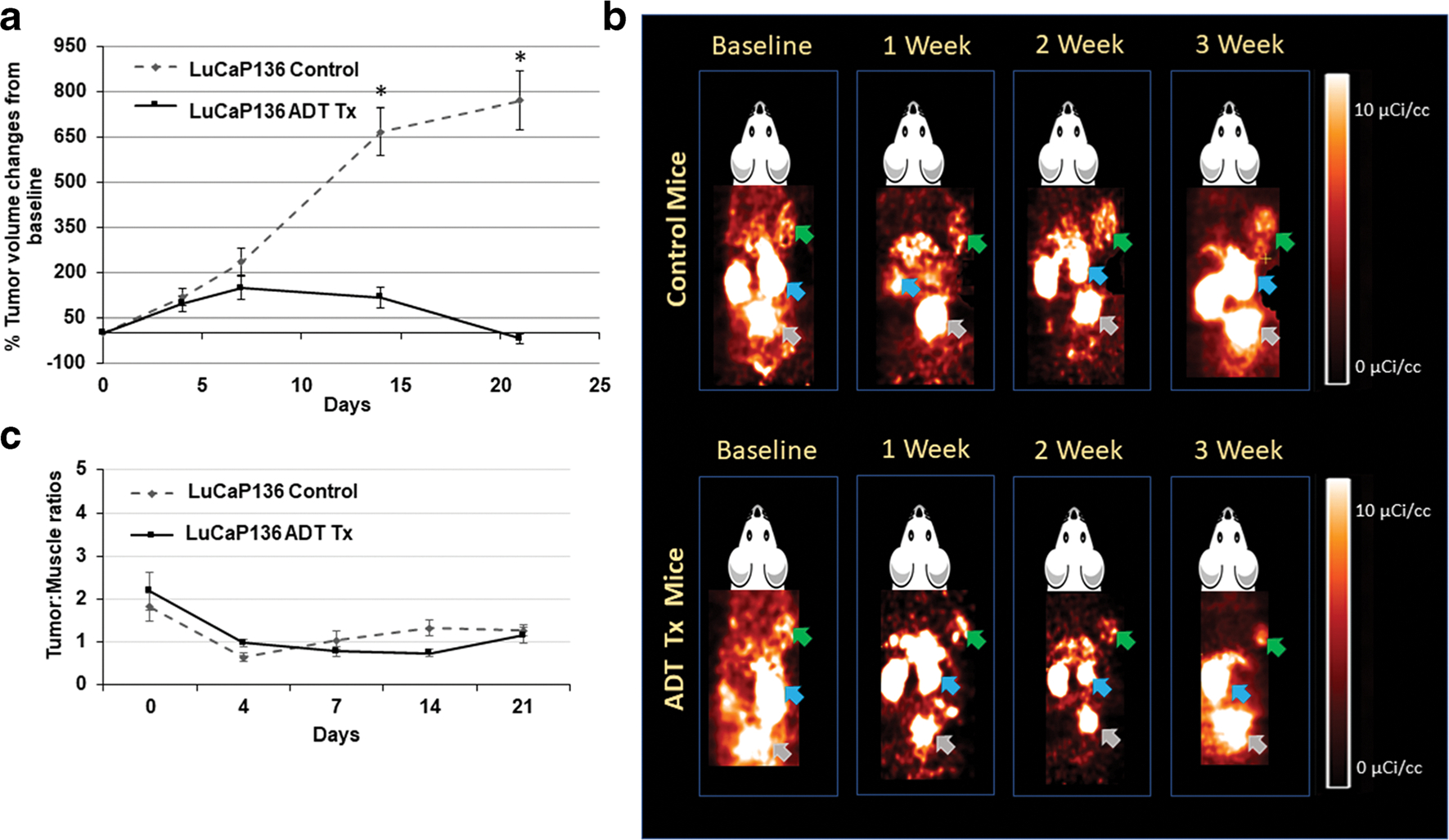
a Comparison of % changes in tumor volumes from baseline in control and ADT-treated LuCaP136 tumor-bearing mice at baseline (0), 2, 7, 14, and 21 days (mean ± SE, n = 5–7 mice; *significant difference between groups, P < 0.05). b Representative [18F]DCFPyL PET images (60 min post-injection) in LuCaP136 control and ADT-treated tumor-bearing mouse over the course of therapy. Green, blue, and grey arrows indicate tumor, kidneys, and bladder respectively. c LuCaP136 tumor:muscle ratios calculated from PET image analysis in control and treated groups (mean tumor:muscle ratios ± SE, n = 5–7 mice; *significant difference between groups, P < 0.05).
Fig. 4.
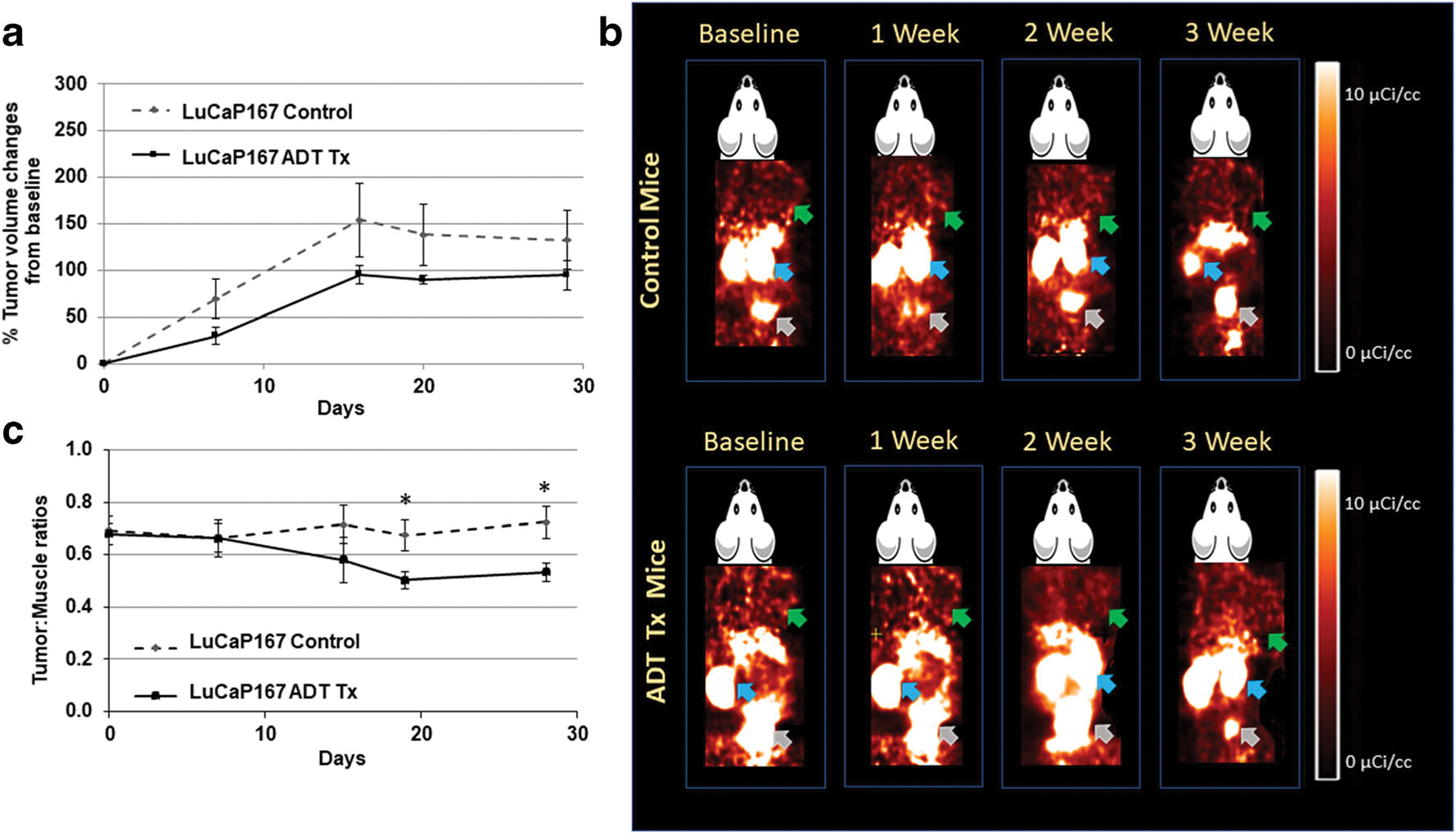
a Comparison of % changes in tumor volumes from baseline in control and ADT-treated LuCaP167 tumor-bearing mice at baseline (0), 2, 7, 14, and 21 days (mean ± SE, n = 5–7 mice; *significant difference between groups, P < 0.05). b Representative [18F]DCFPyL PET images (60 min post-injection) in LuCaP167control and ADT-treated tumor-bearing mouse over the course of therapy. Green, blue, and grey arrows indicate tumor, kidneys, and bladder respectively. c LuCaP167 tumor:muscle ratios calculated from PET image analysis in control and treated groups (mean tumor:muscle ratios ± SE, n = 5–7 mice; *significant difference between groups, P < 0.05).
In vivo PET Imaging and Biodistribution of [18F]DCFPyL
Control and treated LuCaP PDXs were imaged with [18F]DCFPyL to assess changes in PSMA expression over the ADT treatment time course; in addition, PC3(+) xenografts were also imaged (SI Fig. 2). [18F]DCFPyL images (Figs. 2b, 3b, and 4b) revealed high kidney uptake in all groups and differential tumor uptakes in ADT-treated groups compared to controls. LuCaP73 and LuCaP136 tumors were readily visualized on PSMA PET, whereas LuCaP167 tumors showed lower uptake on PET, which was consistent with the rank order of PSMA levels determined in vitro. Quantitative analysis (tumor:muscle ratio; T:M) of PET images of the two CS tumor xenograft models showed significant increases of 3–5-fold (P < 0.05) from 7 to 21 days in LuCaP73 treated vs control tumors (Fig. 2c), while T:M ratios for LuCaP136 (CS) treated tumors were not different from control tumors (Fig. 3c). In contrast, LuCaP167 (CR) treated tumor T:Ms exhibited a significant decrease (P < 0.05) compared to control tumors (Fig. 4c) which because of the low T:Ms may not be representative of a change in PSMA-targeted binding.
After the final imaging session, biodistributions were performed to examine the accumulation of [18F]DCFPyL in various organs. Biodistributions of [18F]DCFPyL reflected the imaging results for the LuCaP tumors as well as in non-target tissues. The non-target tissue uptakes were consistent with literature reports with the highest uptakes in the kidneys (SI Fig. 3) [28]. ADT-treated LuCaP73 tumors (18.83 ± 0.08 %ID/g) showed an 85 % increase (P < 0.05) in [18F]DCFPyL uptake compared to the control tumors (10.13 ± 0.45 %ID/g; Fig. 5a). [18F]DCFPyL uptakes in LuCaP136 and LuCaP167 tumors were significantly lower (P < 0.05) compared to LuCaP73 tumors for both the groups. Treated and control LuCaP136 tumor uptakes were 0.44 ± 0.76 %ID/g and 0.33 ± 0.07 %ID/g of [18F]DCFPyL, respectively, whereas[18F]DCFPyL uptake in LuCaP167 treated and control tumors were 0.34 ± 0.06 %ID/g and 0.23 ± 0.10 %ID/g respectively. Furthermore tumor:muscle (T:M) and tumor:blood (T:B) ratios were calculated to account for the variations observed in the input functions and pharmacokinetics of [18F]DCFPyL in the treated vs control cohorts. LuCaP73 T:B ratios were significantly increased (61.38 %, P < 0.05) in treated (40.89 ± 1.69) vs control groups (25.34 ± 1.67; Fig. 5b). Similarly, LuCaP73 T:M ratios also significantly increased (65 %, P < 0.05) in treated groups compared to control groups (Fig. 5c). No significant differences in T:B ratios were found in LuCaP136 treated (1.32 ± 0.27) and control (1.32 ± 0.44) groups; however, a modest 30 % decrease in [18F]DCFPyL T:M ratios was detected in ADT-treated (4.25 ± 1.10) vs control tumors (5.72 ± 0.57; Fig. 5b–c). In LuCaP167, a 21 % decrease in T:B ratios was seen in treated tumors (0.28± 0.029) vs control tumors (0.36± 0.049), whereas a ~ 45 % decrease in the T:M ratio was observed for LuCaP167 treated tumors (1.80 ± 0.22) compared to control tumors (0.99 ± 0.18; Fig. 5b–c).
Fig. 5.
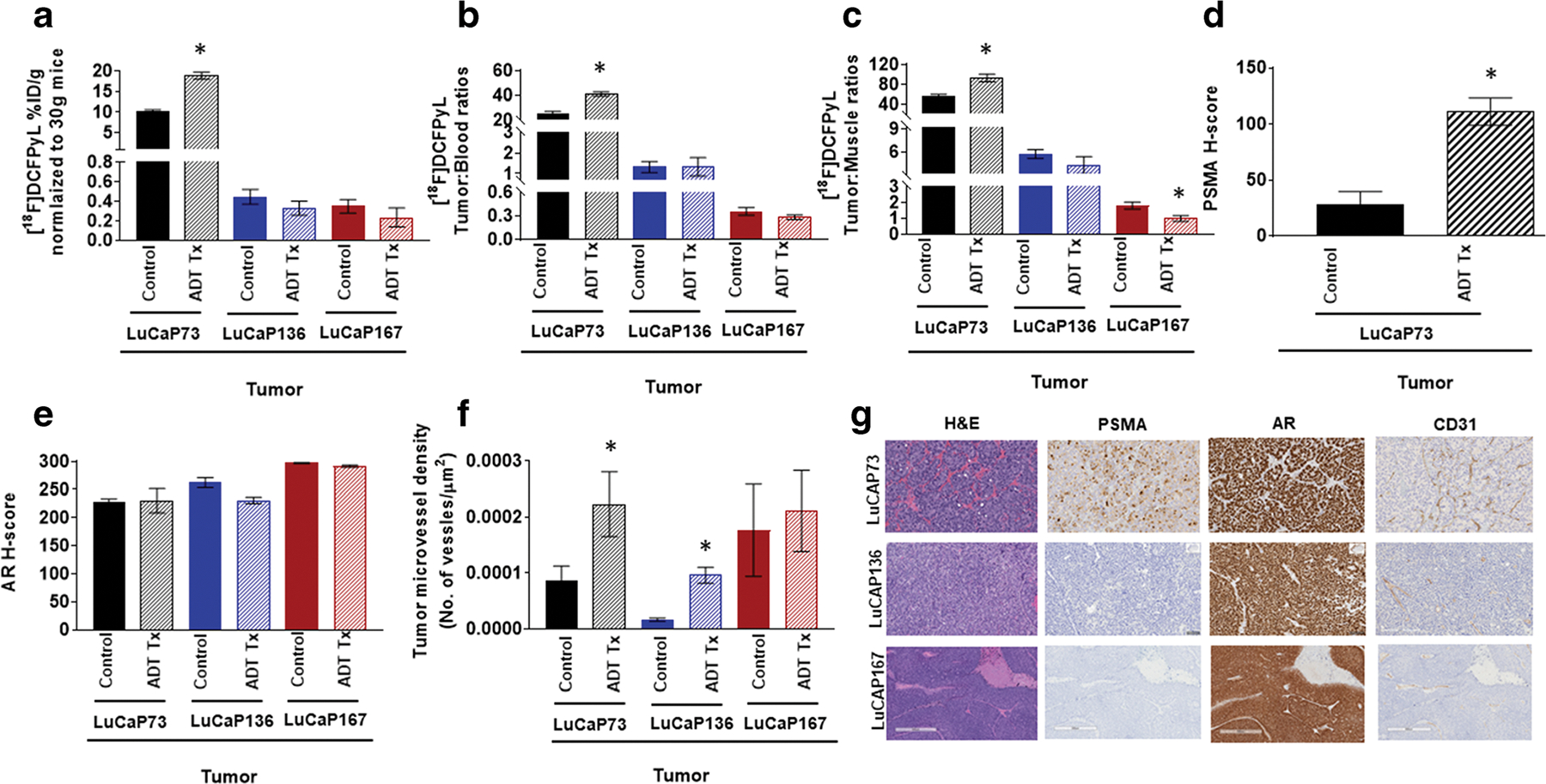
[18F]DCFPyL biodistribution (a), tumor to blood ratios (b; T:B), and tumor:muscle ratios (c; T:M) in control and ADT (degarelix) treated LuCaP73, LuCaP136, and LuCaP167 tumor-bearing mice. Biodistribution was performed at 60 min post [18F]DCFPyL injection. Each bar represents mean %ID/g ± SE (a), mean T:B± SE (b), or T:M ± SE (c); n = 5–7. (d)–(f) Quantitative histological analysis of PSMA (d, H-score), androgen receptor (e, AR, H-score), and CD31 (f, tumor microvessel density). Each bar in (d) and (e) represents mean H-score ± SD. Each bar in (f) represents the number of vessel/μm2 ± SD. (g) Representative H&E and IHC staining of tumor sections showing expression of PSMA, AR, and CD31; *significant difference in the control group versus the treated group (P < 0.05).
Histopathology and Immunohistochemistry
After the final imaging session (day 21), immunohistochemistry for PSMA, AR, and CD31 was obtained from analysis of tumor samples from treated and control PDX xenografts (Fig. 5d–g, Table 1). IHC results indicated a ~ 4-fold increase (P < 0.05) in PSMA levels in LuCaP73 treated tumors compared to control tumors (Fig. 5d). Conversely, in tumors from LuCaP136 and LuCaP167 control and treated xenografts, PSMA protein levels were not detectable by IHC. No significant differences in AR levels were observed among treated vs control tumors for any of the PDXs (Fig. 5e, Table 1). For CD31 staining, among the three tumor types, LuCaP167 control tumors exhibited the highest vessel density compared to control LuCaP73 and LuCaP136 tumors. CD31 levels were significantly increased (P < 0.05) in ADT-treated LuCaP73 and LuCaP136 tumors compared to respective control tumors (Fig. 5f, Table 1). In contrast, no significant change in CD31 levels was observed between the treated and control LuCaP167 tumors.
Gene Expression Analysis
Tumor samples from treated and control xenografts were analyzed by RT-qPCR for changes in gene expression levels (mRNA) of PSMA (FOLH1), AR, and downstream genes (KLK3 and FKBP5; Fig. 6). FOLH1 mRNA expression levels were in the same rank order for the three tumor types as was observed for PSMA PET uptake with LuCaP73 exhibiting the highest FOLH1 levels and LuCaP167 the lowest (Fig. 6a, Table 1). In response to ADT treatment, a 3-fold increase (P < 0.05) in FOLH1 mRNA was observed in LuCaP73 and LuCaP136 compared to controls, whereas no significant change occurred in LuCaP167 treated vs control (Fig. 6a, Table 1).
Fig. 6.
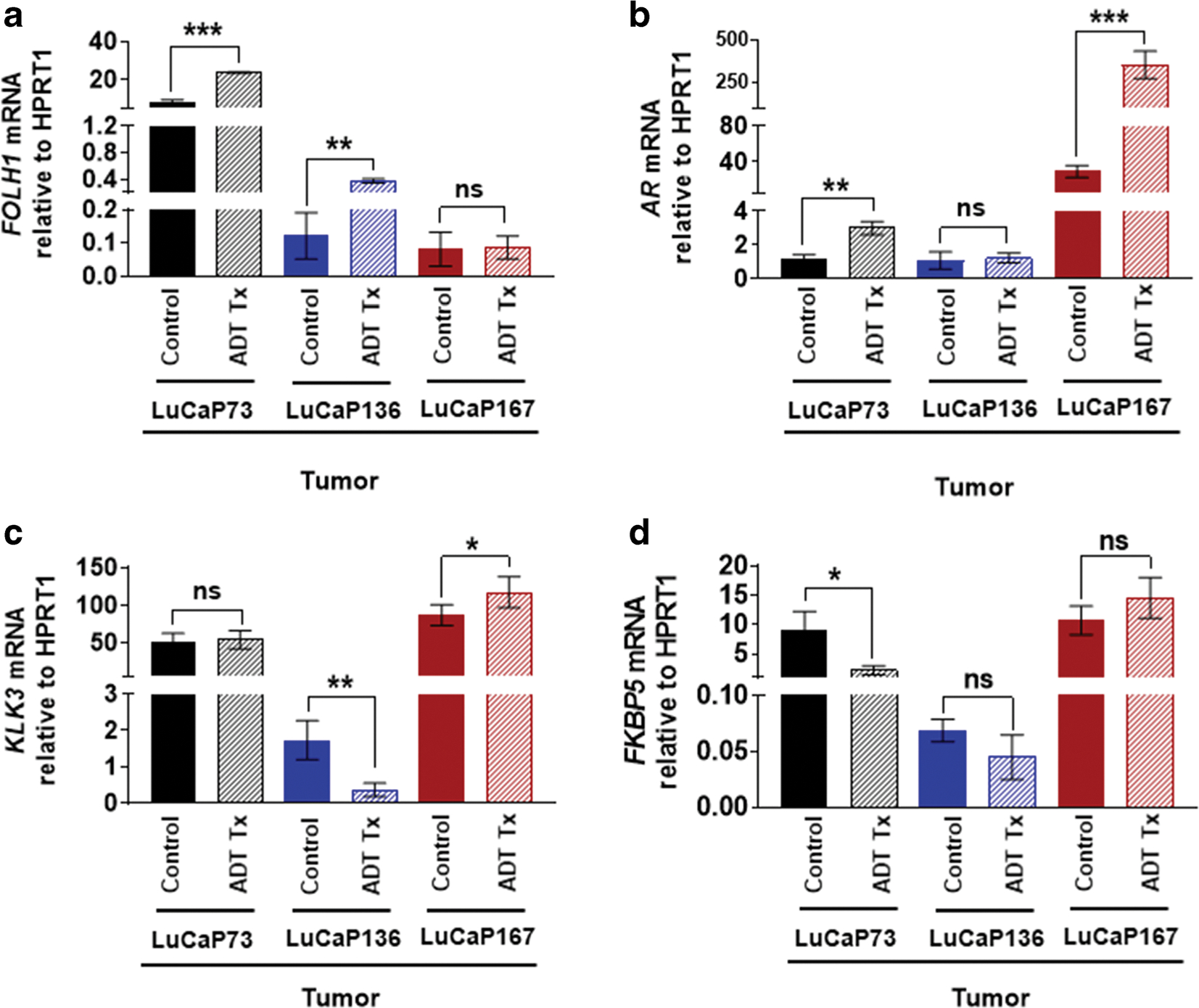
RT-qPCR analysis of FOLH1 (a), AR (b), KLK3 (c), and FKBP5 (d) in LuCaP73, LuCaP136, and LuCaP167 PDXs obtained from control and ADT (degarelix) treated mice; n = 3–4/group; ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001. All the mRNA values were normalized to hypoxanthine-guanine phosphoribosyltransferase 1 (HPRT1).
The AR mRNA levels of control and treated LuCaP167 tumors were 40- to 250-fold higher compared to LuCaP73 and LuCaP136 control and treated tumors, respectively (Fig. 6b, Table 1) and exhibited greater variability across the 3 tumor types compared to the AR protein expression levels determined from IHC (Fig. 5e, Table 1). In response to ADT treatment, AR mRNA levels of both LuCaP73 and LuCaP167 were significantly increased (2–12-fold) compared to controls, while AR mRNA levels of treated LuCaP136 tumors and controls were not significantly different. Although ADT treatment induced compensatory increases in AR for LuCaP73 and LuCaP167, the downstream genes, KLK3 and FKBP5, exhibited differential responses. KLK3 and FKBP5 mRNA levels of LuCaP167 treated tumors were increased compared to controls, but in LuCaP73 treated tumors, KLK3 mRNA levels were similar to controls, and FKBP5 mRNA levels were significantly decreased compared to controls (Fig. 6c–d, Table 1). LuCaP136 treated and control tumors had the lowest AR mRNA levels and exhibited no significant differences in AR mRNA levels (Fig. 6b, Table 1); however, KLK3 and FKBP5 mRNA levels of the treated tumors were decreased (with a significant decrease for KLK3 mRNA) compared to controls (Fig. 6c–d, Table 1).
Discussion
These results show that these LuCaP PDX models which originated from various sites of prostate cancer patients may be useful in providing further insights regarding the variability of PSMA expression associated with ADT treatment responses and progressive disease [29]. In the CS LuCaP models, PSMA levels either increased (LuCaP73) or were unchanged (LuCaP136), whereas in the CR model (LuCaP167), a decrease was observed which may not be attributed to a specific change in PSMA due to low expression levels. It is important to note that the low PSMA expression levels of LuCaP136 and LuCaP167 in a clinical setting would most likely be considered PSMA negative. In these preclinical studies, the tumors have been implanted subcutaneously purposely on the shoulder to facilitate imaging quantitation and minimize background which would not be possible in the clinic. Therefore, preclinical PET imaging would be able to visualize changes in tumors with lower PSMA levels such as LuCaP136, whereas this may not be the case in the clinical setting. Depending upon the location of the tumor within the patient’s body and the associated background as well as clearance from non-target tissues (which is dependent on the time that has elapsed since the injection of the imaging agent), tumors with low PSMA expression levels may be difficult to distinguish from background. Therefore, these PDX models coupled with monitoring PSMA expression levels during a treatment paradigm with PSMA PET imaging may be useful to gain a better understanding of the relationship of PSMA expression in PCa to its genomic signature and therapeutic responses to ADT. Understanding how PSMA expression predicts response to ADT could be useful for future patient management decisions, and having models that reflect the variability in these responses could be particularly useful.
The [18F]DCFPyL imaging and biodistribution results for the CS LuCaP73 tumor model were further confirmed by IHC in which PSMA tumor levels were similarly increased in response to ADT compared to controls. However, for LuCaP136 and LuCaP167, ADT-treated and control tumor samples IHC results were uniformly negative for PSMA, indicating that the low PSMA levels expressed by these two LuCaP models are below the detection limits of the IHC staining. However, this was not the case with the [18F]DCFPyL in vitro saturation studies in which PSMA expression levels were detected in tumor membrane preparations from all three PDXs. In contrast to IHC results, specific binding of [18F]DCFPyL to PSMA was detected in LuCaP136 and LuCaP167 tumor membrane preparations indicating that [18F]DCFPyL is capable of detecting lower levels, which would be expected with this sub-nanomolar affinity ligand and the predicted binding potential (BP) determined from the mathematical binding model, Bmax/Kd (Fig. 1, Table 1) [30, 31]. Sampling error which is inherent with tumor sectioning may also contribute to the differences observed between the PSMA IHC results and in vitro tumor membrane preparation and in vivo results which are representative of the PSMA content of the entire tumor.
The PSMA protein levels as well as T:M and T:B ratios in LuCaP167 tumors were substantially low indicating that the majority of [18F]DCFPyL uptake was not due to specific binding to PSMA but rather non-specific uptake (i.e., tumor blood volume) [32]. LuCaP167 tumors were highly vascularized compared to the CS LuCaP tumors and when treated had similar microvessel densities compared to controls (CD31 IHC; Fig. 5f, Table 1) suggesting that the non-specific uptake may relate to increased tumor blood volume. Thus, a systemic pharmacokinetic alteration of the input function or metabolism of [18F]DCFPyL from ADT treatment would have a greater impact on this non-specific binding compartment resulting in a modest but significant difference in the treated group compared to the controls which may not be attributable to a change in PSMA-targeted binding. Further T:M ratio values < 2 like those observed for LuCaP167 T:M have been shown by statistical analysis to represent primarily non-specific interactions rather than specific targeting [32, 33]. In other biodistribution studies with PC3 tumor xenografts injected with PC3 Wt cells [PSMA negative; no in vitro specific PSMA binding detected (data not shown)], [18F]DCFPyL uptakes in these tumors were comparable and, in some cases, lower than those observed with LuCaP167 (SI Fig. 4). PC3 Wt T:M ratios were slightly lower than LuCaP167 T:M ratios which would further confirm that the majority of the [18F]DCFPyL uptake in LuCaP167 tumors did not represent specific PSMA binding. In contrast, the biodistribution T:M ratios of LuCaP136 were > 2, ~ 5–6, which would be expected to represent PSMA specific binding. Further, these T:M ratios would indicate that LuCaP136 expresses ~3 to 4-fold higher levels of PSMA than LuCaP167 which is in the same rank order as determined in vitro (SI Table 1). Furthermore, because [18F]DCFPyL has high affinity for both human and murine PSMA, the tumor uptakes of [18F]DCFPyL in LuCaP136 and LuCaP167 may be a combination of uptake associated with PSMA expressed on tumor cells as well as tumor neovasculature (although this contribution maybe low) [34, 35].
Similarly, in these LuCaP models, the rank order of the PSMA gene (FOLH1 mRNA) tumor expression levels were the same as PSMA tumor protein levels determined by [18F]DCFPyL, but corresponding changes of mRNA and protein levels in response to ADT differed. Tumor PSMA mRNA levels significantly increased in response to ADT for both CS LuCaP models, whereas only LuCaP73 treated tumor PSMA protein levels significantly increased compared to controls. Errors in the translation of mRNA to a functional protein related to the genomic phenotype may potentially explain this discrepancy between the effect of ADT on PSMA protein and mRNA levels. Similar to the PSMA mRNA results, tumor microvessel densities significantly increased in both CS LuCaPs in response to ADT which may be expected since PSMA has been shown to be associated with tumor neovasculature [35]. The CR LuCaP167 tumor model which had the lowest PSMA expression levels of the three LuCaP models exhibited no change in tumor PSMA mRNA levels in response to ADT treatment which did not parallel the modest significant decrease in [18F]DCFPyL tumor uptake observed in the biodistribution studies which most likely is not representative of a change in specific PSMA uptake.
In contrast to the variability in the LuCaP tumor PSMA levels, androgen receptor tumor levels were similar for the three LuCaP models and not affected by ADT treatment (Fig. 5e). Although the tumor androgen receptor protein levels were unchanged, significant differences were observed in AR (mRNA) and associated downstream genes (Fig. 6b–d). AR mRNA was significantly increased in response to ADT in LuCaP73 and LuCaP167 tumors, whereas for LuCaP136 tumors, AR mRNA was unchanged, but the downstream genes were significantly decreased. LuCaP136, a model with PTEN loss, exhibited repressed AR transcriptional signaling as shown by a minimal baseline gene expression of KLK3 and FKBP5 as compared to PTEN intact LuCaP73 and LuCaP167; upon treatment, the AR that was still present further reduced the levels of KLK3 and FKBP5 gene expression (as evidenced by the lack of effectiveness of ADT on reducing tumor volume compared to LuCaP73) [21]. This reduction of androgen-responsive gene signaling seems to be a characteristic of PTEN loss and has been found to be a frequent genomic alteration observed in castrate-resistant prostate cancer (CRPC) and thus emerged as a potential prognostic biomarker to distinguish indolent tumors from progressive disease by IHC or in situ hybridization of individual tumors [36, 37]. With this example in mind, further PSMA PET imaging studies with additional LuCaP PDXs in a treatment model may serve to establish how changes in PSMA expression levels in response to ADT may be related to the genomic signature and potentially progressive disease.
Although several preclinical studies have reported on the effect of ADT on PSMA, the majority of these studies have used conventional PCa cell lines, which do not represent the genetic diversity observed in PCa patients [18, 20]. Most of these preclinical studies have reported an increase in PSMA levels post-ADT [18, 38, 39]. Collectively, clinical studies indicate that the duration of ADT and type of PCa can have a differential impact on PSMA levels [20, 40–46]. Our results in these PDX models agree with these clinical studies showing variations in response of PSMA expression to ADT. Most PCa patients demonstrate an increase in PSMA expression after short-term ADT and a decrease with long-term ADT; however, a subset of patients do not follow this response pattern [16, 17]. The molecular mechanisms modulating these heterogeneous PSMA responses to ADT has yet to be elucidated, and these genomically well-characterized PDX xenografts models could be important in providing a better understanding of this association and potentially provide clinically relevant information regarding the use of PSMA-targeted therapeutics with or without ADT. For instance, those patients that exhibited increased PSMA levels post-ADT may benefit from a combination therapy including a PSMA-targeted therapeutic such as [177Lu]PSMA-617 which is currently in clinical trials [47]. In contrast, those patients with low to undetectable PSMA levels post-ADT would most likely not benefit from additional PSMA-targeted therapies; however, PET imaging with other agents such as [18F]sodium fluoride, [18F]FAPI, or [18F]fluorodeoxyglucose may provide better guidance towards the most appropriate therapeutics [48–51].
Conclusion
Tumor regression responses to ADT in these LuCaP PDXs were consistent with the spectrum of responses observed in PCa patients ranging from highly sensitive (CS) to resistant (CR). In the CS PDX model with the highest PSMA expression levels, [18F]DCFPyL tumor uptakes were increased in response to ADT indicating an increase in PSMA which is similar to responses to ADT that have been observed clinically in PCa patients. Since these LuCaP PDXs are representative of the molecular heterogeneity of patient PCa tumors, the results obtained using these xenografts may establish the clinical relevance of PSMA expression as a biomarker predictive of ADT response.
Supplementary Material
Funding.
This project was funded by National Cancer Institute, National Institutes of Health. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the US Government.
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
Declarations
Ethics Approval
All animal experiments were conducted in compliance with the protocols approved by Animal Care and Use Committee of National Institutes of Health.
References
- 1.Rawla P (2019) Epidemiology of Prostate Cancer. World J Oncol 10:63–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bravaccini S, Puccetti M, Bocchini M, Ravaioli S, Celli M, Scarpi E, de Giorgi U, Tumedei MM, Raulli G, Cardinale L, Paganelli G (2018) PSMA expression: a potential ally for the pathologist in prostate cancer diagnosis. Sci Rep 8:4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang SS (2004) Overview of prostate-specific membrane antigen. Rev Urol 6(Suppl 10):S13–S18 [PMC free article] [PubMed] [Google Scholar]
- 4.Bouchelouche K, Turkbey B, Choyke PL (2016) PSMA PET and Radionuclide Therapy in Prostate Cancer. Semin Nucl Med 46:522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghosh A, Heston WD (2004) Tumor target prostate specific membrane antigen (PSMA) and its regulation in prostate cancer. J Cell Biochem 91:528–539 [DOI] [PubMed] [Google Scholar]
- 6.Mena E, Lindenberg ML, Shih JH, Adler S, Harmon S, Bergvall E, Citrin D, Dahut W, Ton AT, McKinney Y, Weaver J, Eclarinal P, Forest A, Afari G, Bhattacharyya S, Mease RC, Merino MJ, Pinto P, Wood BJ, Jacobs P, Pomper MG, Choyke PL, Turkbey B (2018) Clinical impact of PSMA-based (18)F-DCFBC PET/CT imaging in patients with biochemically recurrent prostate cancer after primary local therapy. Eur J Nucl Med Mol Imaging 45:4–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rowe SP, Macura KJ, Mena E, Blackford AL, Nadal R, Antonarakis ES, Eisenberger M, Carducci M, Fan H, Dannals RF, Chen Y, Mease RC, Szabo Z, Pomper MG, Cho SY (2016) PSMA-based [F-18]DCFPyL PET/CT is superior to conventional imaging for lesion detection in patients with metastatic prostate cancer. Mol Imaging Biol 18:411–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rahbar K, Afshar-Oromieh A, Seifert R, Wagner S, Schäfers M, Bögemann M, Weckesser M (2018) Diagnostic performance of (18)F-PSMA-1007 PET/CT in patients with biochemical recurrent prostate cancer. Eur J Nucl Med Mol Imaging 45:2055–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sachpekidis C, Kopka K, Eder M, Hadaschik BA, Freitag MT, Pan L, Haberkorn U, Dimitrakopoulou-Strauss A (2016) 68Ga-PSMA-11 dynamic PET/CT imaging in primary prostate cancer. Clin Nucl Med 41:e473–e479 [DOI] [PubMed] [Google Scholar]
- 10.Montorsi F, Fossati N, Gandaglia G, Briganti A (2020) (68)Ga-PSMA-PET/CT scan as primary staging for prostate cancer and its related clinical implications. J Urol:101097JU0000000000001081. [Google Scholar]
- 11.Ekmekcioglu O, Busstra M, Klass ND, Verzijlbergen F (2019) Bridging the imaging gap: PSMA PET/CT has a high impact on treatment planning in prostate cancer patients with biochemical recurrence-a narrative review of the literature. J Nucl Med 60:1394–1398 [DOI] [PubMed] [Google Scholar]
- 12.Perlmutter MA, Lepor H (2007) Androgen deprivation therapy in the treatment of advanced prostate cancer. Rev Urol 9(Suppl 1):S3–S8 [PMC free article] [PubMed] [Google Scholar]
- 13.Boccon-Gibod L, Albers P, Morote J, van Poppel H, de la Rosette J, Villers A, Malmberg A, Neijber A, Montorsi F (2014) Degarelix as an intermittent androgen deprivation therapy for one or more treatment cycles in patients with prostate cancer. Eur Urol 66:655–663 [DOI] [PubMed] [Google Scholar]
- 14.Crawford ED, Heidenreich A, Lawrentschuk N, Tombal B, Pompeo ACL, Mendoza-Valdes A, Miller K, Debruyne FMJ, Klotz L (2019) Androgen-targeted therapy in men with prostate cancer: evolving practice and future considerations. Prostate Cancer P D 22:24–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saranyutanon S, Srivastava SK, Pai S, Singh S, Singh AP (2020) Therapies targeted to androgen receptor signaling axis in prostate cancer: progress, challenges, and hope. Cancers 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ettala O, Malaspina S, Tuokkola T, Luoto P, Löyttyniemi E, Boström PJ, Kemppainen J (2020) Prospective study on the effect of short-term androgen deprivation therapy on PSMA uptake evaluated with Ga-68-PSMA-11 PET/MRI in men with treatment-naive prostate cancer. Eur J Nucl Med Mol I 47:665–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Afshar-Oromieh A, Debus N, Uhrig M, Hope TA, Evans MJ, Holland-Letz T, Giesel FL, Kopka K, Hadaschik B, Kratochwil C, Haberkorn U (2018) Impact of long-term androgen deprivation therapy on PSMA ligand PET/CT in patients with castrationsensitive prostate cancer. Eur J Nucl Med Mol I 45:2045–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soeda F, Watabe T, Naka S et al. (2019) Effect of long-term androgen deprivation therapy (ADE) on [F-18]PSMA-1007 uptake: preclinical study using the prostate cancer xenograft model. J Nucl Med 60 [DOI] [PubMed] [Google Scholar]
- 19.Chang SS, Reuter VE, Heston WD, Hutchinson B, Grauer LS, Gaudin PB (2000) Short term neoadjuvant androgen deprivation therapy does not affect prostate specific membrane antigen expression in prostate tissues. Cancer-Am Cancer Soc 88:407–415 [PubMed] [Google Scholar]
- 20.Hope TA, Truillet C, Ehman EC, Afshar-Oromieh A, Aggarwal R, Ryan CJ, Carroll PR, Small EJ, Evans MJ (2017) Ga-68-PSMA-11 PET imaging of response to androgen receptor inhibition: first human experience. J Nucl Med 58:81–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen HM, Vessella RL, Morrissey C, Brown LG, Coleman IM, Higano CS, Mostaghel EA, Zhang X, True LD, Lam HM, Roudier M, Lange PH, Nelson PS, Corey E (2017) LuCaP prostate cancer patientderived xenografts reflect the molecular heterogeneity of advanced disease and serve as models for evaluating cancer therapeutics. Prostate 77:654–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi CH, Chen X, Fan DX (2019) Development of patient-derived xenograft models of prostate cancer for maintaining tumor heterogeneity. Transl Androl Urol 8:519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basuli F, Zhang X, Woodroofe CC, Jagoda EM, Choyke PL, Swenson RE (2017) Fast indirect fluorine-18 labeling of protein/peptide using the useful 6-fluoronicotinic acid-2,3,5,6-tetrafluorophenyl prosthetic group: A method comparable to direct fluorination. J Labelled Comp Radiopharm 60:168–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakajima T, Mitsunaga M, Bander NH, Heston WD, Choyke PL, Kobayashi H (2011) Targeted, activatable, in vivo fluorescence imaging of prostate-specific membrane antigen (PSMA) positive tumors using the quenched humanized J591 antibody-indocyanine green (ICG) conjugate. Bioconjug Chem 22:1700–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guyader C, Ceraline J, Gravier E et al. (2012) Risk of hormone escape in a human prostate cancer model depends on therapy modalities and can be reduced by tyrosine kinase inhibitors. PLoS One 7:e42252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Pinieux G, Legrier ME, Poirson-Bichat F, Courty Y, Bras-Gonçalves R, Dutrillaux AM, Némati F, Oudard S, Lidereau R, Broqua P, Junien JL, Dutrillaux B, Poupon MF (2001) Clinical and experimental progression of a new model of human prostate cancer and therapeutic approach. Am J Pathol 159:753–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herraiz JL, Espana S, Vaquero JJ, Desco M, Udias JM (2006) FIRST: fast iterative reconstruction software for (PET) tomography. Phys Med Biol 51:4547–4565 [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Pullambhatla M, Foss CA, Byun Y, Nimmagadda S, Senthamizhchelvan S, Sgouros G, Mease RC, Pomper MG (2011) 2-(3-{1-Carboxy-5-[(6-[18F]fluoro-pyridine-3-carbonyl)-amino]-pentyl}-ureido)-pen tanedioic acid, [18F]DCFPyL, a PSMA-based PET imaging agent for prostate cancer. Clin Cancer Res 17:7645–7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaz S, Hadaschik B, Gabriel M, Herrmann K, Eiber M, Costa D (2020) Influence of androgen deprivation therapy on PSMA expression and PSMA-ligand PET imaging of prostate cancer patients. Eur J Nucl Med Mol I 47:9–15 [DOI] [PubMed] [Google Scholar]
- 30.Eckelman WC, Kilbourn MR, Mathis CA (2009) Specific to nonspecific binding in radiopharmaceutical studies: it’s not so simple as it seems! Nucl Med Biol 36:235–237 [DOI] [PubMed] [Google Scholar]
- 31.Eckelman WC, Mathis CA (2006) Targeting proteins in vivo: in vitro guidelines. Nucl Med Biol 33:161–164 [DOI] [PubMed] [Google Scholar]
- 32.Jagoda EM, Lang L, Tokugawa J, Simmons A, Ma Y, Contoreggi C, Kiesewetter D, Eckelman WC (2006) Development of 5-HT1A receptor radioligands to determine receptor density and changes in endogenous 5-HT. Synapse 59:330–341 [DOI] [PubMed] [Google Scholar]
- 33.Jagoda EM, Vaquero JJ, Seidel J, Green MV, Eckelman WC (2004) Experiment assessment of mass effects in the rat: implications for small animal PET imaging. Nucl Med Biol 31:771–779 [DOI] [PubMed] [Google Scholar]
- 34.Roy J, Warner BM, Basuli F, Zhang X, Wong K, Pranzatelli T, Ton AT, Chiorini JA, Choyke PL, Lin FI, Jagoda EM (2020) Comparison of prostate-specific membrane antigen expression levels in human salivary glands to non-human primates and rodents. Cancer Biother Radiopharm 35:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang SS, Reuter VE, Heston WD, Bander NH, Grauer LS, Gaudin PB (1999) Five different anti-prostate-specific membrane antigen (PSMA) antibodies confirm PSMA expression in tumor-associated neovasculature. Cancer Res 59:3192–3198 [PubMed] [Google Scholar]
- 36.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, le C, Koutcher J, Scher H, Scardino PT, Rosen N, Sawyers CL (2011) Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 19:575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wise HM, Hermida MA, Leslie NR (2017) Prostate cancer, PI3K, PTEN and prognosis. Clin Sci (Lond) 131:197–210 [DOI] [PubMed] [Google Scholar]
- 38.Murga JD, Moorji SM, Han AQ, Magargal WW, DiPippo VA, Olson WC (2015) Synergistic co-targeting of prostate-specific membrane antigen and androgen receptor in prostate cancer. Prostate 75:242–254 [DOI] [PubMed] [Google Scholar]
- 39.Evans MJ, Smith-Jones PM, Wongvipat J, Navarro V, Kim S, Bander NH, Larson SM, Sawyers CL (2011) Noninvasive measurement of androgen receptor signaling with a positron-emitting radiopharmaceutical that targets prostate-specific membrane antigen. P Natl Acad Sci USA 108:9578–9582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright GL, Grob BM, Haley C et al. (1996) Upregulation of prostate-specific membrane antigen after androgen-deprivation therapy. Urology 48:326–334 [DOI] [PubMed] [Google Scholar]
- 41.Onal C, Guler OC, Torun N, Reyhan M, Yapar AF (2020) The effect of androgen deprivation therapy on (68)Ga-PSMA tracer uptake in non-metastatic prostate cancer patients. Eur J Nucl Med Mol Imaging 47:632–641 [DOI] [PubMed] [Google Scholar]
- 42.Zacho HD, Petersen LJ (2018) Bone flare to androgen deprivation therapy in metastatic, hormone-sensitive prostate cancer on 68Gaprostate-specific membrane antigen PET/CT. Clin Nucl Med 43:e404–e406 [DOI] [PubMed] [Google Scholar]
- 43.Leitsmann C, Thelen P, Schmid M, Meller J, Sahlmann CO, Meller B, Trojan L, Strauss A (2019) Enhancing PSMA-uptake with androgen deprivation therapy - a new way to detect prostate cancer metastases? Int Braz J Urol 45:459–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aggarwal R, Wei X, Kim W, Small EJ, Ryan CJ, Carroll P, Cooperberg M, Evans MJ, Hope T (2018) Heterogeneous flare in prostate-specific membrane antigen positron emission tomography tracer uptake with initiation of androgen pathway blockade in metastatic prostate cancer. Eur Urol Oncol 1:78–82 [DOI] [PubMed] [Google Scholar]
- 45.Meller B, Bremmer F, Sahlmann CO et al. (2015) Alterations in androgen deprivation enhanced prostate-specific membrane antigen (PSMA) expression in prostate cancer cells as a target for diagnostics and therapy. EJNMMI Res 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emmett L, Yin C, Crumbaker M, Hruby G, Kneebone A, Epstein R, Nguyen Q, Hickey A, Ihsheish N, O’Neill G, Horvath L, Chalasani V, Stricker P, Joshua AM (2019) Rapid modulation of PSMA expression by androgen deprivation: serial Ga-68-PSMA-11 PET in men with hormone-sensitive and castrate-resistant prostate cancer commencing androgen blockade. J Nucl Med 60:950–954 [DOI] [PubMed] [Google Scholar]
- 47.Hofman MS, Violet J, Hicks RJ, Ferdinandus J, Thang SP, Akhurst T, Iravani A, Kong G, Ravi Kumar A, Murphy DG, Eu P, Jackson P, Scalzo M, Williams SG, Sandhu S (2018) [(177)Lu]-PSMA-617 radionuclide treatment in patients with metastatic castration-resistant prostate cancer (LuPSMA trial): a single-centre, single-arm, phase 2 study. Lancet Oncol 19:825–833 [DOI] [PubMed] [Google Scholar]
- 48.Khreish F, Rosar F, Kratochwil C, Giesel FL, Haberkorn U, Ezziddin S (2020) Positive FAPI-PET/CT in a metastatic castration-resistant prostate cancer patient with PSMA-negative/FDG-positive disease. Eur J Nucl Med Mol Imaging 47:2040–2041 [DOI] [PubMed] [Google Scholar]
- 49.Harmon SA, Bergvall E, Mena E, Shih JH, Adler S, McKinney Y, Mehralivand S, Citrin DE, Couvillon A, Madan RA, Gulley JL, Mease RC, Jacobs PM, Pomper MG, Turkbey B, Choyke PL, Lindenberg ML (2018) A prospective comparison of (18)F-sodium fluoride PET/CT and PSMA-targeted (18)F-DCFBC PET/CT in metastatic prostate cancer. J Nucl Med 59:1665–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Letellier A, Johnson AC, Kit NH, Savigny JF, Batalla A, Parienti JJ, Aide N (2018) Uptake of radium-223 dichloride and early [(18)F]NaF PET response are driven by baseline [(18)F]NaF parameters: a pilot study in castration-resistant prostate cancer patients. Mol Imaging Biol 20:482–491 [DOI] [PubMed] [Google Scholar]
- 51.Bauckneht M, Capitanio S, Donegani MI et al. (2019) Role of baseline and post-therapy 18F-FDG PET in the prognostic stratification of metastatic castration-resistant prostate cancer (mCRPC) patients treated with radium-223. Cancers (Basel) 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


