Abstract
PlasmoDB (http://PlasmoDB.org) is the official database of the Plasmodium falciparum genome sequencing consortium. This resource incorporates finished and draft genome sequence data and annotation emerging from Plasmodium sequencing projects. PlasmoDB currently houses information from five parasite species and provides tools for cross-species comparisons. Sequence information is also integrated with other genomic-scale data emerging from the Plasmodium research community, including gene expression analysis from EST, SAGE and microarray projects. The relational schemas used to build PlasmoDB [Genomics Unified Schema (GUS) and RNA Abundance Database (RAD)] employ a highly structured format to accommodate the diverse data types generated by sequence and expression projects. A variety of tools allow researchers to formulate complex, biologically based queries of the database. A version of the database is also available on CD-ROM (Plasmodium GenePlot), facilitating access to the data in situations where Internet access is difficult (e.g. by malaria researchers working in the field). The goal of PlasmoDB is to enhance utilization of the vast quantities of data emerging from genome-scale projects by the global malaria research community.
SCOPE
The disease malaria is caused by members of the genus Plasmodium. There are more than 100 recognized species of Plasmodium, four of which infect humans. One species, Plasmodium falciparum, is the causative agent of cerebral malaria and is responsible for the deaths of more than 1 million people each year, mostly children in sub-Saharan Africa. The P.falciparum genome project was initiated in 1996 by an international consortium (1), and PlasmoDB, the official database of this consortium, went live in June 2000 (2).
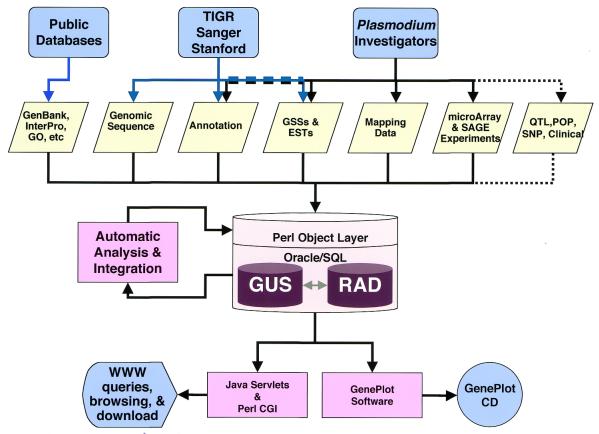
While the data sources and available tools at PlasmoDB have increased dramatically over the last year, the philosophy behind PlasmoDB has remained unchanged. Our goals, as illustrated in Figure 1, are: (i) to provide a single, user-friendly point of access for the data emerging from the three genome sequencing centers involved in the sequencing of Plasmodium genomes (Sanger, Stanford and TIGR/NMRC) and from the malaria research community; (ii) to provide tools for viewing and querying both finished and unfinished genomic sequence data and ancillary information; (iii) to integrate genomic data with other types of data, including (but not limited to) gene expression data and mapping data; (iv) to provide a database with the resources and tools to handle cross-species comparisons as data from other Plasmodium species become available; and (v) to ensure that scientists everywhere, including researchers in the field and other Internet-limited areas, have access to the data via an easily-obtainable, user-friendly and query-capable CD-ROM version of the database, Plasmodium GenePlot.
Figure 1.
Data for PlasmoDB is gathered from several sources and stored in relational databases, GUS and RAD, before it is integrated and processed, then served to users on the World Wide Web or on CD. Dashed lines indicate items that will be added in the future; QTL, quantitative trait loci; POP, populations studies; SNP, single nucleotide polymorphisms.
CONTENT OF THE CURRENT RELEASE
Data
Version 3.2 of the database contains sequence (Genomic, GSS and EST) and mapping data (microsatellite and optical), official gene annotations and expression data (EST, SAGE and microarray). Additional data sets are created from the genomic sequence data in PlasmoDB via a series of automated analyses, including gene predictions (using various algorithms), protein feature predictions (signal peptides, transmembrane domains, secondary structure, hydrophobicity plots, AA content, Pfam or ProSite domains, etc.) and Gene Ontology (GO) function predictions.
The genomic sequence data available in the current release of PlasmoDB (v3.2) consists of finished and unfinished sequences for five Plasmodium species (P.falciparum, P.yoelii, P.chabaudi, P.berghei and P.vivax). In the October 14, 2001 release, 7 of the 14 P.falciparum chromosomes are complete, and the entire genome is represented by 1183 contiguous sequences (contigs). Data for five additional Plasmodium species (including 5-fold genomic coverage for P.yoelii, 3-fold coverage for P.knowlesi, and various GSS, YAC and EST sequences for these speicies, P.berghei, P.chabaudi and P.vivax) are also available, and have been mapped to the P.falciparum genomic sequence. Additional data from ongoing sequencing projects focused on several Plasmodium species will be incorporated into PlasmoDB as they become available.
Official annotations, automated predictions, mapping information and expression data are available for P.falciparum. BLAST similarities to the NCBI non-redundant protein database are available for P.falciparum and P.yoelii, as are similarity comparisons between these two species (Fig. 2). All sequence information is available as BLAST-searchable databases, and sequence data along with any accompanying official annotations or automated analyses are available for download in FASTA, GenBank and EMBL file formats.
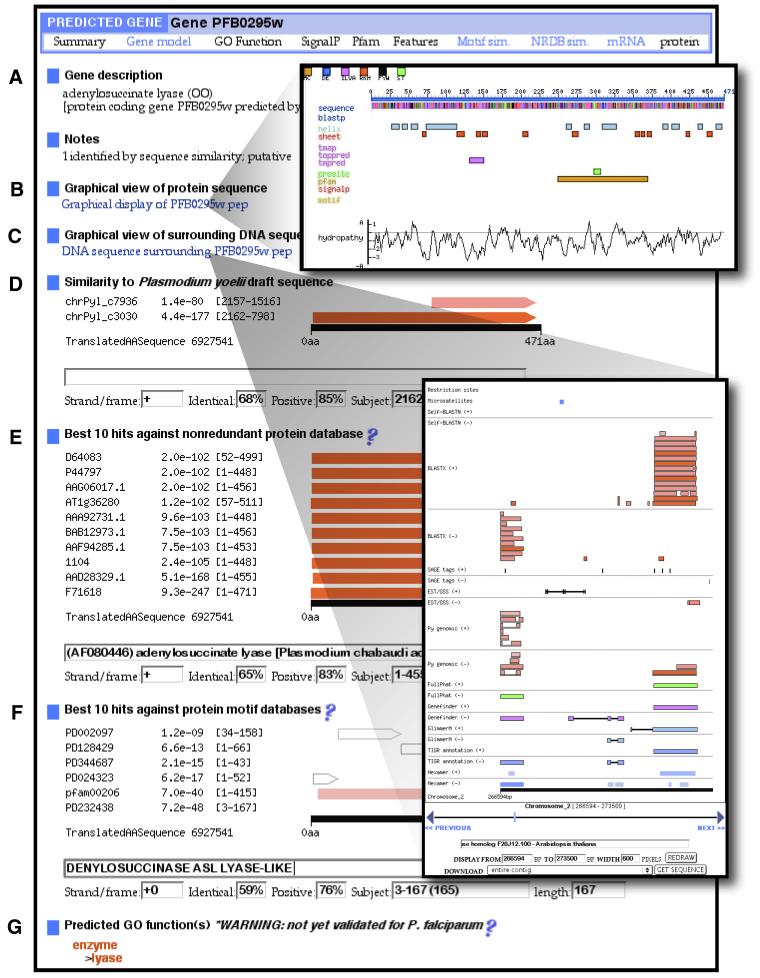
Figure 2.
Composite of three screen shots demonstrating PlasmoDB features for the adenylosuccinate lyase gene, PFB0295w, located on Chr2. The official annotation from TIGR is shown in (A). A graphical view of protein features is available from the link in (B), which shows BLAST hits, predicted secondary structure, domains and motifs and signal peptides. A graphical view of the genomic sequence surrounding the gene is available from the link in (C) which shows BLASTX similarities to the non-redundant protein database (NRDB; the quality of the hit similarity is indicated by the color of the box) proteins and the location of SAGE tags, ESTs and GSSs, automated gene predictions and official annotation. Similarities to P.yoelii are shown in (D) as well as BLAST hits to the NRDB (E), predicted protein motifs (F) and predicted GO functions (G). Each part of the display is a link to further information.
Tools/queries
In addition to the usual data-browsing interface (much of which is accessible from the front page), PlasmoDB offers a powerful, yet easy-to-use, query interface for identifying genes of interest. Basic queries on sequence information stored in GUS (3) cover text keyword matching, sequence similarities, intron/exon structure, protein domains, location, prediction method, predicted function and expression data stored in RAD (4). A history mechanism tracks queries executed in each session allowing users to combine query results, revisit searches or download sequences. A BLAST server is available for both DNA and predicted amino acid sequences.
Plasmodium GenePlot
The Plasmodium GenePlot CD-ROM is a stand-alone platform-independent compilation of all available finished and unfinished DNA sequences from the P.falciparum genome. The data are accessible online, or via CD-ROM. Data are organized by chromosome, and the results of three different gene prediction algorithms and BLAST analysis against the GenBank NRDB are presented graphically, along with official annotations (where available). Annotations, BLAST hits and gene predictions are dynamically searchable by chromosome. Features and annotations can be reviewed via mouse pointer, and features are retrievable by clicking on the image. Tools provided on the CD allow the user to manipulate (translate, reverse complement, convert file format, etc.) any DNA sequence, either from the CD or provided by the user. The Plasmodium GenePlot CD can be used on any computer platform with a compatible web browser (Netscape Communicator 4.7 or Internet Explorer 5.0).
Tutorials and information on how to use the database
In an effort to provide better guidance for the use of tools and database queries that may be new to users, a graphical web-based tutorial has been established. This tutorial is located on the front page of the database and registration is not required for its use. A ‘NEWS’ link has also been added to explain features, data and tools added since the last release, and news stories are archived under ‘What’s new at PlasmoDB’. A ‘?’ link has been added to the menus to provide information on each of the available tools/queries.
FUTURE PLANS
1. PlasmoDB will incorporate official gene and other sequence annotation released by the sequencing centers, while retaining automated predictions to provide alternative views.
2. GenePlot CDs will be linked to entries in PlasmoDB (when Internet access is available) and PlasmoDB will be linked to other databases as these become available.
3. We anticipate rapid growth in the volume of gene expression data and sequence information on related species.
4. New data types to be incorporated include population polymorphisms (including SNPs), QTL data and metabolic pathways.
5. Finally, we look forward to feedback from the malaria community to guide the development and prioritization of new features and data.
DATA ACCESS AND DATA DEPOSITION
PlasmoDB is located at http://PlasmoDB.org and the Plasmodium GenePlot CD is available via the same URL (interactively or as a downloadable image) or as a stand-alone CD-ROM. To obtain the GenePlot/WHO malaria CD-ROM set, contact Ross Coppel, Department of Microbiology, Monash University, Clayton, Victoria 3800, Australia; Email: ross.coppel@med.monash.edu.au. To report errors, submit suggestions or contribute data to the database, contact PlasmoDB at help@PlasmoDB.org or contact the corresponding author.
ADDITIONAL INFORMATION
1. Data release policies for P.falciparum (http://PlasmoDB.org/drpf.shtml) and P.yoelii (http://PlasmoDB.org/drpy.shtml) genomic sequence data. Acceptance of these policies is required for full access to PlasmoDB.
2. Sources. Links to the various data sets that are available on PlasmoDB with links to the original data source and statistics (number of nucleotides, number of contigs, release date, contact name, etc.); http://PlasmoDB.org/bdbs.shtml.
3. Status. Statistics on progress of the P.falciparum genome sequencing effort. Links to each of the original data sources are provided along with statistics on the estimated size of each chromosome, release date for current data, status of sequence assembly and number of nucleotides available; http://PlasmoDB.org/about_data.shtml.
4. Tutorial. Walk through a graphic tutorial on the use of PlasmoDB; http://PlasmoDB.org/Tutorial.shtml.
Acknowledgments
ACKNOWLEDGEMENTS
Financial support for PlasmoDB was provided by the Burroughs Wellcome Fund. Additional computational support was provided by Rahul Dave, Haralabos Tikos and the Eniac2000 project (supported in part through equipment donations from Intel and by the University of Pennsylvania Genomics Institute).
We thank the numerous researchers who have collaborated with, and contributed to, PlasmoDB by depositing data (both published and unpublished) in the database, by making software available and by making useful suggestions on how to improve this community resource.
We wish to thank the scientists and funding agencies comprising the international Malaria Genome Project for making sequence data from the genome of P.falciparum (3D7) public prior to publication of the completed sequence. The Sanger Centre (UK) provided sequences for chromosomes 1, 3–9 and 13, with financial support from the Wellcome Trust. A consortium composed of The Institute for Genome Research (TIGR) along with the Naval Medical Research Center (USA), sequenced chromosomes 2, 10, 11 and 14, with support from NIAID/NIH, the Burroughs Wellcome Fund and the Department of Defense. The Stanford Genome Technology Center (USA) sequenced chromosome 12, with support from the Burroughs Wellcome Fund.
Preliminary sequence and/or preliminary annotated sequence data from the P.yoelii genome was obtained from the TIGR website (www.tigr.org). This sequencing program is carried on in collaboration with the Naval Medical Research Center and is supported by the US Department of Defense. Given that this information is considered preliminary and may contain inaccuracies, it is expected that no one will publish analyses, based on the preliminary data contained in this web site, or of genes on a whole-chromosome or genome scale, without prior permission from TIGR, which plans to publish the completed and annotated sequence in a peer-reviewed journal.
REFERENCES
- 1.Fletcher C. (1998) The Plasmodium falciparum Genome Project. Parasitol. Today, 14, 342–344. [DOI] [PubMed] [Google Scholar]
- 2.The Plasmodium Genome Database Collaborative (2001) PlasmoDB: an integrative database of the Plasmodium falciparum genome. Tools for accessing and analyzing finished and unfinished sequence data. Nucleic Acids Res., 29, 66–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davidson S., Crabtree,J., Brunk,B.P., Schug,J., Tannen,V., Overton,G.C. and Stoeckert,C.J.,Jr (2001) K2/Klesli and GUS: experiments in integrated access to genomic data sources. IBM Systems J., 40, 512–531. [Google Scholar]
- 4.Stoeckert C., Pizarro,A., Manduchi,E., Gibson,M., Brunk,B., Crabtree,J., Schug,J., Shen-Orr,S. and Overton,C.G. (2001) A relational schema for both array-based and SAGE gene expression experiments. Bioinformatics, 17, 300–308. [DOI] [PubMed] [Google Scholar]