Significance
Nanoparticles (NPs) have shown great potential for cancer diagnosis and treatment. However, poor delivery efficiency of NPs has been proved in the tumor extracellular matrix (ECM) in vivo. Though various methods (e.g., modulation of NP size, shape, surface charge, rigidity, or tumor microenvironment) have been developed to enhance NP diffusion, the efficiency is still unsatisfying and the underlying mechanism of the influence of tumor ECM physical properties on NPs diffusion remains unexplored. In this work, we performed molecular dynamics (MD) simulations and in vitro experiments to explore the diffusion mechanism of NPs in tumor ECM with different density, stiffness, and structure. And, we proposed an estimation matrix to predict NP diffusion efficiency according to clinical tumor physical properties.
Keywords: physical microenvironment, tumor extracellular matrix, nanoparticle diffusion
Abstract
Nanoparticles (NPs) are confronted with limited and disappointing delivery efficiency in tumors clinically. The tumor extracellular matrix (ECM), whose physical traits have recently been recognized as new hallmarks of cancer, forms a main steric obstacle for NP diffusion, yet the role of tumor ECM physical traits in NP diffusion remains largely unexplored. Here, we characterized the physical properties of clinical gastric tumor samples and observed limited distribution of NPs in decellularized tumor tissues. We also performed molecular dynamics simulations and in vitro hydrogel experiments through single-particle tracking to investigate the diffusion mechanism of NPs and understand the influence of tumor ECM physical properties on NP diffusion both individually and collectively. Furthermore, we developed an estimation matrix model with evaluation scores of NP diffusion efficiency through comprehensive analyses of the data. Thus, beyond finding that loose and soft ECM with aligned structure contribute to efficient diffusion, we now have a systemic model to predict NP diffusion efficiency based on ECM physical traits and provide critical guidance for personalized tumor diagnosis and treatment.
Nanoparticles (NPs), as an effective platform to deliver medical agents to tumor tissues, have shown great potential for cancer diagnostics and treatment (1–3). However, the therapeutic efficacy in vivo is limited by the poor efficiency of NPs’ targeted delivery. A large meta-analysis showed that only 0.7% of the administered dose of NPs can accumulate in solid tumors (1) and experimental results showed that even fewer NPs (~0.0014%) could reach tumor cells (4). Therefore, improving NP transport in tumor tissues is crucial for cancer therapy. NPs undergo complex transport processes in vivo to enter target cells (5), while tumor microenvironment (TME) brings diverse barriers to NP transport. For instance, excessive proliferation of cells and production of extracellular matrix (ECM) components lead to direct steric obstacles for NP transport (5, 6). Furthermore, immature and leaky tumor microvessels, collapsed lymphatics, and, consequently, elevated interstitial fluid pressure induce an impaired or even disappeared convention in the stromal space; thus, NPs can only rely on diffusion in such a dense stroma (5, 7, 8). Additionally, compression and heterogeneity of tumor vasculature give rise to insufficient blood perfusion to certain tumor regions, which causes heterogeneity of NP diffusion and distribution in the TME (5, 9–11).
Among various barriers, tumor ECM forms a main steric obstacle for NP diffusion. In vivo studies have demonstrated that a majority of NPs are restricted near the perivascular region and trapped in the tumor ECM instead of diffusing successfully into the deep tumor area (4, 12). Tumor ECM is a complex, porous, and highly adhesive network composed of a variety of biological macromolecules (e.g., collagens, proteoglycans, and hyaluronan) (13, 14). Compared with normal tissues, tumor ECM is associated with a series of complex and gradual pathological changes (15, 16). For instance, tumor ECM shows an excessive deposition due to the loss of homeostasis of ECM component production and degradation, leading to significantly increased ECM density and decreased pore size (16, 17). Elevated collagen deposition and fiber cross-linking significantly increase tumor ECM stiffness (16, 18, 19). Besides, tumor ECM shows structural changes as a result of ECM remodeling. Due to the increased cell contraction, fibroblast activation, and collagen cross-linking enzyme expression, fine and twisted collagen fibers are gradually reorganized, forming a highly aligned and elongated architecture (16, 20–23). These ECM physical features have recently been recognized as new hallmarks of tumors (24). More importantly, abnormal ECM properties are closely related to tumor progression. The tumor ECM presents as a lax and porous structure with few collagen fibers and a minimal increase in stiffness in the early stage. Increased collagen deposition and cross-linking of reticular fibers induce enhanced stiffness with tumor growth, while a rigid and aligned ECM is generated in the late grade (25). All the changes in tumor ECM physical properties give rise to inevitable resistance to NP diffusion. Therefore, it is of great importance to understand the effect of tumor ECM physical properties on NP diffusion in solid tumors.
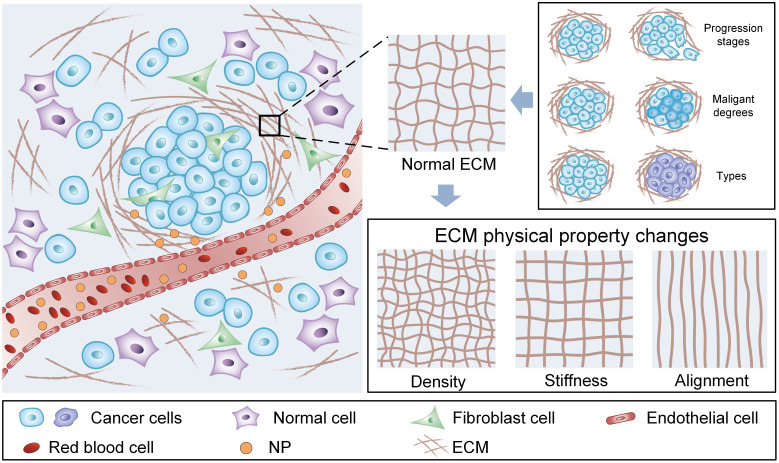
A variety of methods have been developed to enhance NP diffusion in solid tumors. For instance, effective enhancement of NP diffusion in tumor ECM has been obtained by modulating the physiochemical properties of NPs, such as particle size (26, 27), shape (28), surface charge (29, 30), and rigidity (31, 32). In addition, with the understanding of tumor pathology, an increasing number of studies have focused on the modulation of the TME (33). Physical and chemical methods have been used to degrade tumor ECM. For example, NPs diffuse into deep tumor tissues more efficiently through surface modification with functionalized collagenase or hyaluronidase to degrade the tumor ECM (34, 35). The pulsed high-intensity focused ultrasound (36) and hyperthermia (37) are also used to damage ECM components, especially collagen, to improve the NP diffusion efficiency. However, due to the complicated tumor structure, various tumor types, and different growth stages and physiological states, the underlying NP diffusion mechanism in tumor ECM remains elusive (Fig. 1).
Fig. 1.
Extracellular matrix (ECM) physical properties govern the diffusion of NPs in the tumor microenvironment. Schematic illustration of tumor ECM physical properties. Tumor ECM shows different densities, stiffness, and structure in different tumor progression stages, malignant degrees, and tumor types.
In this study, we characterized the clinical gastric tumor samples to observe the pathological and physical properties of tumor ECM, and then we performed experiments on NP distribution in decellularized clinical tumor tissues to confirm the limited NP transport. Furthermore, we performed molecular dynamics (MD) simulations (38, 39) and in vitro single-particle tracking experiments in collagen to investigate the NP diffusion mechanism in tumor ECM with different physical properties. Finally, we developed an estimation matrix model with evaluation scores of NP diffusion through comprehensive analyses of parameters. We aimed to illuminate how ECM physical properties influence particle diffusion performance, reveal the underlying motion mechanism, and predict NP diffusion ability to guide personalized tumor diagnosis and treatment.
Results
Inefficient NP Diffusion in Decellularized Clinical Tumor Tissues.
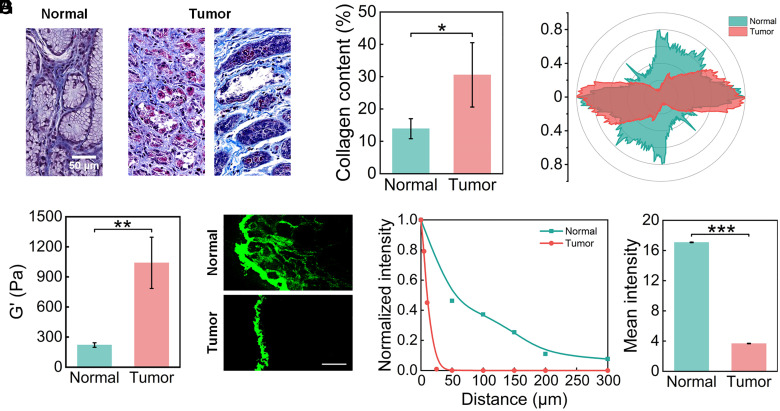
To identify the changes in ECM physical properties, we characterized the density and structure of normal and tumor tissues from clinical samples. Masson’s trichrome staining was performed in both gastric cancer tissues and paracancerous tissues to highlight ECM components especially collagen fibers (Fig. 2A). We observed loose and regular collagen fibers that form network structures in normal tissues while dense and disordered collagen fibers in tumor tissues. In particular, we found that collagen fibers tend to rearrange themselves into one direction and have an aligned appearance. To further quantify the results, we calculated the collagen content (Fig. 2B) of normal and tumor samples from five different patients and data from six visual fields were averaged for each sample. We observed elevated collagen content in tumor tissues (30.54 ± 9.96%) compared with normal tissues (13.90 ± 3.10%). The collagen fiber orientation distribution in tumor tissues shows stronger directionality than that in normal tissues (Fig. 2C). We also characterized the stiffness of normal and tumor tissues by a rheometer. Normal and tumor tissue samples from four different patients were analyzed, and we repeated the tests three times for each sample. The storage modulus (G’) of tumor tissues (1040.6 ± 255.5 Pa) is 4.7-fold higher than that of normal tissues (221.4 ± 22.7 Pa) (Fig. 2D). These results indicate, as expected, that tumor ECM exhibits significant changes in physical properties, including increased density, elevated stiffness, and enhanced alignment compared with normal tissues.
Fig. 2.
Characterization of NP diffusion in normal and tumor tissues from clinical samples. (A) Characterization of normal and gastric tumor tissues through Masson staining. (B) Collagen content of normal and tumor tissues (N = 5). (C) Representative collagen fiber orientation distribution of normal and tumor tissues. Stronger directionality of the tumor tissue indicates a more aligned ECM. (D) Stiffness of normal and tumor tissues (N = 4). (E) Fluorescence images exhibit the diffusion of NPs in decellularized normal and tumor tissues over 48 h. (Scale bar, 100 μm) (F) The fluorescence intensity distribution of NPs in decellularized normal and tumor tissues from the tissue periphery to center in E. (G) The mean fluorescence intensity of NPs in decellularized normal and tumor tissues in E. (One-way ANOVA, ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001).
To further explore whether these physical property changes could affect NP diffusion, NP distribution in decellularized normal and tumor tissue matrices from clinical gastric tumor samples (40) was visualized. In order to exclude the effect of the vascularity, we characterized the vascularity of the normal and tumor tissues before the experiments, and then we selected the tumor tissue samples with vascularity similar to that of the normal tissues (SI Appendix, Fig. S1) to perform further experiments. Polystyrene (PS) fluorescent NPs with a diameter of 80 nm were used in our experiments, whose morphology was shown in transmissionelectron microscopy (TEM) images (SI Appendix, Fig. S2A). The average hydrodynamic diameter of the PS NPs is 112.2 ± 0.62 nm and the average zeta-potential is −28.2 ± 0.09 mV. The fluorescence intensity in tumor tissue was much weaker than that in normal tissue (Fig. 2E), indicating limited NP diffusion in tumor tissues. The result was further confirmed by fluorescence intensity analysis as a function of distance along the direction of from tissue surface to center (Fig. 2F). The diffusion ability of NPs in normal tissue is significantly greater than that in tumor tissue. Fluorescent NPs can penetrate into normal tissue at a depth of about 300 μm while fluorescence could only be observed on the periphery in tumor tissue. In addition, the overall mean fluorescence intensity of the whole tumor tissue area is significantly decreased from 17.08 ± 0.04 to 3.68 ± 0.03, compared with the normal tissue (Fig. 2G). Thus, ECM physical property changes in tumors surely have a great effect on the NP diffusion, but their respective influence, as well as the mechanism, remain to be investigated.
Dense ECM Limits NP Diffusion.
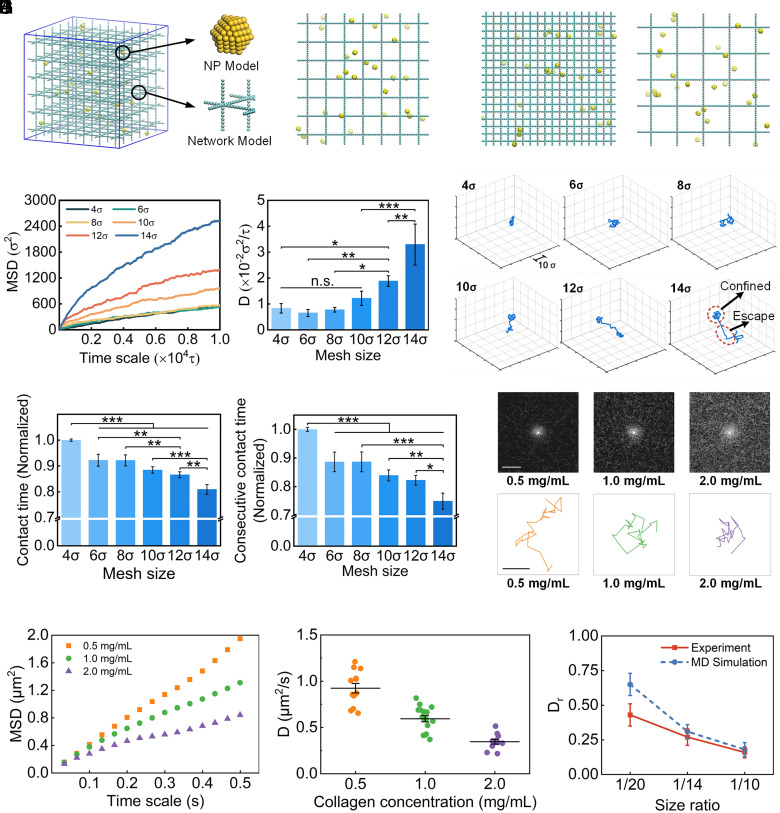
Coarse-grained MD (CGMD) simulations were performed to explore how ECM density influences the NP diffusion. We constructed a model system composed of NPs and cross-linked polymer network and the NPs were randomly distributed in the network (38, 39) (Fig. 3A). According to the pathological characteristics of tumor ECM, the cross-linked networks with different densities were built by adjusting their mesh sizes. We constructed networks with mesh sizes of 4, 6, 8, 10, 12, and 14 (1.6-fold to 5.6-fold of NP size), imitating the change from dense to sparse ECM with tumor progression. The representative network densities (4 and 14) are shown in Fig. 3B. The mean square displacement (MSD) and diffusion coefficient (D) of NPs in a network with different densities were calculated to describe NP diffusion ability.
Fig. 3.
Effect of ECM density in tumor microenvironment on NP diffusion. (A) The molecular model in MD simulation. The enlarged drawing shows details of the NP model and cross-linked ECM network model. (B) Representative network densities with mesh sizes of 4σ and 14σ. (C) The MSD values and calculated diffusion coefficient of NPs in networks with different mesh sizes. (D) Representative trajectories of NPs in networks with increased mesh sizes. (E) The normalized contact time of NPs and consecutive contact time in the network during simulation. (F) Reflectance confocal images of collagen I with increased concentrations. (Scale bar, 50 μm) (G) Typical trajectories of NPs in collagen I with increased densities. (Scale bar, 1 μm) (H) MSD values of NPs as a function of time in collagen I. (I) Diffusion coefficient values of NPs in collagen I on a time scale of 0.5 s. (J) The comparison of the relative diffusion coefficient () for the same size ratios (NP/Pore) in experiment and MD simulation. (One-way ANOVA with Tukey’s post hoc test, n.s., no significance, P > 0.05; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001).
The MSDs of NPs in different networks increase with decreasing ECM density, though there is no significant difference in cases with high density (4, 6, and 8) (Fig. 3C). We obtained the corresponding diffusion coefficients of NPs by linear fitting the MSDs versus the time scale from 2,000 to 10,000. Similarly, the results showed low diffusion coefficients in dense networks (4, 6, and 8). However, with a further increase in the mesh size (or the decrease of density), the diffusion coefficients increase significantly, indicating enhanced diffusion ability for NPs in a loose network. In particular, the diffusion coefficients in network-10, network-12, and network-14 are about 1.5-fold, 2.3-fold, and 4.0-fold higher than those in network-4, respectively. The simulation results demonstrate a negative relationship between NP diffusion ability and network density. A similar phenomenon has also been observed in a previous study where the diffusion coefficients of nanospheres and nanorods both increased with the increase in mesh size (38). The results also suggest that NP diffusion ability will increase significantly once the pore size of ECM reaches a certain value that is three times larger than the NP size in our model. This implies that a minimum pore size (at least four times of the NP size) of ECM is required to effectively enhance NP diffusion.
We further tracked the NP trajectory during the simulation to analyze the mechanism. We observed two main motion states for NPs in networks, which could be summarized as “confined” and “escape”. When NPs interact with networks, they are trapped in meshes and attached to polymer chains or nodes, swinging or rotating around the networks. Once getting rid of the confinement, NPs show long-distance diffusion to pass through the meshes. The representative trajectories of NPs (Fig. 3D) reveal that NPs experience a longer constraint and more limited diffusion regions in dense networks while they escape from confinement more easily in larger meshes. Thus, we hypothesized that increased network density induces increased contact frequency between NPs and networks, which results in a stronger impediment for NP diffusion. To test this, we calculated the average contact time between NPs and network structure (Fig. 3E). The “contact time” is defined as when the distance between the centroid of NPs and any network bead is smaller than 2σ. Furthermore, to describe the network confinement to NP diffusion, we calculated the consecutive contact time of NPs and network structure for three or more continuous contact frames (Fig. 3E). We observed that the contact time between NPs and network is shorter when the mesh size of the network is larger with the same stiffness and structure. For example, the normalized contact time and the consecutive contact time decrease from 1.0 to about 0.8 and 0.75, respectively, when mesh size increases from 4σ to 14σ. The results indicate that NPs tend to be impeded and blocked in the dense network due to frequent interactions between NPs and the network.
To verify the simulation results, NP diffusion in the hydrogel was examined in vitro using collagen I with different densities. The morphology of PS fluorescent NPs (~200 nm in diameter) was revealed in TEM images (SI Appendix, Fig. S2B) and their average hydrodynamic diameter (267.2 ± 3.97 nm) and zeta-potential (−28.3 ± 1.04 mV) were also characterized. Through reflectance confocal images, we observed that the density significantly increases with increasing collagen concentration (Fig. 3F). To investigate the diffusion behavior of NPs, their trajectories were obtained by tracking the movement of the single NP in collagen under a confocallaser scanning microscope (CLSM) (41, 42). The representative trajectories during a period of around 1.0 s for each case are shown in Fig. 3G. The time scale-dependent MSD values were quantified to evaluate the NP diffusion ability (Fig. 3H). The higher MSD values across all time scales in 0.5 mg/mL collagen indicate the stronger diffusion ability of NPs. Diffusion coefficients of individual NP were also calculated (Fig. 3I). We observed relatively high diffusion coefficients in less dense collagen (0.5 mg/mL) with a mean value of approximately 0.92 ± 0.18 μm2/s. The diffusion coefficients decrease with increasing collagen concentrations, with an average of 0.59 ± 0.13 μm2/s and 0.35 ± 0.08 μm2/s in collagen with concentration of 1.0 mg/mL and 2.0 mg/mL, respectively.
To further validate our models with significantly smaller NP sizes, we performed additional diffusion experiments in collagen I hydrogels with smaller NPs (RITC-labeled mesoporous silica NPs, size of 52.06 ± 3.60 nm) (SI Appendix, Fig. S3A). NPs were allowed to diffuse into the collagen hydrogels with concentrations of 0.7 mg/mL, 1.0 mg/mL, and 1.5 mg/mL in μ-slide channels. The diffusion of NPs at a fixed time (2 h) was imaged using CLSM, and we observed decreased penetration depth in the collagen channel with increasing concentration from 0.7 mg/mL to 1.5 mg/mL (SI Appendix, Fig. S3B). The mean fluorescence intensity of NPs along penetration depth is 2.4 fold and 1.9 fold higher in 0.7 mg/mL and 1.0 mg/mL collagen hydrogels at penetration depth of 50 μm than that in 1.5 mg/mL collagen hydrogel, respectively (SI Appendix, Fig. S3C). Besides, NPs in 0.7 mg/mL collagen hydrogel are able to diffuse deeper than 300 μm, while there is almost no NPs at a depth of 200 μm for 1.0 mg/mL and 1.5 mg/mL collagen hydrogels. NPs show a diffusion coefficient (D) of 2.67 μm2/s in 0.7 mg/mL collagen hydrogel compared to 0.92 μm2/s in 1.0 mg/mL collagen hydrogel and 0.52 μm2/s in 1.5 mg/mL collagen hydrogel (SI Appendix, Fig. S3D). The experimental results indicate that the diffusion ability of NPs increases with increasing ECM pore size (or the decrease of density), which matches well with the simulation results.
We further set up a framework to quantitatively compare the diffusion coefficients in simulation and experiments (SI Appendix, Table S1). Since direct comparison is challenging because all qualities are unitless in simulations (43), the relative diffusion coefficient (), which is defined as the diffusion coefficient of NPs in polymer network over the diffusion coefficient of NPs in water, has been widely used to compare the results between simulation and experiments (44, 45). We calculated the size ratio and in the experiments and simulations, respectively. The pore size is about ~4 μm, ~2.8 μm, and ~2 μm for the collagen with a concentration of 0.5 mg/mL, 1.0 mg/mL, and 2.0 mg/mL, respectively, as reported by a previous study (46). The NP/pore size ratios are thus 1/20, 1/14, and 1/10 correspondingly. We then performed additional simulations with network pore sizes of 50σ, 36σ, and 24σ to obtain the same size ratio with the experiments.
Under the same size ratio, in the simulation is 0.65 ± 0.08, 0.31 ± 0.05, and 0.18 ± 0.05, which is 1.5 fold, 1.1 fold, and 1.1 fold higher than that in the experiments (0.43 ± 0.08, 0.27 ± 0.06, and 0.16 ± 0.04), respectively (Fig. 3J). There are two reasons that may contribute to this slight difference. First, the polymer networks in simulations are regular ones with uniform mesh sizes, while the collagen hydrogels in experiments are characterized by heterogeneously distributed fibers and pore sizes. NPs are more likely to be blocked in the tiny pores (47). Thus, NP diffusion ability is stronger in the ideal regular network in simulations. Second, collagen used in the experiments is a highly hydrophilic biomaterial. The collagen fibers show affinity to water molecules to decrease their mobility, which may induce the decreased NP activity (48, 49). Thus, the NP diffusion coefficients in the collagen in experiments are lower.
Deformation of Softer ECM Enhances NP Diffusion.
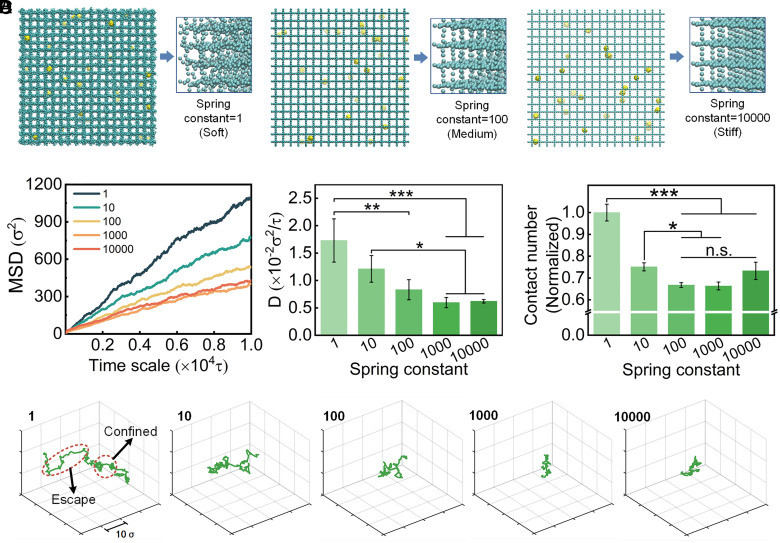
The increase in ECM network stiffness is one hallmark of many types of tumor tissues (50) and is associated with the tumor progression (51, 52). The stiffened ECM, mainly due to the increased collagen deposition and cross-linking (53, 54), has also been shown to promote malignant transition of normal epithelial cells and disease progression (6, 55, 56). The physiological range of the ECM network stiffness in tumor is from below 0.5 kPa to over 10 GPa (57–65), varying by tumor location [tumor core vs. tumor periphery (52, 58)], type [breast cancer vs. lung cancer, (57, 66)], and even disease progression [earlier stage vs. late stage (53)] due to the changes in collagen compositions and architectures. To assess the effect of ECM stiffness on NP diffusion, we built cross-linked networks with different stiffnesses by tuning the spring constant (1, 10, 100, 1,000, and 10,000). Soft networks are more relaxed and deformable during simulation, while stiff networks almost keep their initial construction, with each CG bead confined to its initial position (Fig. 4A). To characterize the polymer chain stiffness, we calculated the persistence length with the following equation:
| [1] |
Fig. 4.
Effect of ECM stiffness in tumor microenvironment on NP diffusion. (A) Representative network stiffness with spring constant of 1ε/σ2, 100ε/σ2, and 10,000ε/σ2. The enlarged 3D drawing shows decreased deformation of network chains in simulation. (B) The MSD values and calculated diffusion coefficient of NPs in networks with different stiffness. (C) The normalized contact numbers of NPs with the network during simulation. (D) Representative trajectories of NPs in networks with increased spring constant. (One-way ANOVA with Tukey’s post hoc test, n.s., no significance, P > 0.05; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001).
where is the bond length, and is the bond angle (67). The results of polymer chains with increasing spring constants in our simulations are shown in SI Appendix, Fig. S4. The persistence length of polymer chains increases by three orders of magnitude from 3.20 ± 0.39 σ to 1698.36 ± 292.19 σ. The persistence length covers a large range from soft to rigid polymer chains (68, 69), which is consistent with the wide stiffness range of tumor ECM.
We observed that with increasing network stiffness, MSD and D of NP diffusion gradually decrease but tend to be constant when stiffness increases to a certain degree. NP diffusion coefficients decrease from 1.73 × 10−2 to 1.21 × 10−2 , 0.83 × 10−2 , 0.60 × 10−2 , and 0.62 × 10−2 , when the spring constant increases from 1 to 10 , 100 , 1,000 , and 10,000 , respectively (Fig. 4B). The results indicate a negative correlation between NP diffusion ability and ECM network stiffness. It also suggests that NP diffusion is sensitive to stiffness changes when ECM fibers are relatively soft while the impact gradually decreases if the stiffness increases constantly.
The experiments of NP diffusion in collagen hydrogels with different stiffness were performed to verify our simulations. We tuned the collagen hydrogel stiffness from 56.0 ± 1.0 Pa to 137.1 ± 8.6 Pa by incubation with 100 mM ribose using a previously reported method (70) (SI Appendix, Fig. S5A). The diffusion of NPs at a fixed time (10 h) was imaged using CLSM, and the fluorescent images indicated the decreased penetration depth in the stiff collagen hydrogel (SI Appendix, Fig. S5B). The mean fluorescence intensity of NPs along the penetration depth was also analyzed to examine the diffusion ability (SI Appendix, Fig. S5C). The intensity is higher in the soft collagen hydrogel than that in the stiff collagen hydrogel, within the range of the penetration depth from 25 to 150 μm. NPs in the stiff collagen hydrogel are hardly observed deeper than 100 μm, while they can diffuse to a depth of 200 μm in the soft collagen hydrogel. The diffusion coefficient (D) is 0.95 μm2/s in the soft collagen hydrogel compared to 0.42 μm2/s in the stiff collagen hydrogel (SI Appendix, Fig. S5D). The results indicate that the soft collagen hydrogel can enhance the NP diffusion, which matches well with the simulation results qualitatively.
To further investigate the underlying mechanism of this phenomenon, we tracked the network behavior during the simulation. We found obvious deformation in softer networks which would cause increased contact areas between NPs and networks; thus, we defined contact number as the number of network beads that contact with one NP during the time scale of simulation. The results show that the contact numbers decrease with increasing network stiffness (Fig. 4C). In addition, the contact numbers show a slowing rate of decline when the networks become stiff, which is consistent with the MSD and D results. Softer networks tend to perform larger deformation and more contact sites with NPs than stiffer networks due to the network deformability. A greater contact area induced by network deformation in softer networks tends to promote NP diffusion and enables them to avoid trapping by the network. We found from the motion trajectories (Fig. 4D) that NPs are also either confined by meshes or escape from them to walk a longer distance. The network deformation would give rise to fluctuations in the mesh space and interactions between NPs and networks. As a result, NPs in soft networks show a larger motion extent, and NPs more easily escape even when they are confined around polymer chains and nodes. We observed a slight increase of the contact number when spring constant increase from 1,000 to 10,000. To explain this, additional MD simulations with greater spring constant were performed and the contact number and contact time were also calculated (SI Appendix, Fig. S6). For the contact time, there is an obvious increase when spring constant increases from 100 to 10,000. The NPs are more easily confined in the stiff network based on the NP motion trajectories (Fig. 4D), which increases the contact time between the NPs and network. Although the contact number is affected mainly by the deformation of soft network when the spring constant is from 1 to 1,000, the contact number is an averaged value for the whole simulation process that the influence of contact time is also included. With the continuous increase of spring constant, the increase of contact time cannot be ignored, which induces an overall increase in contact number when spring constant increases from 1,000 to 10,000.
ECM Alignment Improves NP Diffusion Significantly by Increasing the Directionality.
The changes in ECM microstructures have been widely acknowledged as a physical hallmark of cancer (71–73). Especially, the previous study has observed and defined three tumor-associated collagen signatures (TACS) to characterize the collagen density and alignment during mammary tumor progression (20, 74). Dense collagen fibers are straightened parallel to the tumor boundary in the TACS-2 and aligned ultimately perpendicular to the tumor boundary in the TACS-3. The radial alignment of collagen fibers facilitates local tumor cell invasion during tumor progression. Thus, the changes in ECM microstructures are a potential factor to promote NP diffusion.
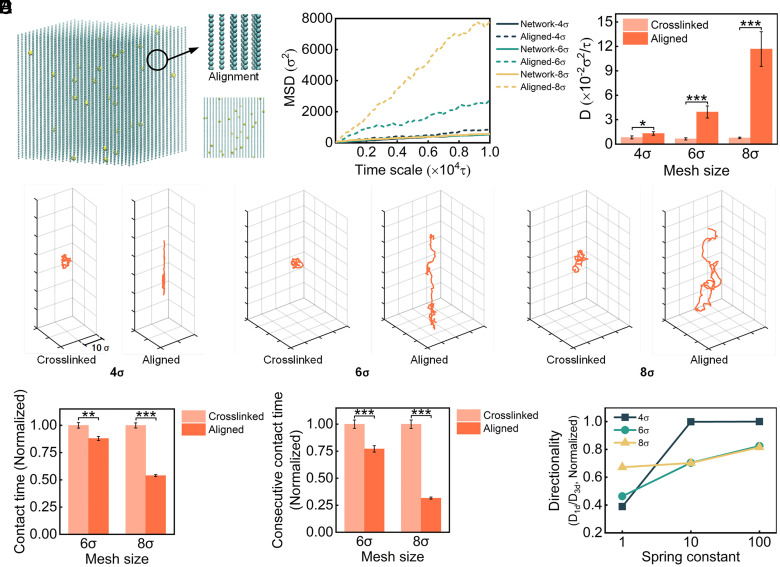
To investigate the effect of network structure on NP diffusion, we built aligned networks with densities similar to network-4, network-6, and network-8 (SI Appendix, Table S2 and Fig. 5A). Compared with a cross-linked network structure, the MSDs and diffusion coefficients in an aligned structure greatly increase (Fig. 5B), indicating an improved NP diffusion ability. In addition, with the decrease in network density, the enhancement increases significantly. For instance, when the effective mesh size is 4, the NP diffusion coefficient of aligned structure is 1.6-fold higher than that of network structure. However, when the effective mesh size reaches 6 and 8, the NP diffusion coefficients of the aligned structure are sixfold and 15-fold higher than those of the network structure. The results suggest that the aligned ECM structure plays a major role in NP diffusion behavior and this influence is simultaneously related to structure density.
Fig. 5.
Effect of ECM alignment in tumor microenvironment on NP diffusion. (A) Representative network with an aligned structure, and it has an effective mesh size of 4σ. (B) The MSD values and calculated diffusion coefficient of NPs in networks with cross-linked and aligned structures. (C) Representative trajectories of NPs in cross-linked and aligned networks. (D) The normalized contact time of NPs and consecutive contact time in the network during simulation. (E) The directionality of NP motion in the aligned network. (One-way ANOVA with Tukey’s post hoc test, n.s., no significance, P > 0.05; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001).
To verify the ECM alignment simulation results, we constructed collagen hydrogels with aligned fibers following previous protocol (75) and performed NP diffusion experiments (SI Appendix, Fig. S7A). The diffusion of NPs at a fixed time (6 h) was imaged using CLSM. The fluorescent images demonstrated that NPs penetrate deeper in the aligned collagen hydrogels than that in the nonaligned collagen network (SI Appendix, Fig. S7B). The mean fluorescence intensity of NPs along penetration depth was also analyzed to examine the diffusion ability (SI Appendix, Fig. S7C). The intensity is higher in the aligned collagen than that in the nonaligned collagen network. In addition, NPs in aligned collagen are able to diffuse deeper than 250 μm while they are hardly observed at a depth of 150 μm in the collagen network. Compared to 0.10 μm2/s in the collagen network, the diffusion coefficient of NPs increases to 0.30 μm2/s in the aligned collagen (SI Appendix, Fig. S7D). The results indicate that the aligned collagen can enhance the NP diffusion, which matches well with the simulation results qualitatively.
To understand why the network structure can modulate NP diffusion, we analyzed the motion trajectories of NPs and revealed the different NP diffusion behavior in cross-linked and aligned networks (Fig. 5C). NPs in cross-linked networks are restricted by tightly arranged meshes and nodes. The isotropic structure provides NP motion with a relatively equal probability in all directions but limited accessible regions. For NPs in aligned networks, the direction of the path becomes evident. We then calculated the average contact time and the consecutive contact time (Fig. 5D). For the same effective density, NPs will undergo fewer interactions with aligned structures compared with the cross-linked structure. In addition, the difference becomes more significant with the increase in network mesh size, further confirming the changes in diffusion coefficient in Fig. 5B. Thus, the aligned structure plays a positive role in NP diffusion because of considerably decreased interactions between NPs and the network, resulting in reduced restrictions for NPs.
To elucidate how the aligned structure influences NP diffusion behavior, we calculated the diffusion coefficients of NPs displayed in one dimension (the same direction as aligned network chains):
| [2] |
The ratio of the one-dimensional diffusion coefficient () to the three-dimensional diffusion coefficient was defined as the directionality of NP diffusion. We observed that an aligned structure could affect the directionality of diffusion, which depends on both network density and stiffness (Fig. 5E). For a certain density, directionality generally increases with increasing stiffness; for instance, the green line in Fig. 5E. However, the influence of density on directionality is different in ECM with low and high stiffness. At a relatively high stiffness (spring constant = 10 and 100 ), the morphology of the grid is relatively stable; therefore, density effects dominate. NPs obtain a stronger directionality in the dense network (e.g., network-4). At low stiffness (e.g., spring constant = 1 ), the network deformation increases and stiffness effects dominate. 1D diffusion is limited by network deformation, especially in a dense network. As a result, the directionality increases with increasing mesh size.
The Estimation Matrix Model to Predict NP Diffusion Efficiency.
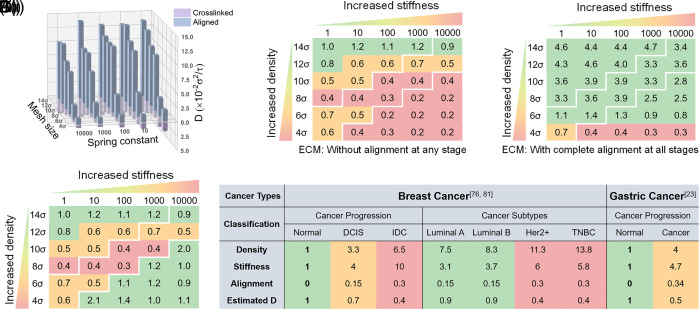
The ECM’s physical properties, such as density, stiffness, and structure, not only undergo continuous changes with the development of tumor progression stages and degrees of malignancy but also show differences in different tumor types. In addition, these properties are supposed to impact each other and change simultaneously. To obtain a comprehensive analysis of ECM physical property effects, we assessed the overall influence of these factors (Fig. 6A). The results revealed that whether in a cross-linked or aligned network, the diffusion coefficient of NPs increases with the decrease of ECM density and stiffness. In addition, the alignment significantly enhances NP diffusion which becomes more evident with the decrease of density and stiffness by comparing the cross-linked and aligned networks.
Fig. 6.
Comprehensive analyses of tumor ECM physical properties on NP diffusion. (A) The overall presentation of NP diffusion coefficient with the influence of tumor physical properties. (B) Integrated influence matrix of NP diffusion ability for a given tumor physical property. All scores were obtained by normalizing diffusion coefficients to the value when density and stiffness are 14 and 1 , respectively, in B(i). B(iii) Scores were calculated based on B(i) by integrating the effect of alignment at tumor late stages according to Eq. 3. (C) Application of the estimation matrix. The diffusion coefficient was estimated according to ECM physical properties in the literature. (Breast cancer: ECM density and stiffness values are from ref. 76, and alignment values are from ref. 81; Gastric cancer: ECM density and alignment values are from ref. 23, and stiffness values are from Fig. 2D. All values are normalized to normal ECM).
To evaluate whether NP diffusion ability could be predicted according to tumor physical characteristics, we proposed an estimation matrix based on previously calculated diffusion coefficients (Fig. 6B). In the estimation matrix, each numerical value can be considered as a score that represents the diffusion ability of NPs in the corresponding tumor conditions. All scores are normalized to the diffusion coefficient value when the density and stiffness are 14 and 1 , respectively, which we considered as the normal ECM. First, the matrix is divided into three regions according to tumor ECM density and stiffness, which are relevant to the early, mid, and late stages of tumors. The three divided regions are distinguished by white lines in our figures. Furthermore, the estimation matrix without alignment influence (Fig. 6 B, i) is determined by normalizing the NP diffusion coefficient, resulting in the cross-linked network. Similarly, the estimation matrix with the complete influence of the aligned structure (Fig. 6 B, ii) is determined by normalizing the diffusion coefficients in the aligned network. However, for tumor tissues, ECM structure changes induced by tumor progression exist to a certain degree instead of ideal and complete alignment. Considering the partial alignment and the notable diffusion ability increase in the aligned network, the effect of alignment could also be integrated into the final matrix by a weight multiplier. Eq. 3 is the summary of the estimation scheme, where is the averaged increase multiples (each diffusion coefficient in the aligned network divided by that in the cross-linked network) of diffusion coefficients in each subregion, represents the weight multiplier in corresponding subregions, and is the normalized value of diffusion coefficient in the cross-linked network.
| [3] |
The results suggest that NP diffusion coefficients are closely related to ECM density and stiffness in ECM without fiber alignment. As a result, NPs exhibit the strongest diffusion ability in the early tumor stage and the weakest diffusion in the late stage. However, NP diffusion ability is greatly affected by the aligned structure once the alignment appears. We found that NPs show significantly increased diffusion ability in the late tumor stage due to 50% alignment (Fig. 6 B, iii). In the late tumor stage with an aligned structure, D increases to three times higher than that in the cross-linked network and is even better than that in the midterm stage. Furthermore, if the tumor ECM is characterized by ideal and complete alignment in structure, there is an overall improvement of NP diffusion ability in all the three stages up to a 15-fold enhancement.
In conclusion, the estimation matrix provides an evaluation method for NP diffusion ability according to tumor ECM physical characteristics. We offered estimated relative diffusion coefficients of different tumor progression and tumor types of clinical specimens in the literature to explain the application (Fig. 6C). Taking breast cancer as an example, we assumed that the density, stiffness, and alignment in normal tissues are 1, 1, and 0, respectively, which represent ECM with normalized density and stiffness and without alignment. Then, the dimensionless density and stiffness in different tumor progression stages and tumor subtypes can be obtained according to ECM characterization (76) (SI Appendix, Table S3). Collagen alignment in human breast cancer has been proven by many reports (20, 77–79), but it is challenging to quantify the alignment degree. We observed that the proportion of anisotropic collagen fibers varies from 0.1 to 0.35 (80, 81), so we assigned 0.15 and 0.3 as the alignment degree according to malignant progression and aggression. After that, the respective normalized diffusion coefficients are calculated using our estimation matrix model. Similarly, we also applied our model to gastric cancer according to the characterization results from previous work (density and alignment) (23) and ours (stiffness). As the results show, the estimated D decreases as the tumor progresses and aggression. We suggest that if the estimated D is relatively low (e.g., 0.4 in breast cancer IDC), a pretreatment of ECM is necessary before the drug delivery.
Discussion
Given that NPs show limited delivery efficiency in biological systems, especially in tumor ECM, it is necessary to figure out how to achieve improved NP diffusion ability for cancer diagnosis and treatment (82, 83). Previous works have made great efforts in regulating particle size, shape, surface chemistry, and stiffness although NP design and engineering are complicated processes (84, 85). Moreover, the low delivery efficiency of NPs is still a huge challenge for clinical translation of cancer nanomedicines, although great efforts have been made to regulate NP physicochemical parameters (1). Recently, increasing interest and research on the physical properties of the TME have led us to pay more attention to the tumor ECM (86, 87). The physical abnormalities of tumor ECM induced by tumor development will in turn affect tumor progression and treatment resistance (54, 88). In our study, we explored approaches to improve NP delivery by investigating the influence of tumor ECM physical properties.
We examined the physical property changes of human gastric cancer samples by comparing normal and tumor tissues. It was shown in Masson’s trichrome staining that there is a difference in ECM appearance after tumorigenesis. Further quantitative analysis suggests increased collagen content and elevated stiffness mainly caused by collagen accumulation in tumor regions. The dense ECM composition provides restricted motion space for NPs, implying the role of ECM physical properties in mediating NP diffusion. In addition, tissue carcinogenesis causes structure remodeling of ECM, resulting in aligned collagen fibers. Compared with dispersed network structures, fibers in a similar arrangement may act as convenient access for NP transport. NP diffusion experiment in normal and tumor tissues has shown a large impediment for NP penetration in cancerous tissues, revealing the crucial role of ECM physical properties. MD simulation and in vitro experiments in our work demonstrate the impact of each factors on NP diffusion including ECM density, stiffness, and structural changes. ECM density and stiffness negatively affect NP diffusion ability, while interestingly ECM alignment significantly increases NP diffusion. Generally, ECM is characterized by increased density and stiffness with the pathogenesis and progression of tumors, which suggests the benefit of early cancer diagnosis and treatment in the clinic. The emergence and extent of ECM alignment are not consistent with tumor growth. Thus, when considering alignment, more analysis of each specific condition is required to answer whether this factor influences NP diffusion and how to influence it.
Our findings also inspire us to determine the feasibility of personalized medicine by predicting nano-drug delivery in advance. For this, we developed an evaluation matrix to estimate and predict medication effects from a transport perspective by providing a diffusion score according to ECM conditions. The evaluation matrix would help researchers or clinicians make an early judgment of drug efficiency by determining the tumor ECM’s physical appearance. Through our evaluation matrix obtained based on a series of fixed parameters, a flexible selection of weight coefficients for each factor makes it possible to be applied to different tumor progression and types. Expected practical data are supposed to further improve and complete the evaluation model for broader but more precise utilization. We look forward to the wide clinical application prospects of our evaluation model in guiding the quantitative characterization of tumor ECM and therapeutic strategies.
It should be noted that there are still some limitations associated with our model. A pure diffusion model is used to simulate and predict the NP diffusion efficiency, while other complex transport processes, such as the convention and tissue movement, are not included here (89). The regular cross-linked network in the model is an ideal simplification of the ECM, which is a porous material with heterogeneously distributed pores and multiscale structure in native tumor tissues (47). In addition, the threshold exists for the size ratio of the NP and network mesh. On one side, if the mesh size is equal to the NP size, in which the NP is confined by the mesh, the diffusion coefficient is approximate to zero. With the increase of the mesh size, the effect of the mesh on the NP diffusion decreases. The mesh size can be increased infinitely, but the diffusion coefficient is approximate to that of the NP diffusion in solvents under this condition. Last but not the least, we focus only on the diffusion process of NPs in ECM after the vascular extravasation. The vascularity is not considered in our simulations and the proposed estimation matrix model.
Conclusions
Given that nanomedicine is confronted with limited and disappointing delivery efficiency in tumors clinically, an in-depth understanding of the diffusion mechanism is desperately required. Researchers have made constant efforts to develop NPs of various sizes, shapes, biochemical and mechanical properties. Nevertheless, the TME receives less attention and the role of tumor ECM physical traits in NP diffusion remains largely unexplored. In our study, the physical properties of tumor ECM were characterized by clinical samples, and the decreased penetration depth of NPs in tumor tissues was also confirmed by using decellularized clinical samples. MD simulations and single-particle tracking experiments in collagen were performed to explore the effects of ECM physical properties on NP diffusion. We found that NP diffusion ability decreases with increasing network density and stiffness, while an aligned network enhances NP diffusion up to 15-fold. In vitro experiments in which NPs diffuse in collagen confirmed the negative correlations between NP diffusion coefficient and ECM density. We also developed an estimation matrix model with evaluation scores of NP diffusion through comprehensive analyses of diffusion parameters. Although the efficiency needs to be demonstrated by further implementation, our findings have inspired us to pay more attention to tumor hallmarks in drug delivery. Further investigation of the interaction between particles and ECM will help us to exploit innovative NP designs and TME regulation methods.
Materials and Methods
Reagents and Materials.
Rat tail collagen I was purchased from Corning Incorporated. PS fluorescent NPs were purchased from BaseLine ChromTech Research Centre. The 1× Dulbecco’s phosphate-buffered saline (DPBS, without calcium and magnesium) solution was purchased from Labgic Technology Co., Ltd. Sodium lauryl ether sulfate (SLES, 70%) was purchased from Macklin Biochemical Co., Ltd. Gastric tumor and paracancerous tissues were provided by Honghui Hospital. The paracancerous tissues, which are physically next to the cancerous tissues, are widely used as the normal control to compare with tumor tissues. All samples were deidentified properly prior to use in the study.
MD Simulations.
The coarse-grained (CG) NPs were constructed as a cuboctahedral structure in Materials Studio (MS) by a face-centered cubic lattice (38, 39) with a strong bead–bead harmonic spring force (Eq. 4). was the energy for bond stretching of adjacent beads, and was the spring constant and was the equilibrium bond length. As a result, the CG NP can be considered as a rigid spherical NP with a diameter of approximately 2.5 . The regular polymer network was constructed to simulate the ECM structure (38). Each polymer chain was composed of about 72 CG beads and cross-linked by a node bead in the initial model. The size of the simulation box was 72×72×72 , and periodic boundary condition was used in all the three directions. The bonded interaction between adjacent CG beads within each polymer chain is also described by a harmonic spring force. And, we utilized the Lennard–Jones potential (Eq. 5) to describe the nonbonded interactions between two beads where was the interaction strength, was the distance between two beads, and was the cutoff distance.
| [4] |
| [5] |
The detailed interaction parameters of Eqs. 4 and 5 are listed in SI Appendix, Table S4. The dimensionless units (bead radius), (bead mass), , and are the units of length, mass, time, and energy, respectively. In the simulation, the Velocity-Verlet algorithm was utilized to perform the time integration with a time integration of . We performed constant number–volume–energy integration updates of position and velocity for beads during simulations. A Langevin thermostat was applied to control the system temperature at where is the Boltzmann constant, and is temperature. The total simulation time was 10,000 and four times were repeated for each case, then the mean MSD of the NPs was obtained. The MSD curve was liner fitted from 2,000 to 10,000 and the slope () of the fitting line was calculated. Then, the diffusion coefficient was obtained by . All MD simulations were performed using the Large-scale Atomic/Molecular Massively Parallel Simulator package.
Network density was regulated by adjusting the mesh size which was changed from 4 to 14 (i.e., 4, 6, 8, 10, 12, 14). A spring force was applied to each CG bead of the network to constrain its initial position during simulation, which restricted network fiber deformation and controlled network stiffness. To regulate the stiffness of the ECM network model, the spring constant was set as 1, 10, 100, 1,000, and 10,000 to adjust the network from a considerable soft state to quite stiff. The soft network was more relaxed and deformable during simulation, while the stiff network was restricted and practically maintained its initial construction. We changed the available cross-linked network formation into an aligned and parallel arrangement to modulate the network structure. In an aligned network structure, all polymer chains lined up in the same direction and there were no node beads connecting them. To ensure a density comparable to that of the previous network and keep the system a cube at the same time, we selected proper square numbers as the numbers of polymer chains to adjust them to be as constant as possible with cross-linked networks (SI Appendix, Table S2).
NP Diffusion in Decellularized Tumor Tissues.
Characterization of the gastric tumor and paracancerous tissues.
The gastric tumor and paracancerous (normal) tissues collected from clinical surgery were kept in PBS solution at 4 °C and transferred to the lab within 4 h. Then, they were cut into 3.0 × 3.0 × 3.0 mm3 sections and freeze-sectioned into 10 μm slices for Masson trichrome staining. Collagen content and fiber orientation distribution were analyzed using ImageJ software (SI Appendix, Fig. S8). The image was imported into ImageJ and converted to 8-bit grayscale format. Then, an appropriate threshold was determined to select collagen fiber area as complete as possible. The collagen area was measured and the fraction of the collagen area in the whole image was considered as the collagen content. For fiber orientation distribution, the image to be analyzed was similarly imported into ImageJ and converted to 8-bit grayscale format. The plugin OrientationJ-Distribution was applied to analyze the fiber orientation distribution automatically. The generated data sheet was used to develop the distribution probability–direction profile.
Tissues were cut into 4 mm diameters and 1.5 mm thicknesses for rheology tests using a rheometer (Anton Paar, MCR302, Austria). Frequency tests were performed with a shear strain of 1% and an angular frequency ranging from 0.562 to 100 rad/s for normal and tumor tissues. The storage modulus G’ and loss modulus G’’ were recorded while G’ is related to the Young’s modulus of the tissues. All tests were repeated three times for each sample. The final G’ was calculated by averaging the modulus values of the last three recorded frequencies, with G’ remaining stable.
NP diffusion in decellularized tissues.
Tumor tissues were decellularized following the method described in the literature (40). Briefly, the collected gastric tumor tissues and paracancerous tissues were cut into 3.0 × 3.0 × 3.0 mm3 sections followed by the removal of blood cells in DPBS solution for 4 h. Then, the tissues were transferred into 1% SLES solution with gentle shaking on an orbital shaker for 24 h. After that, the tissues were washed using sterilized water and DPBS solution with gentle shaking for 8 × 2 h to remove residual SLES. Finally, the decellularized tumor tissues were kept in DPBS solution at 4 °C for the following use.
NP size and surface property were characterized before our in vitro experiments. Morphological images were captured using a JEOL JEM-2100Plus TEM while hydrodynamic diameters and zeta-potentials were measured using a Malvern Zetasizer Nano ZSE. For the NP diffusion experiment, 1% fluorescence NPs of 80 nm in diameter were dispersed in PBS for 1 h using an ultrasonic cleaner followed by culture with the decellularized tissues for 48 h. Then, the tissues were freeze-sectioned into 10 μm slices at the center position for imaging. We obtained the fluorescence images of NP diffusion and analyzed the fluorescence intensity and tumor tissue area using the ImageJ software.
Single-Particle Tracking Experiments.
Preparation of collagen hydrogel.
Collagen I hydrogel was prepared according to the product instructions. Briefly, rat tail collagen I was added to a centrifuge tube placed in an ice bath, followed by the addition of deionized water. Then, the mixture was added to 0.2 mol/L NaOH solution and mixed immediately. Then, the 10× PBS was added and fully mixed to form the final mixture. The collagen was transferred to a 96-well plate and put into a 37 °C incubator for 50 min for cross-linking.
Single-particle tracking.
Collagen was prepared as described above. For NP tracking, fluorescent PS (200 nm) particles were added to collagen and incubated for 20 min. Then, a volume of 40 μL collagen-containing NPs was transferred on a glass bottom dish and a coverslip was placed slightly. NP diffusion was observed and measured under a CLSM following the procedure from the literature (41, 42). Briefly, the motions of the particles within the collagen were recorded at a temporal resolution of 33.3 ms for 10 s. Approximately N = 15 particles were tracked for each experiment, and their trajectories were analyzed using ImageJ. MSD and D of these particles were then calculated as follows:
| [6] |
| [7] |
Supplementary Material
Appendix 01 (PDF)
NP diffusion in crosslinked network with mesh size of 4σ and spring constant of 100.
NP diffusion in crosslinked network with mesh size of 14σ and spring constant of 100.
NP diffusion in crosslinked network with mesh size of 4σ and spring constant of 10,000.
NP diffusion in aligned network with mesh size of 8σ and spring constant of 100.
Acknowledgments
This work was financially sponsored by the National Natural Science Foundation of China (12225208, 51806169, 11972280, 12202193, 32271296, 31900939) and Key Research and Development Program of Shaanxi (2020GXLH-Y-018).
Author contributions
X.H., Y.Y., Y.H., and F.X. designed research; Y.Y., Y.H., C.C., Z.Z., C.X., and L.L. performed research; X.H., Y.H., H.G., L.H., N.L., S.H., and F.X. contributed new reagents/analytic tools; X.H., Y.Y., Y.H., C.C., Z.Z., L.L., C.X., H.G., L.W., L.H., Z.Q., N.L., S.H., and F.X. analyzed data; and X.H., Y.Y., Y.H., C.C., Z.Z., L.L., C.X., H.G., L.W., L.H., Z.Q., N.L., S.H., and F.X. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Na Liu, Email: liuna1@xjtu.edu.cn.
Shuang Han, Email: shuanghamy@163.com.
Feng Xu, Email: fengxu@mail.xjtu.edu.cn.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Wilhelm S., et al. , Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 1, 16014 (2016). [Google Scholar]
- 2.Wang A. Z., Langer R., Farokhzad O. C., Nanoparticle delivery of cancer drugs. Annu. Rev. Med. 63, 185–198 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Dang Y., Guan J., Nanoparticle-based drug delivery systems for cancer therapy. Smart Mater. Med. 1, 10–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai Q., et al. , Quantifying the ligand-coated nanoparticle delivery to cancer cells in solid tumors. ACS Nano 12, 8423–8435 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Chauhan V. P., Stylianopoulos T., Boucher Y., Jain R. K., Delivery of molecular and nanoscale medicine to tumors: Transport barriers and strategies. Annu. Rev. Chem. Biomol. Eng. 2, 281–298 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Spill F., Reynolds D. S., Kamm R. D., Zaman M. H., Impact of the physical microenvironment on tumor progression and metastasis. Curr. Opin. Biotechnol. 40, 41–48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stylianopoulos T., Munn L. L., Jain R. K., Reengineering the physical microenvironment of tumors to improve drug delivery and efficacy: From mathematical modeling to bench to bedside. Trends Cancer 4, 292–319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nizzero S., Ziemys A., Ferrari M., Transport barriers and oncophysics in cancer treatment. Trends Cancer 4, 277–280 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Overchuk M., Zheng G., Overcoming obstacles in the tumor microenvironment: Recent advancements in nanoparticle delivery for cancer theranostics. Biomaterials 156, 217–237 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Dewhirst M. W., Secomb T. W., Transport of drugs from blood vessels to tumour tissue. Nat. Rev. Cancer 17, 738 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jain R. K., Stylianopoulos T., Delivering nanomedicine to solid tumors. Nat. Rev. Clin. Oncol. 7, 653 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perrault S. D., Walkey C., Jennings T., Fischer H. C., Chan W. C. W., Mediating tumor targeting efficiency of nanoparticles through design. Nano lett. 9, 1909–1915 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Grady M. E., et al. , Intracellular nanoparticle dynamics affected by cytoskeletal integrity. Soft Matter 13, 1873–1880 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Theocharis A. D., Skandalis S. S., Gialeli C., Karamanos N. K., Extracellular matrix structure. Adv. Drug Deliv. Rev. 97, 4–27 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Nicolas-Boluda A., Silva A. K. A., Fournel S., Gazeau F., Physical oncology: New targets for nanomedicine. Biomaterials 150, 87–99 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Malandrino A., Mak M., Kamm R. D., Moeendarbary E., Complex mechanics of the heterogeneous extracellular matrix in cancer. Extreme Mech. Lett. 21, 25–34 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sykes E. A., et al. , Tailoring nanoparticle designs to target cancer based on tumor pathophysiology. Proc. Natl. Acad. Sci. U.S.A. 113, E1142–E1151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohan V., Das A., Sagi I., Emerging roles of ECM remodeling processes in cancer. Semin. Cancer Biol. 62, 192–200 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Rice A. J., et al. , Matrix stiffness induces epithelial-mesenchymal transition and promotes chemoresistance in pancreatic cancer cells. Oncogenesis 6, e352 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Provenzano P. P., et al. , Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 4, 38 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drifka C. R., et al. , Periductal stromal collagen topology of pancreatic ductal adenocarcinoma differs from that of normal and chronic pancreatitis. Mod. Pathol. 28, 1470–1480 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Drifka C. R., et al. , Highly aligned stromal collagen is a negative prognostic factor following pancreatic ductal adenocarcinoma resection. Oncotarget 7, 76197–76213 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Z. H., et al. , Reorganized collagen in the tumor microenvironment of gastric cancer and its association with prognosis. J. Cancer 8, 1466–1476 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nia H. T., Munn L. L., Jain R. K., Physical traits of cancer. Science 370, eaaz0868 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgos-Panadero R., et al. , The tumour microenvironment as an integrated framework to understand cancer biology. Cancer Lett. 461, 112–122 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Sykes E. A., Chen J., Zheng G., Chan W. C., Investigating the impact of nanoparticle size on active and passive tumor targeting efficiency. ACS Nano 8, 5696–5706 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Wang J., et al. , The role of micelle size in tumor accumulation, penetration, and treatment. ACS Nano 9, 7195–7206 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Chauhan V. P., et al. , Fluorescent nanorods and nanospheres for real-time in vivo probing of nanoparticle shape-dependent tumor penetration. Angew. Chem. Int. Ed. Engl. 50, 11417–11420 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao K., et al. , The effect of surface charge on in vivo biodistribution of PEG-oligocholic acid based micellar nanoparticles. Biomaterials 32, 3435–3446 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han D., et al. , The effects of surface charge on intra-tumor penetration of drug delivery vehicles along with tumor progression. J. Mater. Chem. B 6, 3331–3339 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Guo P., et al. , Nanoparticle elasticity directs tumor uptake. Nat. Commun. 9, 130 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng H., et al. , Modulating rigidity of nanoparticles for tumor penetration. Chem. Commun. 54, 3014–3017 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Sun Q., Ojha T., Kiessling F., Lammers T., Shi Y., Enhancing tumor penetration of nanomedicines. Biomacromolecules 18, 1449–1459 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuhn S. J., Finch S. K., Hallahan D. E., Giorgio T. D., Proteolytic surface functionalization enhances in vitro magnetic nnoparticle mobility through extracellular matrix. Nano Lett. 6, 306–312 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Zhou H., et al. , Hyaluronidase embedded in nanocarrier PEG shell for enhanced tumor penetration and highly efficient antitumor efficacy. Nano Lett. 16, 3268–3277 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Lee S., et al. , Extracellular matrix remodeling in vivo for enhancing tumor-targeting efficiency of nanoparticle drug carriers using the pulsed high intensity focused ultrasound. J. Control. Release 263, 68–78 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Raeesi V., Chan W. C., Improving nanoparticle diffusion through tumor collagen matrix by photo-thermal gold nanorods. Nanoscale 8, 12524–12530 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Yu M., et al. , Rotation-facilitated rapid transport of nanorods in mucosal tissues. Nano Lett. 16, 7176–7182 (2016). [DOI] [PubMed] [Google Scholar]
- 39.He X., et al. , Tailoring patchy nanoparticle design to modulate serum albumin adsorption and membrane interaction. Soft Matter 17, 2071–2080 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Qin J., Gang L., Li S., Yuan H., Ma Y., Decellularized breast matrix as bioactive microenvironment for in vitro three-dimensional cancer culture. J. Cell. Physiol. 234, 3425–3435 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Han Y. L., et al. , Cell swelling, softening and invasion in a three-dimensional breast cancer model. Nat. Phys. 16, 101–108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu M., et al. , Rapid transport of deformation-tuned nanoparticles across biological hydrogels and cellular barriers. Nat. Commun. 9, 2607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu M., et al. , Temperature- and rigidity-mediated rapid transport of lipid nanovesicles in hydrogels. Proc. Natl. Acad. Sci. U.S.A. 116, 5362–5369 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cai L. H., Panyukov S., Rubinstein M., Mobility of nonsticky nanoparticles in polymer liquids. Macromolecules 44, 7853–7863 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai L. H., Panyukov S., Rubinstein M., Hopping diffusion of nanoparticles in polymer matrices. Macromolecules 48, 847–862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lang Nadine R., et al. , Estimating the 3D pore size distribution of biopolymer networks from directionally biased data. Biophys. J. 105, 1967–1975 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu H., Schwartz D. K., Nanoparticle tracking to probe transport in porous media. Acc. Chem. Res. 53, 2130–2139 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Ferreira A. M., Gentile P., Chiono V., Ciardelli G., Collagen for bone tissue regeneration. Acta Biomater. 8, 3191–3200 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Liu X., Zheng C., Luo X., Wang X., Jiang H., Recent advances of collagen-based biomaterials: Multi-hierarchical structure, modification and biomedical applications. Mater. Sci. Eng. C Mater. Biol. Appl. 99, 1509–1522 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Giussani M., Merlino G., Cappelletti V., Tagliabue E., Daidone M. G., Tumor-extracellular matrix interactions: Identification of tools associated with breast cancer progression. Semin. Cancer Biol. 35, 3–10 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Levental K. R., et al. , Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139, 891–906 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Payen T., et al. , Harmonic motion imaging of pancreatic tumor stiffness indicates disease state and treatment response. Clin. Cancer Res. 26, 1297–1308 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burgos-Panadero R., et al. , The tumour microenvironment as an integrated framework to understand cancer biology. Cancer Lett. 461, 112–122 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Mohan V., Das A., Sagi I., "Emerging roles of ECM remodeling processes in cancer" in Seminars in cancer biology (Elsevier; ), pp 192–200. [DOI] [PubMed] [Google Scholar]
- 55.Acerbi I., et al. , Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr. Biol. 7, 1120–1134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alonso-Nocelo M., et al. , Matrix stiffness and tumor-associated macrophages modulate epithelial to mesenchymal transition of human adenocarcinoma cells. Biofabrication 10, 035004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lartey F. M., et al. , Dynamic CT imaging of volumetric changes in pulmonary nodules correlates with physical measurements of stiffness. Radiother. Oncol. 122, 313–318 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mieulet V., et al. , Stiffness increases with myofibroblast content and collagen density in mesenchymal high grade serous ovarian cancer. Sci. Rep. 11, 4219 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rezk R., Marín-García R., Gad A. K. B., The fibrillar matrix: Novel avenues for breast cancer detection and treatment. Engineering 7, 1375–1380 (2021). [Google Scholar]
- 60.Jang M., et al. , Matrix stiffness epigenetically regulates the oncogenic activation of the yes-associated protein in gastric cancer. Nat. Biomed. Eng. 5, 114–123 (2021). [DOI] [PubMed] [Google Scholar]
- 61.Bunevicius A., Schregel K., Sinkus R., Golby A., Patz S., REVIEW: MR elastography of brain tumors. Neuroimage Clin. 25, 102109 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stewart D. C., Rubiano A., Dyson K., Simmons C. S., Mechanical characterization of human brain tumors from patients and comparison to potential surgical phantoms. PLoS One 12, e0177561 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kawano S., et al. , Assessment of elasticity of colorectal cancer tissue, clinical utility, pathological and phenotypical relevance. Cancer Sci. 106, 1232–1239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang X., et al. , A multi-level comparative analysis of human femoral cortical bone quality in healthy cadavers and surgical safe margin of osteosarcoma patients. J. Mech. Behav. Biomed. Mater. 66, 111–118 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Gohil V. P., Canavan P. K., Nayeb-Hashemi H., “Effect of bone tumor and osteoporosis on mechanical properties of bone and bone tissue properties: A finite element study” in ASME 2007 International Mechanical Engineering Congress and Exposition. (ASME Press, New York, NY, 2007), pp. 947–949. [Google Scholar]
- 66.Chang J. M., et al. , Stiffness of tumours measured by shear-wave elastography correlated with subtypes of breast cancer. Eur. Radiol. 23, 2450–2458 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Zhang J. Z., et al. , The persistence length of semiflexible polymers in lattice Monte Carlo simulations. Polymers 11, 295 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carrillo J. M. Y., Dobrynin A. V., Polyelectrolytes in salt solutions: Molecular dynamics simulations. Macromolecules 44, 5798–5816 (2011). [Google Scholar]
- 69.Faller R., Kolb A., Müller-Plathe F., Local chain ordering in amorphous polymer melts: Influence of chain stiffness. Phys. Chem. Chem. Phys. 1, 2071–2076 (1999). [Google Scholar]
- 70.Mason B. N., Starchenko A., Williams R. M., Bonassar L. J., Reinhart-King C. A., Tuning three-dimensional collagen matrix stiffness independently of collagen concentration modulates endothelial cell behavior. Acta Biomater. 9, 4635–4644 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Conklin M. W., et al. , Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 178, 1221–1232 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maller O., et al. , Collagen architecture in pregnancy-induced protection from breast cancer. J. Cell Sci. 126, 4108–4110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun X., et al. , Tumour DDR1 promotes collagen fibre alignment to instigate immune exclusion. Nature 599, 673–678 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schedin P., Keely P. J., Mammary gland ECM remodeling, stiffness, and mechanosignaling in normal development and tumor progression. Cold Spring Harb. Perspect. Biol. 3, a003228 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pei D., et al. , Remodeling of aligned fibrous extracellular matrix by encapsulated cells under mechanical stretching. Acta Biomater. 112, 202–212 (2020). [DOI] [PubMed] [Google Scholar]
- 76.Acerbi I., et al. , Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr. Biol. (Camb). 7, 1120–1134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mercatelli R., Triulzi T., Pavone F. S., Orlandi R., Cicchi R., Collagen ultrastructural symmetry and its malignant alterations in human breast cancer revealed by polarization-resolved second-harmonic generation microscopy. J. Biophotonics 13, e202000159 (2020). [DOI] [PubMed] [Google Scholar]
- 78.Conklin M. W., et al. , Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 178, 1221–1232 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Conklin M. W., et al. , Collagen alignment as a predictor of recurrence after ductal carcinoma in situ. Cancer Epidemiol. Biomarkers Prev. 27, 138–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Richinga K. M., et al. , 3D collagen alignment limits protrusions to enhance breast cancer cell persistence. Biophys. J. 107, 2546–2558 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Majeed H., Okoro C., Kajdacsy-Balla A., Toussaint K., Popescu G., Quantifying collagen fiber orientation in breast cancer using quantitative phase imaging. J. Biomed. Opt. 22, 046004 (2017). [DOI] [PubMed] [Google Scholar]
- 82.Henke E., Nandigama R., Ergün S., Extracellular matrix in the tumor microenvironment and its impact on cancer therapy. Front. Mol. Biosci. 6, 160 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saint A., Van Obberghen-Schilling E., The role of the tumor matrix environment in progression of head and neck cancer. Curr. Opin. Oncol. 33, 168–174 (2021). [DOI] [PubMed] [Google Scholar]
- 84.Ding J., et al. , Engineered nanomedicines with enhanced tumor penetration. Nano Today 29, 100800 (2019). [Google Scholar]
- 85.Zhang Y. R., et al. , Strategies to improve tumor penetration of nanomedicines through nanoparticle design. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 11, e1519 (2019). [DOI] [PubMed] [Google Scholar]
- 86.Eble J. A., Niland S., The extracellular matrix in tumor progression and metastasis. Clin. Exp. Metastasis 36, 171–198 (2019). [DOI] [PubMed] [Google Scholar]
- 87.Nallanthighal S., Heiserman J. P., Cheon D. J., The role of the extracellular matrix in cancer stemness. Front. Cell Dev. Biol. 7, 86 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Najafi M., Farhood B., Mortezaee K., Extracellular matrix (ECM) stiffness and degradation as cancer drivers. J. Cell. Biochem. 120, 2782–2790 (2019). [DOI] [PubMed] [Google Scholar]
- 89.Chauhan V. P., et al. , Normalization of tumour blood vessels improves the delivery of nanomedicines in a size-dependent manner. Nat. Nanotechnol. 7, 383–388 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
NP diffusion in crosslinked network with mesh size of 4σ and spring constant of 100.
NP diffusion in crosslinked network with mesh size of 14σ and spring constant of 100.
NP diffusion in crosslinked network with mesh size of 4σ and spring constant of 10,000.
NP diffusion in aligned network with mesh size of 8σ and spring constant of 100.
Data Availability Statement
All study data are included in the article and/or SI Appendix.