Significance
Viruses may downregulate MHC class I expression on infected cells to avoid elimination by cytotoxic T cells. We report that the accessory proteins ORF7a and ORF3a of SARS-CoV-2 mediate this function and delineate the two distinct mechanisms involved. While ORF3a inhibits global protein trafficking to the cell surface, ORF7a acts specifically on MHC-I by competing with β2m for binding to the MHC-I heavy chain. This is the first account of molecular mimicry of β2m as a viral mechanism of MHC-I downregulation to facilitate immune evasion.
Keywords: antigen processing, immune evasion, SARS-CoV-2, molecular mimicry
Abstract
Major histocompatibility complex class I (MHC-I) molecules, which are dimers of a glycosylated polymorphic transmembrane heavy chain and the small-protein β2-microglobulin (β2m), bind peptides in the endoplasmic reticulum that are generated by the cytosolic turnover of cellular proteins. In virus-infected cells, these peptides may include those derived from viral proteins. Peptide-MHC-I complexes then traffic through the secretory pathway and are displayed at the cell surface where those containing viral peptides can be detected by CD8+ T lymphocytes that kill infected cells. Many viruses enhance their in vivo survival by encoding genes that down-regulate MHC-I expression to avoid CD8+ T cell recognition. Here, we report that two accessory proteins encoded by SARS-CoV-2, the causative agent of the ongoing COVID-19 pandemic, down-regulate MHC-I expression using distinct mechanisms. First, ORF3a, a viroporin, reduces the global trafficking of proteins, including MHC-I, through the secretory pathway. The second, ORF7a, interacts specifically with the MHC-I heavy chain, acting as a molecular mimic of β2m to inhibit its association. This slows the exit of properly assembled MHC-I molecules from the endoplasmic reticulum. We demonstrate that ORF7a reduces antigen presentation by the human MHC-I allele HLA-A*02:01. Thus, both ORF3a and ORF7a act post-translationally in the secretory pathway to lower surface MHC-I expression, with ORF7a exhibiting a specific mechanism that allows immune evasion by SARS-CoV-2.
Major histocompatibility complex class I (MHC-I) molecules are key players in mediating adaptive immune responses to viral infections. These molecules are heterodimers of a glycosylated transmembrane heavy chain and the small-protein β2-microglobulin (β2m) that are synthesized and assembled in the endoplasmic reticulum (ER) (1). MHC-I is then loaded with peptides derived from the turnover of cellular proteins, including viral proteins in infected cells. Peptide loading is facilitated by the concerted action of proteins that comprise the peptide loading complex (PLC), namely the transporter associated with antigen processing (TAP), which imports cellular and viral peptides from the cytosol into the ER, the peptide editor tapasin, and the ER chaperone proteins ERp57 and calreticulin (1). Peptide-MHC-I molecules then traffic to the surface of cells where self or virus-derived, nonself-peptides are exhibited to the immune system. This is the antigen-processing and presentation pathway. Components of the adaptive arm of the immune system, particularly CD8+ T cells also known as cytotoxic T lymphocytes or CTLs, identify and kill cells that express nonself-peptides (1).
Viruses down-regulate the expression of MHC-I by various mechanisms to avoid elimination by adaptive immune responses. This can involve transcriptional downregulation of MHC-I genes (2) or posttranslational inhibition of MHC-I function (3, 4). In the latter case, several steps in the antigen processing pathway are targeted by viruses. Examples include the herpes simplex virus ICP47 protein that inhibits TAP to prevent the import of cytosolic peptides, including virus-derived peptides, into the ER (3); the adenovirus E3/19K protein that prevents the interaction of MHC-I with the PLC (3) thereby interfering with effective peptide loading; and human papilloma virus E5 protein that retains MHC-I in the ER (5). Several viruses also encode pore-forming viral proteins or viroporins, which integrate into the host membranes, usually the ER, disrupting ion homeostasis (6) and inhibiting host protein trafficking through the secretory pathway. Examples include the influenza A virus M2 protein, poliovirus 2B and 3A proteins, coxsackievirus 2B protein, and SARS-CoV-1 ORF3a protein (7, 8). Cellular expression of poliovirus 3A viroporin (9) and coxsackievirus 2B viroporin (10) has been shown to reduce antigen presentation by MHC-I.
This study focuses on identifying SARS-CoV-2 gene products that posttranslationally down-regulate MHC-I expression and the mechanisms involved. SARS-CoV-2 is the causative agent of COVID-19, the ongoing pandemic that has afflicted the world for over 2 y, despite the availability of vaccines and emerging antiviral therapies (11). Thus, there remains a need to understand the molecular mechanisms of SARS-CoV-2 pathogenesis to aid the development of more effective, second-generation antiviral therapies. SARS-CoV-2 is a single, positive-stranded, enveloped RNA virus that encodes a number of translatable open reading frames (ORFs). These encode three main categories of proteins—nonstructural, structural, and accessory proteins (Fig. 1A). Infection of cells with SARS-Cov-2 has been reported to down-regulate MHC-I gene expression and reduce the surface expression of a specific MHC-I allele, HLA-A*02:01 (HLA-A2) (12, 13). Two accessory proteins have been implicated in this process. ORF6 suppressed the transcriptional upregulation of MHC-I observed upon viral infection (12), and expression of ORF8 posttranslationally decreased overall levels of MHC-I. ORF8 was shown to interact with HLA-A2 and direct it to autophagosomes for degradation, thus depleting the intracellular pool and in turn lowering surface HLA-A2, leading to reduced recognition and killing of ORF8-expressing cells by CTLs (13).
Fig. 1.
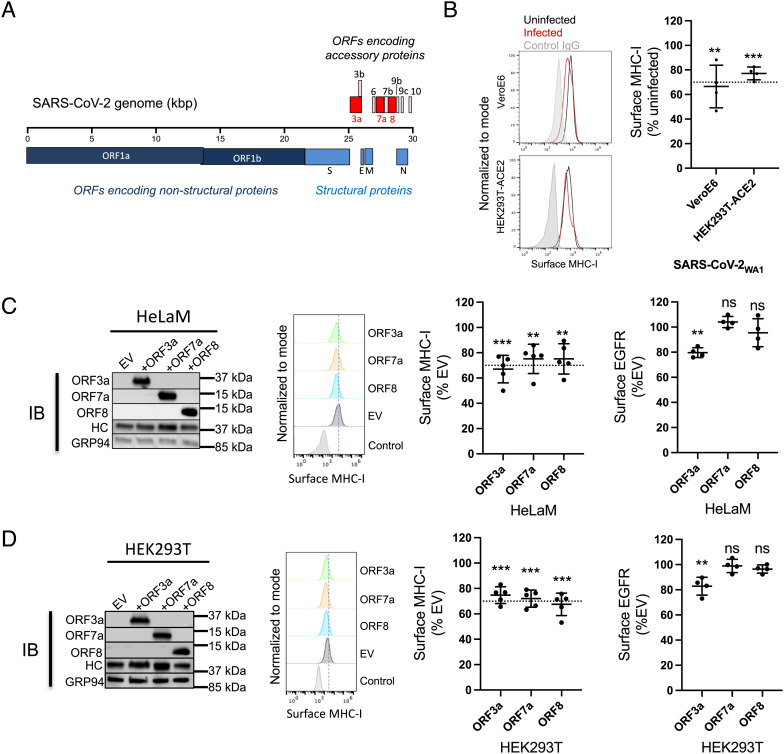
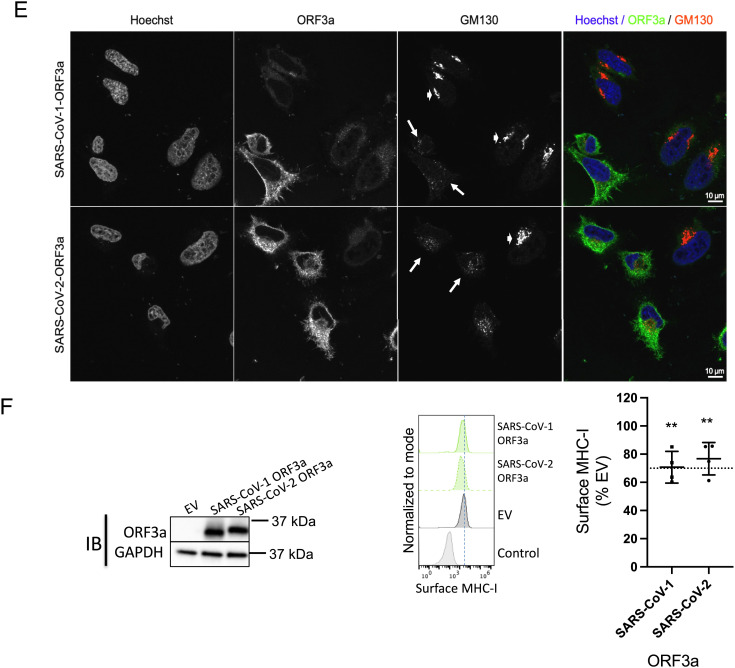
Downregulation of MHC-I by SARS-CoV-2 WA1 and its ER-localized gene products. (A) Genome organization of SARS-CoV-2. SARS-CoV-2 accessory proteins are depicted at the 3′ end of the genome. ORF3a, ORF7a, and ORF8 are highlighted in red. (B) Vero E6 or HEK293T-Ace2 cells were infected with SARS-CoV-2WA1 (MOI = 10). 24 h post infection, cells were collected for flow cytometry analysis for surface MHC-I (n = 4). Histograms (Left) are summarized (Right) where mean fluorescence intensity of MHC-I staining with W6/32 was normalized to uninfected group. (C) Transient expression of ORF3a, ORF7a, and ORF8 in HeLaM cells. Western Blot analysis of expression of accessory proteins, MHC-I heavy chain (HC), or GRP94 as a loading control, 24 h post transfection (Left). Flow cytometric analysis of surface MHC-I (Middle) and EGFR (Right) in cells expressing ORF3a, ORF7a, and ORF8 in HeLaM cells (n = 4). (D) Transient expression of ORF3a, ORF7a, and ORF8 in HEK293T cells. Western Blot analysis of expression of accessory proteins, MHC-I heavy chain (HC), or GRP94 as a loading control 24 h post transfection (Left). Flow cytometric analysis of surface MHC-I (Middle) and EGFR (Right) in cells expressing ORF3a, ORF7a, and ORF8 in HEK293T cells (n = 4). (E) Effect of ORF3a on the Golgi morphology. HeLaM cells were transfected with plasmids encoding SARS-CoV-1 ORF3a or SARS-CoV-2 ORF3a for 24 h. Fixed cells were stained with Hoechst (blue), for ORF3a (green), and anti-GM130 to visualize the Golgi (red). (F) HelaM cells were transfected with plasmids encoding SARS-CoV-ORF3a or SARS-CoV-2-ORF3a for 24 h, followed by analysis of expression by western blotting (Left) or flow cytometric analysis of surface MHC-I (Right, n = 4). Quantitative data shown are mean ± SD (error bars), and the dashed line indicates the 70% mark. Statistical significance was evaluated using the unpaired Student’s t test; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
Our analysis focused specifically on SARS-CoV-2 ORF3a and ORF7a as both of these proteins are predicted to localize to the secretory pathway and could post-translationally impact antigen presentation by MHC-I. ORF3a has a topology that suggests that it can integrate into lipid bilayers (14, 15), and studies in liposomes showed that it is permeable to cations such as calcium ions indicating that it is a viroporin (15). ORF7a has an N-terminal signal peptide and one putative transmembrane domain, indicating it is membrane associated (16). While roles for ORF3a and ORF7a in regulating innate, antiviral immune responses have been demonstrated (17), no role in adaptive immune responses has been described to date. We report here that ORF3a and ORF7a both reduce MHC-I levels to an extent comparable to ORF8 but by distinct mechanisms. ORF3a disrupts protein trafficking along the secretory pathway to inhibit surface expression of MHC-I, while ORF7a specifically interacts with the MHC-I heavy chain and delays its export from the ER.
Results
MHC-I Is Down-Regulated by SARS-CoV-2 Infection or on Expression of Its ER-Localized Accessory Proteins.
To investigate the effect of viral infection on antigen presentation by MHC-I, we first looked at the effect on the surface expression of MHC-I. We infected VeroE6 cells or HEK293T cells expressing human Angiotensin-Converting Enzyme 2 or Ace2, the receptor for SARS-CoV-2, (HEK293T-hAce2) with SARS-CoV-2WA1 at an MOI of 10 for 24 h, followed by estimation of surface MHC-I by flow cytometry. We observed that surface MHC-I was reduced by 30 to 40% in infected cells compared to uninfected cells (Fig. 1B and SI Appendix, Fig. S1). To specifically look at viral proteins that might regulate MHC-I by interfering with steps in the antigen presentation pathway, we focused on those that were likely to enter the ER. These include ORF7a and ORF8, which have N-terminal ER-localization signal sequences. While ORF3a does not have such a sequence, it has been shown to be a viroporin that integrates into lipid membranes (15). These three proteins and their positions in the SARS-CoV-2 genome are highlighted in Fig. 1A. To evaluate the effect on surface MHC-I expression, we transiently expressed them individually in the human cell lines HeLaM or HEK293T and measured surface MHC-I 24 h post transfection. All three proteins reduced surface MHC-I by 25 to 30% (Fig. 1 C and D and SI Appendix, Fig. S2). To ask whether this downregulation was specific to MHC-I or a consequence of an overall reduction of glycoprotein trafficking, we measured the expression levels of epidermal growth factor receptor (EGFR), estimated to have a similar half-life on the surface of cells to MHC-I [~8 to 18 h (18, 19)], and CD47, estimated to have a much longer surface half-life (~72 h) (19). ORF7a or ORF8 did not affect the surface expression of these proteins in either cell line, while ORF3a down-regulated the expression of EGFR and CD47 in both cell lines (Fig. 1 C and D, Right and SI Appendix, Fig. S3). We hypothesized that expression of ORF3a generally impairs protein trafficking through the secretory pathway, while the expression of ORF7a, like ORF8 (13), specifically affects MHC-I expression. We compared the downregulation mediated by ORF7a with another known viral inhibitor of MHC-I function, ICP47 (3) in both HeLaM and Huh7.5 cells, where the latter is known to be permissive for SARS-CoV-2 infection (20). We found that both proteins down-regulated MHC-I to a similar extent in both cell lines, with ICP47 being slightly more effective (SI Appendix, Fig. S4).
Expression of ORF3a Induces Golgi Fragmentation.
After synthesis in the ER, proteins destined for the cell surface usually traverse the Golgi apparatus en route to the plasma membrane. The Golgi structure has a characteristic appearance of a stack of aligned flat cisternae, and the disruption of the Golgi morphology by pathogens can lead to the dispersal or fragmentation of the stacked cisternae and is known to block anterograde protein trafficking (21, 22). In a report that has to date not been peer-reviewed, expression of the E-protein derived from infectious bronchitis virus (IBV-E protein) or the SARS-CoV-1 ORF3a protein was shown to disrupt the Golgi morphology, with a concomitant reduction of protein trafficking to the cell surface (8). We therefore looked at the effect of SARS-CoV-2 ORF3a expression on the architecture of the Golgi apparatus to ask if the same mechanism was involved. We expressed SARS-CoV-1 ORF3a or SARS-CoV-2 ORF3a proteins in HeLaM cells and assessed the Golgi morphology by confocal microscopy. In both instances, we found that the Golgi was fragmented in cells expressing ORF3a, observed as a loss of signal of the Golgi marker GM130 (Fig. 1E, panel 3, long white arrows) compared to cells that did not express ORF3a (Fig. 1E, panel 3, short white arrows). Transient expression of either SARS-CoV-1 or SARS-CoV-2 ORF3a in cells for 24 h led to a ~30% decrease in surface MHC-I levels (Fig. 1F). These data, along with the observed decrease in surface EGFR and CD47 (Fig. 1 C and D, Right and SI Appendix, Fig. S3), lead us to propose that ORF3a down-regulates surface MHC-I by blocking trafficking of glycoproteins to the cell surface by disrupting the Golgi architecture.
ORF7a Slows the Rate of Export of MHC-I from the ER.
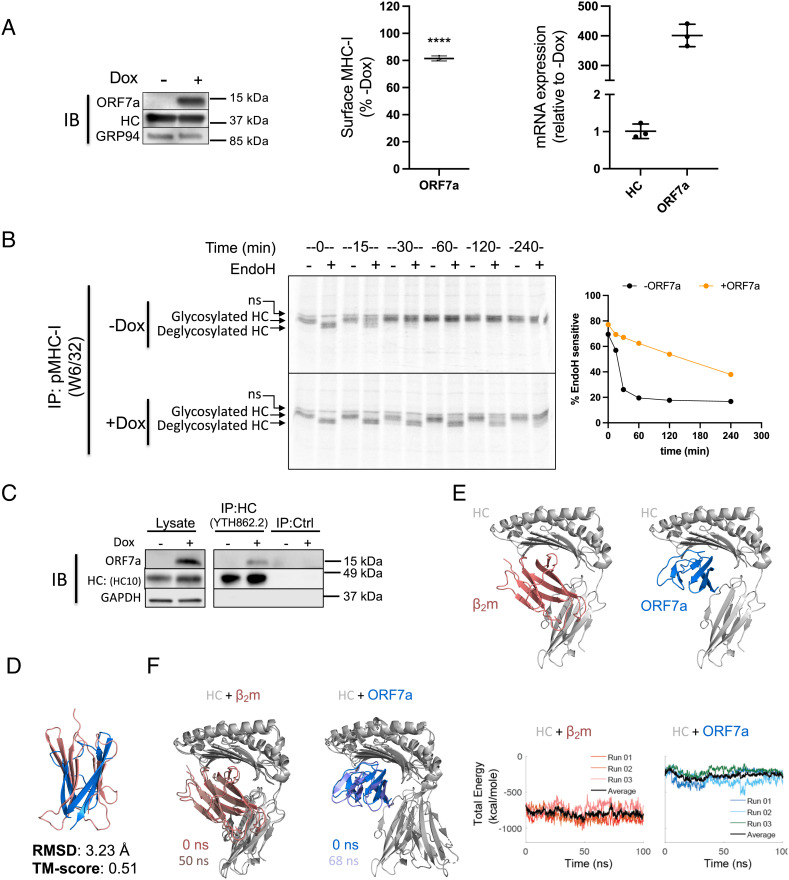
We have successfully used HeLaM cells in the past to understand the interactions between MHC-I and the PLC (23). Therefore, to evaluate the mechanism of ORF7a-mediated MHC-I downregulation, we generated a HeLaM cell line that allowed the inducible expression of ORF7a (HeLaM-iORF7a) on treatment with doxycycline (Dox). After ORF7a induction for 24 h, we observed a 20% decrease in surface MHC-I expression (Fig. 2 A, Left and Middle). There was no change in MHC-I mRNA levels, confirming that the downregulation was not transcriptional (Fig. 2 A, Right). Since there was no overall reduction in MHC-I protein levels in HeLaM cells expressing ORF7a compared to control cells in cell lysates (Figs. 1C and 2A), we asked if the intracellular distribution of MHC-I, specifically if its trafficking through the secretory pathway, was affected by ORF7a. We took advantage of the single N-linked glycan on the heavy chain of MHC-I to assess this. As newly synthesized N-linked glycoproteins traverse the secretory pathway, glycans are modified and rendered resistant to the enzyme Endoglycosidase H (EndoH) (24). We therefore carried out a pulse–chase experiment, followed by immunoprecipitation of MHC-I and EndoH treatment to assess progress through the secretory pathway.
Fig. 2.
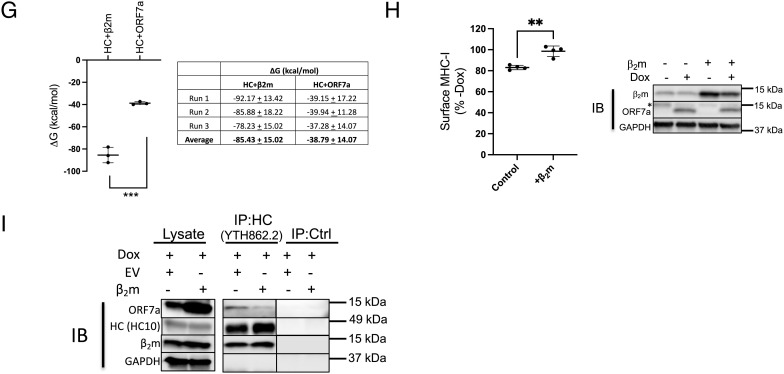
ORF7a slows the export of MHC-I from the ER by interacting with the heavy chain of MHC-I. (A) Stable expression of a doxycycline (Dox)-inducible construct of ORF7a in HeLaM cells was verified in uninduced and induced (+Dox for 24 h) cells by western blotting (Left), and the effect on surface MHC-I was assayed by flow cytometry (Middle, n = 3). MHC-I and ORF7a mRNA levels were assessed by qPCR and relative expression was normalized to uninduced (−Dox) cells (n = 3). (B) Uninduced (-Dox, Upper panel) or induced (+Dox for 24 h, Lower) cells were labeled with [35S]Met for 15 min followed by immunoprecipitation of MHC-I complexes from the lysates at the indicated time points of chase, followed by treatment with Endoglycosidase H (EndoH). Samples were visualized after separation on nonreducing SDS-polyacrylamide gels by autoradiography, and the intensity of the EndoH sensitive heavy chain relative to total heavy chain overtime has been plotted (Right). (C) Interaction between ORF7a and MHC-I was assessed by immunoprecipitation of total heavy chain (free and complexed) from uninduced or induced (+Dox for 24 h) cells lysed in digitonin using normal IgG as a control (IP:Ctrl) or α-HLA-A/B/C, clone YTH862.2 (IP: HC:YTH862.2), followed by western blot analysis to detect ORF7a, MHC-I heavy chain (HC), or GAPDH as loading control. (D) Structural analysis of ORF7a and MHC-I. Alignment of the crystal structures human β2m in red (PDBID: 2D4F) and SARS-CoV-2 ORF7a in blue (PDBID: 6W37); the rmsd and TM-Score (template modeling score) are also reported. (E) Cartoon representations of the crystal structure of HLA-A2 (gray) in complex with β2m (red) (Left) and the predicted structure of HLA-A2 (gray) complexed with ORF7a (blue) as modeled by ClusPro. (F) Cartoon representations showing a comparison of the original (0 ns) and a low-energy conformer obtained from MD simulation runs (run 01, 50 ns for HC+β2m and run 02, 68 ns HC + ORF7a). Graphs represent total energies (van der Waal + electrostatic) of the HC+β2m (red, Left) and HC+ORF7a (blue, Right) complexes over the course of three independent MD simulation runs. Average energies are shown in black in each case. (G) Plot of the free energy changes of binding (ΔG) of the HC+β2m and HC+ORF7a complexes. (H) HeLaM-iORF7a cells were transiently transfected with control plasmid or plasmid encoding human β2m for 8 h, followed by the induction of ORF7a expression (+Dox for 24 h) and effect on surface MHC-I was assayed by flow cytometry (Left, n = 3). Expression of ORF7a and β2m was assessed by western blotting, with GAPDH as loading control (Right). (I) Interaction between ORF7a and MHC-I in the presence of overexpressed human β2m was assessed by coimmunoprecipitation analysis. Human β2m or empty vector was transiently transfected into HeLaM-iORF7a cells followed by the induction of ORF7a expression for 24 h. Cells lysates were prepared in 1% digitonin and immunoprecipitation using normal IgG as a control (IP:Ctrl) or α-HLA-A/B/C, clone YTH862.2 (IP: HC: YTH862.2) was carried out, followed by western blot analysis to detect ORF7a, MHC-I heavy chain (HC), β2m, or GAPDH as loading control. Quantitative data shown are mean ± SD (error bars). Statistical significance was evaluated using the unpaired Student’s t test; *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
Doxycycline-treated and untreated HeLaM-iORF7a cells were pulse-labeled with 35S-methionine followed by a chase for up to 4 h. We immunoprecipitated the MHC-I complex at different time points and incubated it with or without EndoH followed by SDS-PAGE and autoradiography. MHC-I retained in the ER is sensitive to EndoH action and treatment results in the generation of a lower molecular weight, deglycosylated heavy chain. We found that in the absence of ORF7a (−Dox), the glycan was initially sensitive to EndoH action, but acquired resistance within 30 min (Fig. 2 B, Upper). However, in the presence of ORF7a (+Dox), EndoH resistance was delayed by up to 4 h (Fig. 2 B, Lower). This delay was specific to MHC-I as the EndoH-sensitivity pattern of the total radiolabeled glycoprotein pool, bound by the lectin Concanavalin A, was relatively unchanged by ORF7a induction after 4 h in a similar experiment (SI Appendix, Fig. S5).
ORF7a Competes with the β2m Interaction with the MHC-I Heavy Chain.
We then asked whether ORF7a was interacting with MHC-I or any components of the peptide-loading machinery that might interfere with the antigen processing pathway and cause a delay in ER export. We immunoprecipitated assembled peptide-MHC-I complexes (using W6/32 or α-β2m); tapasin [using PaSta1 (25), which is known to coimmunoprecipitate components of the PLC]; free heavy chains (using HC10) (26); or both free and complexed heavy chains (using the commercially available antibodies YTH862.2 from Novus and ɑ-rab-15240 from Proteintech) to look for associated ORF7a. We found ORF7a coimmunoprecipitated only with the latter two antibodies (Fig. 2C and SI Appendix, Fig. S6), implying that ORF7a is associated with the heavy chain outside of the PLC and with heavy chains that are not associated with β2m. Immunoprecipitation of ORF7a to detect associated heavy chain was unsuccessful, perhaps because the ORF7a epitope recognized by the monoclonal antibody used is blocked by heavy-chain interaction. We inferred from these results that the heavy chain can associate with either ORF7a or β2m but not both, suggesting that ORF7a expression may interfere with β2m association. We therefore analyzed the available structures of ORF7a, β2m, and the peptide-MHC-I complex to look for a potential interaction of ORF7a with the heavy chain. Both β2m and ORF7a contain an immunoglobulin (Ig)-like fold so we asked if the two are indeed structurally similar. A structural alignment using the Pairwise Structure Alignment tool provided by RCSB PDB showed a weak structural similarity, inferred by a higher rmsd value of 3.23 Å and a modest template modeling score (TM-score) of 0.51 (Fig. 2D). However, this TM-score also indicates that they do have the same protein fold. We then wanted to compare the structure of the heavy chain–β2m complex (HC-β2m) with that of a likely heavy chain–ORF7a complex (HC-ORF7a). While the crystal structure is available for the former (Fig. 2 E, Left), no structural information is available for the latter. We therefore modeled the interaction of ORF7a and the heavy chain of MHC-I using the ClusPro protein–protein docking server. The predicted structure indicated that ORF7a interacts with the heavy chain in the same region as β2m (Fig. 2 E, Right). This is compatible with our data indicating that either ORF7a or β2m, but not both, can interact with the heavy chain.
We then performed all atom, explicit solvent molecular dynamics (MD) simulations to determine the free energy changes (ΔG) of binding of β2m and ORF7a to the heavy chain using the structural models. Analysis of the MD simulation trajectories revealed a greater Cα atom rmsd and root mean squared fluctuation (RMSF) for the heavy chain when in complex with ORF7a compared to β2m (SI Appendix, Fig. S7 A–C). This is reflected as a greater change in the structure and higher total energy of HC-ORF7a compared to HC-β2m over time during the MD simulations (Fig. 2F). The most stable conformation of the complex observed during the simulation vs. the original structure has been superimposed (Fig. 2 F, Left), which is at 50 ns for HC-β2m and 68 ns for HC-ORF7a. Trajectory analysis revealed a lower total energy of HC-β2m compared to HC-ORF7a (Fig. 2 F, Right), which is driven largely by electrostatic interactions (SI Appendix, Fig. S7 D and E). In agreement with this, calculated ΔG values for the two complexes revealed a ~twofold difference, which was significant (Fig. 2H). Based on this information, we asked if the intracellular association of ORF7a and the heavy chain could be outcompeted by overexpressing β2m to rescue surface MHC-I. We transiently expressed a control plasmid or a plasmid encoding β2m in the HeLaM-iORF7a cells, followed by induction of ORF7a expression. Cells transfected with the control plasmid resulted in the expected decrease in MHC-I surface in Dox-treated compared to untreated cells, but this was eliminated upon expression of β2m (Fig. 2H), accompanied by a decreased association of ORF7a with the heavy chain (Fig. 2I). Thus, excess β2m can overcome the action of ORF7a.
Localization of ORF7a to the ER Determines Retention of MHC-I.
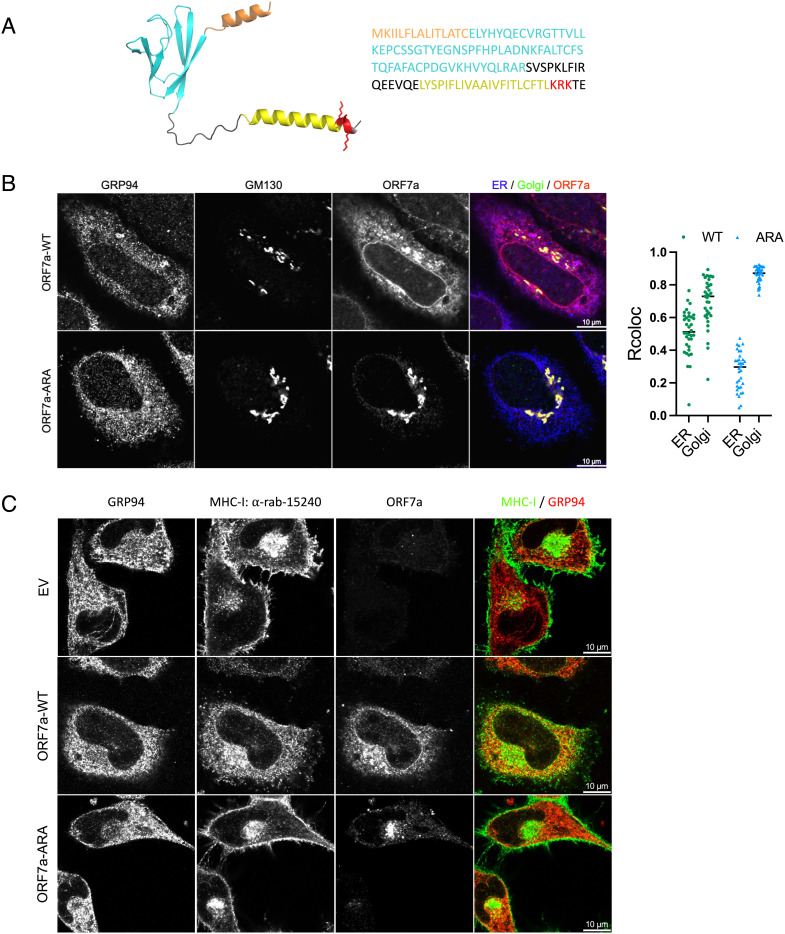
Our data so far indicate that the effect of ORF7a on MHC-I occurs in the ER. However, previous studies have found that ORF7a is primarily localized in the Golgi (16, 17, 27), despite harboring a dibasic amino acid sequence in its cytosolic C-terminal tail, which is a likely ER-retrieval sequence (28) (Fig. 3A). We therefore examined the localization pattern of the untagged ORF7a construct used throughout this study. Confocal analysis of HeLaM cells transiently expressing ORF7a showed that it localized to both the ER and Golgi (Fig. 3B). We generated a variant, ORF7a-ARA, where the lysine residues in the potential dibasic ER-retrieval sequence in the C-terminus of ORF7a were mutated to alanine and found that it was localized primarily in the Golgi (Fig. 3B). In agreement with our pulse–chase analysis, we saw that MHC-I was retained in the ER in the presence of ORF7a but this was not the case in the presence of ORF7a-ARA (Fig. 3C), where MHC-I localization was similar to mock-transfected cells. We then investigated whether ORF7a and ORF7a-ARA impact surface MHC-I levels and antigen presentation.
Fig. 3.
Localization of ORF7a to the ER determines retention of MHC-I. (A) A structural model of the full-length ORF7a as generated by AlphaFold (Left) and amino acid sequence of the protein (Right). Color coding indicates functional features of the protein- orange, ER-targeting signal sequence; cyan, Ig-like fold; yellow, transmembrane domain; red, the ER-retrieval motif (KRK) in the cytosolic C-terminal tail with the side chains shown. (B) Localization of ORF7a by confocal analysis. HeLaM cells were transfected with plasmids encoding ORF7a-WT or ORF7a-ARA and 24 h post transfection cells were fixed and stained with GRP94 (ER, blue), GM130 (Golgi, green), and ORF7a, red (Scale bar, 10 µm.) to determine colocalization (n = 35). (C) Localization of MHC-I in the presence of empty vector, ORF7a-WT, or ORF7a-ARA was determined by transfecting HeLaM cells with the corresponding plasmids for 24 h followed by confocal analysis of ORF7a, MHC-I (clone 15240, green), and GRP94 (ER, red) (Scale bar, 10 µm.).
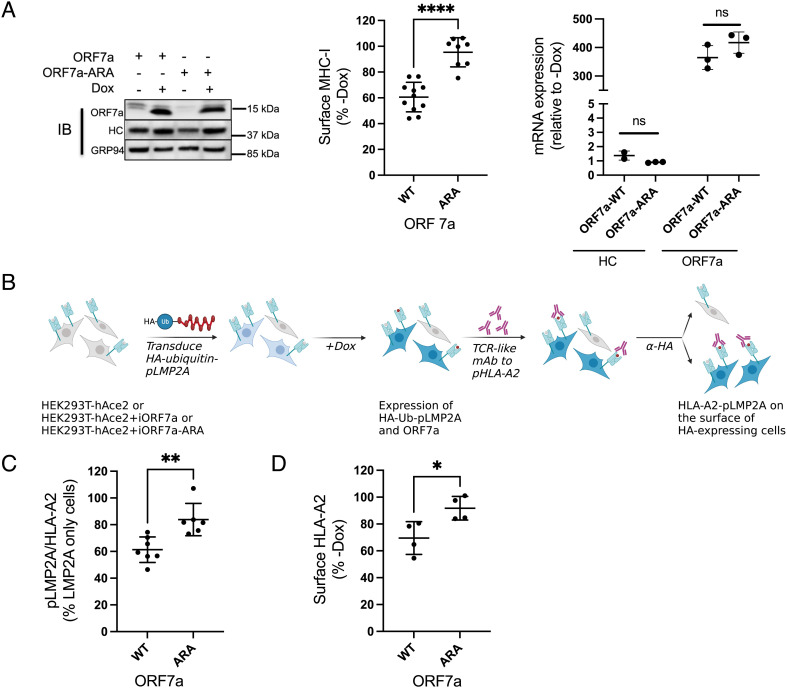
ORF7a-Mediated Downregulation of Antigen Presentation by MHC-I Is Dependent on the ER Retrieval Signal.
We generated HEK293T-hAce2 cells that expressed Dox-inducible versions of ORF7a or ORF7a-ARA (with the mutated ER-retrieval sequence) to assess the effect on surface MHC-I as well as antigen presentation by HLA-A*02:01 (HLA-A2), an allele that is not expressed in HeLaM cells. Following the induction of ORF7a or ORF7a-ARA expression for 24 h (Fig. 4 A, Left), MHC-I expression on the cell surface was determined. We observed that expression of ORF7a induced an approximately 40% reduction of surface MHC-I, which was not seen upon ORF7a-ARA induction (Fig. 4 A, Middle). There was no change in MHC-I mRNA levels as assessed by qPCR analysis (Fig. 4 A, Right). We found no differences in the expression of other surface glycoproteins such as EGFR and CD47 (SI Appendix, Fig. S8), in keeping with our previous finding that ORF7a does not affect overall glycoprotein trafficking (Fig. 1 C and D, Right and SI Appendix, Figs. S3–S5).
Fig. 4.
ORF7a-mediated downregulation of antigen presentation by MHC-I is dependent on the ER retrieval signal. (A) Stable expression of a doxycycline (Dox)-inducible construct of ORF7a-WT and ORF-7a-ARA in HEK293T-hAce2 cells was verified in uninduced and induced (+Dox for 24 h) cell by western blotting (Left), and the effect on surface MHC-I was assayed by flow cytometry (Middle, n = 4). MHC-I and ORF7a mRNA levels were assessed by qPCR and relative expression was normalized to uninduced (−Dox) cells (n = 3). (B) Outline of the experimental strategy employed to determine the effect of ORF7a on peptide presentation by HLA-A2. (C) The amount of pLMP2A presented by HLA-A2 was determined by flow cytometric analysis of induced cells (+Dox for 24 h) that expressed the fusion protein using a TCR-like monoclonal antibody specific to this complex (n = 6). (D) Flow cytometric analysis of surface HLA-A2 in cells expressing ORF7a-WT and ORF-7a-ARA (n = 4). Statistical significance was evaluated using the unpaired Student’s t test; *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
To assess whether the ORF7a-mediated decrease in MHC-I expression led to a decrease in antigen presentation, we evaluated the surface expression of a specific highaffinity peptide-MHC-I complex. The peptide we made use of is derived from the Epstein–Barr virus protein latent membrane protein 2A (LMP2A), which is presented efficiently by HLA-A2 and binds to it with highaffinity (29). We assessed the presentation of this peptide using a monoclonal antibody that mimics T cell receptors (TCRs) on the surface of CTLs that recognize this specific peptide-HLA-A2 complex (29). This flow cytometry-based assay provides a more quantitative assessment of antigen presentation than measuring cytotoxic activities of CTLs (29). We transduced HEK293T-hAce2 cells as well as these cells expressing inducible ORF7a or ORF7a-ARA (Fig. 4 B and C) with an inducible HA-tagged ubiquitin-fusion construct encoding the LMP2A-derived peptide (pLMP2A: CLGGLLTMV). The cytosolic ubiquitin-peptide fusion protein is degraded by cellular ubiquitin hydrolases, ensuring efficient presentation of the peptide (30), and the HA-tag allows the detection of cells expressing the construct (Fig. 4B and SI Appendix, Fig. S9). On induction with Dox, both ORF7a and pLMP2A were expressed, mimicking viral infection of cells with pLMP2A serving as a proxy for newly generated viral peptides. We found that in the HA-positive cells, expression of ORF7a but not ORF7a-ARA significantly reduced the presentation of pLMP2A (Fig. 4C). This decrease is likely due to the reduction of surface HLA-A2 mediated by ORF7a (Fig. 4D).
Discussion
The SARS-CoV-2 virus is a successful pathogen that can evade both innate and adaptive immunity. Adaptive immunity includes activation of CTLs in response to viral peptides presented by MHC-I on the surface of infected cells, leading to the elimination of these cells and restriction of viral replication. Here, we report two SARS-CoV-2 accessory proteins that can down-regulate MHC-I levels and demonstrate that they do so by two distinct mechanisms. ORF3a is a viroporin with three transmembrane domains that can oligomerize to form an ion channel, allowing cation permeability (15). This property allows SARS-CoV-1 to disrupt the architecture of the Golgi (8) (Fig. 1F), and Golgi fragmentation is accompanied by a reduction of protein trafficking through the secretory pathway and a decrease in the levels of surface proteins (8). We propose this is also the case for SARS-CoV-2 ORF3a, given their similar effects on the Golgi and their high degree of similarity (85%) with 73% identity.
There is no natural virus isolate that lacks ORF3a expression, but the contribution of ORF3a to the pathogenicity of SARS-CoV-2 has been evaluated in the K18 human ACE2 transgenic mouse model (31). In this mouse model of SARS-CoV-2 infection, the pathogenicity of infectious recombinant viral particles (rSARS-CoV-2) that lacked each of the accessory proteins was evaluated. Mice infected with ΔORF3a rSARS-CoV-2 showed significantly less morbidity and mortality (75% survival) and started to recover from the virus within 5 d of infection, while mice infected with the intact virus died within 6 d. Viral loads in the nasal turbinates and lungs were also significantly lower. ORF3a plays many roles that enhance viral pathogenicity in addition to the MHC-I downregulation described here, including inhibition of general surface protein trafficking, induction of apoptosis (32), inhibition of autophagy (17), and attenuating innate immune responses (17, 33). Whether a specific process or a combination of these processes reduce pathogenicity by ΔORF3a rSARS-CoV-2 remains to be evaluated.
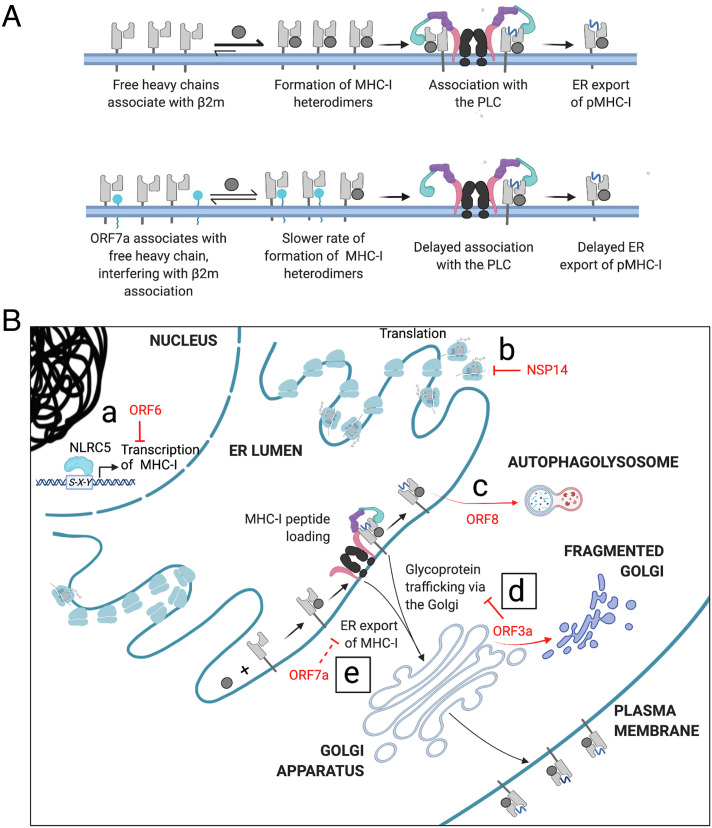
While the downregulation of MHC-I by ORF3a seems to be a consequence of a general inhibition of protein trafficking, MHC-I downregulation by ORF7a is specific. We found that ORF7a interacts with MHC-I heavy chains in the ER and slows the export of peptide–MHC-I complexes. We were unable to detect any ORF7a interacting with tapasin, under solubilization conditions that maintain tapasin association with the other components of the PLC, or with the assembled p–MHC-I complex. We speculate that newly synthesized, free heavy chains are the targets of interaction of ORF7a. The partial structure of ORF7a has been determined [PDBID: 6W37, (34)], and it is composed of an Ig-like domain. The human protein most structurally similar to ORF7a is intercellular adhesion molecule 3 (ICAM3), a potent immune signaling molecule. It was proposed that ORF7a may thus interact with partners of ICAM3 to modulate the immune response (35). Indeed, cell-free ORF7a can associate with the surface of uninfected CD14-positive cells (monocytes), implying a role in immune modulation although its interacting partner has not been identified (34) and how ORF7a, a transmembrane protein, might exit the cell is unclear. Human β2m is also an Ig-like molecule, and there is some structural overlap between β2m and ORF7a. Molecular modeling and MD simulation analysis of the complexes indicate that the interaction between ORF7a and MHC-I heavy chain is energetically favorable (even if it is less stable than the β2m and heavy-chain interaction) and that the projected interaction is broadly in the same region as β2m. Our data also indicate that ORF7a and β2m compete to interact with the heavy chain as overexpressed β2m decreases the association of ORF7a with the heavy chain and rescues the ORF7a-mediated downregulation of MHC-I. This suggests that ORF7a is a molecular mimic of β2m. Viral mimics of MHC-I heavy chain have been described, which permit infected cells to evade detection by natural killer cells that target cells with low MHC-I expression, as in the case for rodent herpesvirus Peru (36), or sequester viral peptides to prevent their effective presentation, as in the case for human cytomegalovirus (37). However, this report is the first description of a viral protein mimicking β2m, to the best of our knowledge.
Unlike β2m, ORF7a is membrane anchored and contains an ER-retrieval sequence, and further structural and biophysical characterization of the interactions between full-length ORF7a and the heavy chain as well as in vitro analysis of the kinetics of binding will aid in teasing out the details of this mimicry. It is known that β2m facilitates the folding of newly synthesized heavy chain (38), and the kinetics of heavy-chain synthesis in the presence of ORF7a as well as the interaction with other immune proteins that associate with β2m such as CD1d are important questions that arise from our finding that ORF7a is a molecular mimic of β2m. Based on our current findings, we propose that ORF7a interacts with the heavy chain to interfere with β2m association, which delays the formation of heavy-chain–β2m heterodimers, impairs their recruitment to the PLC, and reduces viral peptide loading and ER export (Fig. 5A). We have no evidence to suggest that ORF7a directly interferes with peptide loading; the reduced surface presentation of the LMP2A-derived peptide by HLA-A2 may simply be a consequence of the allele’s reduced surface expression. However, if mutation of the ER-retrieval sequence results in OR7a–heavy chain complex formation elsewhere in the secretory pathway or on the surface of cells and whether this impacts antigen presentation and T cell recognition remains to be assessed. A comprehensive analysis of the immunopeptidome of cells expressing ORF7a vs ORF7a-ARA will properly evaluate the full impact on peptide presentation.
Fig. 5.
Mechanisms by which SARS-CoV-2 evades adaptive immune responses mediated by MHC-I (A) Proposed mechanism by which ORF7a slows the export of MHC-I from the ER. In uninfected cells, the free heavy chain associates with β2m to form the MHC-I heterodimer. This associates with the PLC and MHC-I molecules loaded with peptide (pMHC-I) are released for export from the ER en route to the cell surface (Upper). ORF7a interacts with free heavy chain, interfering with its association with β2m and thus the peptide-loading complex, delaying exit from the ER (Lower). (B) Various gene products of SARS-CoV-2 down-regulate MHC-I-mediated immune responses in an infected cell. ORF6 prevents the transcriptional upregulation of MHC-I following infection (a), while NSP14 globally down-regulates translation (b). ORF8 interacts with HLA-A2 and targets it to the autophagosome where it is destined to be degraded on fusion with lysosomes to form the autophagolysosome (c). We report that the expression of ORF3a causes Golgi fragmentation, inhibiting the trafficking of MHC-I and other surface proteins (d) and that ER-localized ORF7a impedes the export of MHC-I from the ER, down-regulating surface MHC-I (e).
The ORF7a-induced downregulation of surface MHC-I and antigen presentation was alleviated when the ER-retrieval sequence of ORF7a was mutated, causing it to localize to the Golgi. In all our experiments, we expressed untagged ORF7a and found that it localized to both the ER and the Golgi by confocal microscopy. This is in contrast to three previous studies that indicated that ORF7a was primarily localized to the Golgi apparatus (16, 17, 27). The ORF7a constructs used in those studies were tagged C-terminally with either a small FLAG-tag (8 aa), 2x StrepII tag (30 aa), or mCherry-FLAG (244 aa). The function of the dibasic ER retrieval signal is likely to be masked by C-terminal extensions, resulting in a cellular distribution similar to the ORF7a-ARA mutant. One study of untagged SARS-CoV-1 ORF7a indicated that it localized to the Golgi and weakly to the ER in Vero cells (39, 40), which is more in keeping with our observations. The two CoV-1 and CoV-2 proteins show a sequence identity of 87%, and the ER retention sequence is conserved between them (34). Given the functional consequence of localization of ORF7a we have observed, we suggest that studies employing C-terminally tagged ORF7a need to be interpreted with caution. However, a recent report that has not been peer-reviewed found that C-terminally tagged ORF7a was capable of down-regulating MHC-I in HEK293T cells (41) and that the C-terminal tail was not necessary for this function (42). While we expressed ORF7a-ARA in an inducible manner, the ORF7a in these two studies was transiently expressed, and we are investigating whether this or other mechanisms account for this difference. In addition to the altered localization of ORF7a-ARA, we are in the process of characterizing any other functional consequences of this mutation. For example, ORF7a is known to be ubiquitinated on K119, (KRK119TE), which plays a role in antagonizing host innate immune responses (43) and would be lost. What impact this may have directly or indirectly on regulating MHC-I function remains to be determined.
Unlike ORF3a, SARS-CoV-2 variants lacking functional ORF7a appear to exist in nature. A viral strain with a 392 nucleotide deletion resulted in the expression of the signal sequence of ORF7a in fusion with ORF8, and another strain carrying a deletion of 227 nucleotides resulted in the fusion of ORF7a and ORF7b (44). Thus, both strains are effective ORF7a knockouts, but no study to date has determined if this changes disease severity. However, a strain that carried a truncated ORF7a caused by a 115 nucleotide deletion exhibited impaired in vitro growth and led to elevated interferon-mediated responses. In addition, it died out in the immunocompetent population 1.5 mo after it was detected (45). Another study identified a mutation in the transmembrane domain of ORF7a that improved its stability and caused severe infection despite low viral loads (46) although the mechanism behind this has not been determined. A mouse model of COVID-19 using ΔORF7a rSARS-CoV-2 showed that this virus had a replication defect in vitro and was cleared more efficiently from the nasal turbinates than an intact virus, and lung lesions caused by it were also resolved sooner (31). However, although the mice began to recover earlier than the mice infected with the intact virus, only 25% survived. This is in contrast to the 75% that survived when ORF3a is deleted. As with ORF3a, it is difficult to parse out which function of ORF7a is responsible for the amelioration of symptoms on infection with ΔORF7a rSARS-CoV-2. As a cautionary note, a similar analysis with ORF8 indicated that ΔORF8 rSARS-CoV-2 was as virulent as the WT virus in mice, but population studies in humans of SARS-CoV-2 with a 382 nucleotide deletion in ORF8 show that it results in milder infection and is not fixed in the population (47).
We assessed the effect of ORF7a on antigen presentation by detecting the specific HLA-A2/pLMP2A complex and found that it was down-regulated in the presence of ORF7a but not ORF7a-ARA. On inducing the expression of the LMP2A-derived peptide at the same time as ORF7a expression, we are mimicking the events of an infection where ORF7a is expressed along with pLMP2A, which serves as a proxy for viral peptides that may be presented by the cell. The interaction of ORF7a with free heavy chains likely delays the loading of MHC-I viral peptides by the PLC and enables the virus to evade immune detection. To evaluate the effect of ORF7a on MHC-I in the context of infection, we used the recombinant SARS-CoV-2 mNeonGreen (CoV-2-mNG) virus, where ORF7a has been replaced with the fluorescent protein, rendering it a knockout for ORF7a (48). However, we found no difference in surface MHC-I levels in cells infected with this virus compared to the wild type (SI Appendix, Fig. S10). There are multiple factors that may explain this. First, many SARS-CoV-2 proteins contribute to the downregulation of MHC-I on infection: transcriptional suppression of MHC-I expression upon infection by accessory protein ORF6 (12) and inhibition of global translation by the nonstructural protein NSP14 (49); ORF3a and ORF8 (13) also contribute to post translational downregulation of MHC-I. Second, ORF7a is expressed abundantly early in infection [within 3 h of the 8-h replication cycle (50)], presumably targeting nascent MHC-I molecules to prevent the presentation of peptides produced early in the viral infection, while our analysis was carried out 24 h post infection, perhaps missing the window of ORF7a-specific effects. Finally, rather than the levels of surface MHC-I, antigen presentation needs to be assessed at the early time point to see if there is indeed an inhibition of viral peptide presentation, as is suggested by our data in Fig. 4E.
Given the success of SARS-CoV-2 as a pathogen, it is unsurprising that it has evolved multiple strategies for evading immune responses. Many studies have focused on the role of SARS-CoV-2 proteins in regulating innate immunity (17, 51) and the mechanisms of how adaptive immune responses, specifically T cell responses, are modulated by the manipulation of MHC-I and are now coming to light. There are several regulatory points in the antigen processing pathway where MHC-I expression may be altered by SARS-CoV-2. Here, we describe two additional viral proteins that manipulate MHC-I levels posttranslationally by distinct mechanisms. There is a global decrease in the trafficking of surface proteins caused by ORF3a, and a specific retention of MHC-I in the ER by ORF7a. Both events delay MHC-I trafficking and impair antigen presentation (Fig. 5B). As with many viruses, these accessory proteins are multifunctional. A deeper understanding of the interactions of SARS-CoV-2 proteins with host proteins that manipulate immune responses will help in elucidating the mechanisms that contribute to pathogenicity and provide new avenues for the development of therapeutics, as well as aid in our understanding of any related emergent pathogens in the future.
Materials and Methods
Plasmids.
pLEX307-ACE2-blast was a gift from Alejandro Chavez and Sho Iketani (Addgene plasmid # 158449). pCAGGS vector encoding untagged viral proteins ORF3a, ORF7a, ORF8, and ORF7a-ARA were generated by Gibson assembly by PCR amplification from their respective HA-tagged constructs in pCAGGS (49). Plasmid pTRIPZ-puro-ORF7a-WT or ARA was generated by Gibson assembly by the PCR amplification of the genes of interest from their respective pcAGGS constructs and pTRIPZ-puro vector backbone digested with AgeI and EcoRI according to the manufacturer’s instructions. pTRIPZ-puro-HA-Ub was generated by Gibson assembly by PCR amplification of HA-tagged ubiquitin gene from the plasmid HA-Ubiquitin which was a gift from Edward Yeh (Addgene plasmid # 18712) and pTRIPZ-puro digested with AgeI and XhoI. This plasmid was digested with EcoRI and was used along with single-stranded DNA primers encoding pLMP2A with 20 bp overlap with the vector in a Gibson assembly reaction according to the manufacturer’s instructions to generate pTRIPZ-puro-HA-Ub-pLMP2A. The plasmid encoding ICP47 with a GFP reporter pMSCV-ICP47-IRES-GFP (available in the lab) was used to express the TAP inhibitor. All constructs were verified by sequencing. The details of primers used are listed in SI Appendix, Table S1.
Antibodies.
Rat anti-human GRP94 (ADI-SPA-850) was purchased from Enzo Life Sciences. The anti-human tapasin antibodies PaSta1 (25), free heavy chain antibody (HC10) (26), and the anti-HLA-A2 antibody BB7.2 (52) have been reported previously. We used mouse anti-human HLA-ABC (W6/32) conjugated to Alexa Fluor® 647 (MCA81A647, Bio-Rad), which is known to also react with MHC-I of African Green monkey from which Vero E6 cells are derived (53) for flow cytometric analysis. Mouse anti-human EGFR antibody conjugated to Alexa Fluor® 647(352917), mouse anti-human CD47 antibody conjugated to APC (323123), and anti-HA Alexa Fluor® 488 anti-HA.11 Epitope Tag (901509) were purchased from Biolegend. The antibody to ORF3a was a gift from Prof. Carolyn E. Machamer (8), and antibodies to ORF7a (GTX632602) and ORF8 (GTX135591) were purchased from GeneTex. We used a mouse monoclonal antibody to GM130 (BDB610822, from BD Sciences) and a rabbit polyclonal antibody to GM130 (11308 from Proteintech) for Golgi staining. The HLA-ABC antibody (clone YTH862.2) was purchased from Novus for immunoprecipitation, and the rabbit polyclonal antibody (15240) was purchased from Proteintech for immunoprecipitation and immunofluorescence imaging of MHC-I heavy chains. The fluorescently labeled spike protein antibody was purchased from eBioscience (53-6491-82). The TCR-like mAb that detects HLA-A2-pLMP2A (CLGGLLTMV) complexes by flow cytometry was a gift from Prof. Paul A. MacAry (41). All secondary antibodies used for flow cytometry and immunofluorescence imaging were purchased from Invitrogen, and those for immunoblotting that were coupled to horseradish peroxidase were purchased from Jackson ImmunoResearch.
Reagents.
Transit®-2020 and Transit®-Lenti (Mirus), polybrene (Millipore), Triton X-100 (AmericanBio), digitonin (Calbiochem), EDTA-free protease inhibitor mixture (Roche Applied Science), L-[35S]methionine, and L-[35S]cysteine (PerkinElmer Life Sciences), protein molecular weight markers, [methyl-14C] methylated (PerkinElmer Life Sciences), TRIzol (Invitrogen), ConcanavalinA-Sepharose 4B, Restore™ PLUS Western blotting stripping buffer (Thermo Scientific), and doxycycline hyclate (Sigma).
Transfection and Transduction.
For DNA plasmid transfection, cells were plated one day before and transfected using Transit®-2020 according to the manufacturer's instructions. Lentiviral particles were generated in HEK293T cells transfected with the packaging vector pCL-Ampho (Imgenex) and the construct of interest using Transit®-Lenti (Mirus) in Opti-MEM (Invitrogen), followed by two rounds of transduction of the recipient cell line with 8 µg/mL Polybrene (Millipore). Cells stably expressing the desired construct were obtained by selection in a complete medium supplemented with 2 µg/mL puromycin (Sigma) or 10 µg/mL blasticidin.
Cell Culture.
VeroE6, HeLaM, and HEK293T cells and derivatives were grown in Dulbecco’s modified Eagle’s medium (Sigma) supplemented with 10% fetal calf serum (HyClone) and 1% penicillin/streptomycin (Invitrogen).
Cell Lines Generated.
HEK293T cells expressing human Ace2 (HEK293T-hAce2) were generated by lentiviral transduction of cells with pLEX307-ACE2-blast, the vector encoding human ACE2, followed by selection with blasticidin (10 μg/mL). Expression of hAce2 was confirmed by western blotting. Inducible cell lines: HeLaM cells expressing inducible ORF7a (HeLaM-iORF7a) or HEK293T-hAce2 cells expressing inducible proteins (HEK293T-hAce2+iORF7a or HEK293T-hAce2+iORF7a-ARA) were generated by lentiviral transduction of the plasmid encoding the protein of interest followed by puromycin selection (2 μg/mL). HEK293T-hAce2 cell line and its variants were again transduced with a plasmid encoding the fusion protein of HA-tagged ubiquitin and LMP2A derived peptide (HA-Ub-pLMP2A) to generate HEK293T-hAce2+ipLMP2A, HEK293T-hAce2+iORF7a+ipLMP2A, and HEK293T-hAce2+iORF7a-ARA+ipLMP2A. Since this construct also carried the puromycin marker, transduced cells expressing the HA-Ub-pLMP2A construct were identified by intracellular staining with anti-HA as outlined in Fig. 4B. In all instances, induction of protein expression was carried out using 5 µg/mL doxycycline for 24 h.
SARS-Cov-2 Infection.
High-titer stocks of SARS-CoV-2 virus (isolate USA-WA1/2020, CoV-2 WA1) obtained from BEI reagent repository and SARS-CoV-2-mNG (48) were obtained by passage in Vero E6 cells. Viral titers were determined by plaque assay on Vero E6 cells. Virus was cultured exclusively in a biosafety level 3 facility. Vero E6 cells were either mock infected or infected at a multiplicity of infection (MOI) of 10 in serum-free DMEM for 1 h. Unbound virus was washed off, and cells were maintained in 2% fetal bovine serum in Dulbecco's modified Eagle's medium at 37 °C for 24 h, followed by staining for surface MHC-I for flow cytometric analysis. Post-staining cells were fixed using Cytofix Cytoperm, and intracellular fluorescence-activated cell sorting (FACS) was carried out where indicated.
Flow Cytometry.
1Cells were detached with trypsin and sequentially stained with primary and secondary antibodies (unless the primary antibody was fluorescently labeled) in FACS staining buffer (PBS + 0.5% BSA + 0.02% sodium azide) for 1 h at 4 °C. Cells were washed three times with FACS buffer after incubation with primary and secondary antibodies and analyzed using FACSCalibur (BD Biosciences) and FlowJo software (Tree Star). Staining with propidium iodide was carried out to distinguish dead from live cells in unfixed cells, and the Live/Dead Fixable Red Dead Cell kit (L34971, Invitrogen) was used in experiments that required fixing and intracellular staining. To distinguish transfected vs. untransfected cells, intracellular staining for the protein of interest was carried out after surface staining for MHC-I. Cells were fixed with 4% PFA (or CytofixCytoperm for virus-infected cells), followed by cell permeabilization with 0.4% TritonX-100 in in intracellular FACS buffer (TBS + 2.5% BSA + 0.1% Tween20), followed by staining with antibodies in intracellular FACS buffer overnight at 4 °C. Cells were then washed with TBS followed by staining with secondary antibodies or analysis as mentioned above.
Coimmunoprecipitation.
HeLaM cells were lysed in TBS, pH7.4, containing1% digitonin + protease inhibitors + 2 mM CaCl2 at 4 °C for 1 h. Clarified lysates were then either prepared for SDS-PAGE by the addition of Laemmli sample buffer or immunoprecipitated with indicated antibodies plus protein G-Sepharose for 2 h at 4 °C. After three washes in 0.1% digitonin lysis buffer and elution in nonreducing sample buffer, immunoprecipitates were separated by SDS-PAGE under nonreducing conditions followed by western blotting.
Immunoblotting.
Cell lysates were prepared by directly lysing an equal number of cells in 2% Laemmli sample buffer. Proteins were separated by SDS-PAGE followed by Western blot analysis. After probing with primary antibodies in TBST (TBS+ 0.1% Tween20) + 2.5% BSA overnight at 4 °C, blots were incubated with secondary horseradish peroxidase-conjugated antibodies in TBST + 0.5% BSA for 1 h at RT, and analyzed by the Chemidoc MP Imaging System (Bio-Rad). Blots were stripped and reprobed following treatment with Restore™ PLUS Western blotting stripping buffer according to the manufacturer’s instructions. Data are representative of experiments carried out with at least three biological replicates.
Immunofluorescence Assay.
Visualization of proteins of interest was carried out by confocal microscopy. HeLaM cells were plated on coverslips 24 h before transfection with plasmids encoding DNA of interest and processed 24 h post transfection. Cells were washed with PBS and fixed with 4% PFA for 15 min at room temperature, rinsed thrice with TBS + 100 mM glycine, and permeabilized with 0.4% TritonX-100 in TBST + 2.5% BSA for 10 min. Antibody staining was carried out in intracellular FACS buffer overnight at 4 °C in a humidified chamber, followed by three washes with TBST and incubation with secondary antibodies conjugated to fluorescent dyes and Hoechst 33342 stain for 30 min at 4 °C and three more washes with TBST. Coverslips were then mounted on slides using ProLongGold mountant according to the manufacturer’s instructions followed by imaging on the Leica STELLARIS 8. Images are representative of at least three biological replicates and at least 10 fields of view for each. Any postacquisition processing was kept consistent across images. The images were analyzed by Fiji (54). By applying the Polygon Selection function to GRP94 stained images, the cells were segmented to define the region of interest. Colocalization of ORF7a with GRP94 and GM130, respectively, was analyzed by the Colocalization Threshold function of Fiji. The Rloc values which represent the Pearson’s coefficient above the threshold were plotted.
Quantitative RT-PCR.
Cells were harvested in TRIzol, and RNA was extracted using the Direct-zol RNA MiniPrep kit (Zymo Research). cDNA synthesis was carried out using the iScript™ cDNA synthesis kit (Bio-Rad). PerfeCTa® SYBR® Green SuperMixMaster Mix (QuantaBio) was used for qPCR analysis according to the manufacturer’s instructions, and the reactions were set up by the epMotion M5073 robot (Eppendorf) and carried out in the CFX384 Touch™ real-time PCR detection system (BioRad). The details of the primers used are listed in SI Appendix, Table S2.
Metabolic Labeling and Immunoprecipitation.
HeLaM-iORF7a cells were incubated in cysteine- and methionine-free Dulbecco’s modified Eagle’s medium supplemented with 5% dialyzed fetal bovine serum (Invitrogen), 1% Glutamax, and 1% penicillin/streptomycin for 1 h, followed by labeling with [35S]methionine/cysteine (PerkinElmer Life Sciences) in the same medium for 15 min. Cells were then spun down and resuspended in complete medium, and cells were collected at indicated times of chase. Cell lysates were prepared in 1% Triton X-100 in TBS (10 mM Tris, pH 7.4, 150 mM sodium chloride) + protease inhibitors and precleared with normal rabbit IgG, followed by immunoprecipitation with W6/32 and protein G-Sepharose. The immunoprecipitates were washed three times with TBS + 0.5% Triton X-100, and proteins were visualized by autoradiography after separation on nonreducing SDS-polyacrylamide gels.
Structural Analysis.
The crystal structures deposited in the protein data bank (PDB) of human β2m (PDBID: 2D4F) (55), ORF7a (PDBID: 6W37) (56), and HLA-A2/β2m/peptide complex (PDBID: 1DUY) (57) were used in the analysis. The structural alignment of hβ2m and ORF7a was carried out using the Pairwise Structure Alignment tool provided by RCSB PDB (58, 59), using the TM-align parameter. The molecular docking of ORF7a and HLA-A2 heavy chain was carried out using ClusPro (60–63), and analysis of the interactions was carried out using the PRODIGY server (64, 65). The model of the full length of ORF7a was generated using AlphaFold (66).
MD Simulations and Binding Free Energy Analysis.
Nanoscale Molecular Dynamics (NAMD) V2.13 software (67) and CHARMM36 force field (68) were utilized to perform the MD simulations as described previously (69–71). The simulation systems consisting of the biomolecular complexes of HC+β2m and HC+ORF7a described above were generated using the QwikMD Toolkit (72) available as a plugin in Visual Molecular Dynamics (VMD) software V1.9.3 (73). Briefly, TIP3P (74) cubic water box was used to explicitly solvate the proteins. Sodium chloride (NaCl) at a concentration of 0.15 M in the explicit solvent was used for charge neutralization with Periodic Boundary Conditions applied. Prior to performing MD production runs, energy minimization was carried out on the biomolecular simulation systems for 2,000 timesteps (0.004 ns) followed by an annealing step for 0.25 ns where the temperature was elevated at 1 K increments from 60 to 310 K. Temperature and pressure were then held constant at 310 K using Langevin temperature control and at 1.0 atm using Nose-Hoover Langevin piston control, respectively, followed by 1 ns equilibration step where backbone atoms of proteins were constrained through harmonic potential. Finally, three independent 100-ns runs were performed for each of the HC+ β2m and HC+ORF7a complex biomolecular simulation systems. A 2 fs integration time step was selected for all simulations where trajectory frames were saved every 10,000 steps. A 12 Å cut-off with 10 Å switching distance was chosen to handle short-range nonbonded interactions, while long-range nonbonded electrostatic interactions were handled using the Particle-mesh scheme at 1 Å PME grid spacing. Analysis of interaction energy (vdW and electrostatic) throughout the 100-ns simulation was performed using the “NAMD Energy” analysis tool available as part of VMD (68). Binding free energy changes (ΔG) were estimated through molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) method (70) using the CaFE 1.0 tool (71) combined with VMD (68) over the course of entire 100-ns MD simulation runs. All structures were visualized in PyMOL and images were created in PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Dr. Carolyn Machamer of Johns Hopkins University School of Medicine for advice on the function of ORF3a and for a critical reading of the manuscript. M.L.-R. was supported by grant number KL2 TR001862, J.P. was supported by 5-T32 HL 7975-19, and J.C.-C. was supported by a Cancer Research-Irvington Postdoctoral Fellowship. The work was supported by NIH grant RO1 AI059167 to P.C. W.S.A. is supported by a scholarship from the College of Health & Life Sciences, Hamad Bin Khalifa University, a member of the Qatar Foundation. Some of the computational research work reported in the manuscript was performed using high-performance computer resources and services provided by the Research Computing group in Texas A&M University at Qatar. Research Computing is funded by Qatar Foundation for Education, Science and Community Development (http://www.qf.org.qa).
Author contributions
N.A., K.H.B., and P.C. designed research; N.A., M.L.-R., W.S.A., S.M.M., J.P., and P.C. performed research; J.C.-C.H. contributed new reagents/analytic tools; N.A., W.S.A., D.S., K.H.B., and P.C. analyzed data; and N.A. and P.C. wrote the paper.
Competing interest
The authors have organizational affiliations to disclose, Debrup Sengupta, now an employee of Corner Therapeutics, Cambridge, MA.
Footnotes
Reviewers: P.J.L., University of Cambridge; and S.S., Jacobs University Bremen.
Data, Materials, and, Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Blum J. S., Wearsch P. A., Cresswell P., Pathways of antigen processing. Annu. Rev. Immunol. 31, 443–473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Menachery V. D., et al. , MERS-CoV and H5N1 influenza virus antagonize antigen presentation by altering the epigenetic landscape. Proc. Natl. Acad. Sci. U.S.A. 115, E1012–E1021 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van de Weijer M. L., Luteijn R. D., Wiertz E. J., Viral immune evasion: Lessons in MHC class I antigen presentation. Semin. Immunol. 27, 125–137 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Hewitt E. W., The MHC class I antigen presentation pathway: Strategies for viral immune evasion. Immunology 110, 163–169 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashrafi G. H., Brown D. R., Fife K. H., Campo M. S., Down-regulation of MHC class I is a property common to papillomavirus E5 proteins. Virus Res. 120, 208–211 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Gargan S., Stevenson N. J., Unravelling the immunomodulatory effects of viral ion channels, towards the treatment of disease. Viruses 13, 2165 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sze C. W., Tan Y. J., Viral membrane channels: Role and function in the virus life cycle. Viruses 7, 3261–3284 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzales R. R., Machamer C. E., The SARS CoV-1 3a protein disrupts Golgi complex morphology and cargo trafficking. bioRxiv [Preprint] (2021). 10.1101/2021.04.19.440492. [DOI]
- 9.Deitz S. B., Dodd D. A., Cooper S., Parham P., Kirkegaard K., MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proc. Natl. Acad. Sci. U.S.A. 97, 13790–13795 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kemball C. C., et al. , Coxsackievirus B3 inhibits antigen presentation in vivo, exerting a profound and selective effect on the MHC class I pathway. PLoS Pathog. 5, e1000618 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hebbani A. V., et al. , COVID-19: Comprehensive review on mutations and current vaccines. Arch. Microbiol. 204, 8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoo J. S., et al. , SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the STAT1-IRF1-NLRC5 axis. Nat. Commun. 12, 6602 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y., et al. , The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Iota. Proc. Natl. Acad. Sci. U.S.A. 118, e2024202118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janoschke M., et al. , Efficient integration of transmembrane domains depends on the folding properties of the upstream sequences. Proc. Natl. Acad. Sci. U.S.A. 118, e2102675118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kern D. M., et al. , Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs. Nat. Struct. Mol. Biol. 28, 573–582 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J. G., et al. , Characterization of SARS-CoV-2 proteins reveals Orf6 pathogenicity, subcellular localization, host interactions and attenuation by Selinexor. Cell Biosci. 11, 58 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayn M., et al. , Systematic functional analysis of SARS-CoV-2 proteins uncovers viral innate immune antagonists and remaining vulnerabilities. Cell Rep. 35, 109126 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sorkin A., Duex J. E.. Quantitative analysis of endocytosis and turnover of epidermal growth factor (EGF) and EGF receptor. Curr. Protoc. Cell Biol. 15, 15.14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen W., Smeekens J. M., Wu R., Systematic study of the dynamics and half-lives of newly synthesized proteins in human cells. Chem. Sci. 7, 1393–1400 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dittmar M., et al. , Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-Cov-2. Cell Rep. 35, 108959 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Midgley R., et al. , A role for endoplasmic reticulum exit sites in foot-and-mouth disease virus infection. J. Gen. Virol. 94, 2636–2646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aistleitner K., Clark T., Dooley C., Hackstadt T., Selective fragmentation of the trans-Golgi apparatus by Rickettsia rickettsii. PLoS Pathog. 16, e1008582 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panter M. S., Jain A., Leonhardt R. M., Ha T., Cresswell P., Dynamics of major histocompatibility complex class I association with the human peptide-loading complex. J. Biol. Chem. 287, 31172–31184 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bieberich E., Synthesis, processing, and function of N-glycans in N-glycoproteins. Adv. Neurobiol. 9, 47–70 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dick T. P., Bangia N., Peaper D. R., Cresswell P., Disulfide bond isomerization and the assembly of MHC class I-peptide complexes. Immunity 16, 87–98 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Stam N. J., Spits H., Ploegh H. L., Monoclonal antibodies raised against denatured HLA-B locus heavy chains permit biochemical characterization of certain HLA-C locus products. J. Immunol. 137, 2299–2306 (1986). [PubMed] [Google Scholar]
- 27.Zhang J., et al. , A systemic and molecular study of subcellular localization of SARS-CoV-2 proteins. Signal Transduct. Target. Ther. 5, 269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao C., et al. , Retention mechanisms for ER and Golgi membrane proteins. Trends Plant Sci. 19, 508–515 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Sim A. C., et al. , Defining the expression hierarchy of latent T-cell epitopes in Epstein-Barr virus infection with TCR-like antibodies. Sci. Rep. 3, 3232 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolf B. J., Princiotta M. F., Viral and bacterial minigene products are presented by MHC class I molecules with similar efficiencies. Mol. Immunol. 48, 463–471 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Silvas J. A., et al. , Contribution of SARS-CoV-2 accessory proteins to viral pathogenicity in K18 human ACE2 transgenic mice. J. Virol. 95, e0040221 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren Y., et al. , The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell Mol. Immunol. 17, 881–883 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia H., et al. , Evasion of type I interferon by SARS-CoV-2. Cell Rep. 33, 108234 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Z., et al. , Structural insight reveals SARS-CoV-2 ORF7a as an immunomodulating factor for human CD14(+) monocytes. iScience 24, 102187 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ongaro A., Oselladore E., Memo M., Ribaudo G., Gianoncelli A., Insight into the LFA-1/SARS-CoV-2 Orf7a complex by protein-protein docking, molecular dynamics, and MM-GBSA calculations. J. Chem. Inf. Model 61, 2780–2787 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X., et al. , A herpesvirus encoded Qa-1 mimic inhibits natural killer cell cytotoxicity through CD94/NKG2A receptor engagement. Elife 7, e38667 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Z., Bjorkman P. J., Structure of UL18, a peptide-binding viral MHC mimic, bound to a host inhibitory receptor. Proc. Natl. Acad. Sci. U.S.A. 105, 10095–10100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H., Capps G. G., Robinson B. E., Zuniga M. C., Ab initio association with beta 2-microglobulin during biosynthesis of the H-2Ld class I major histocompatibility complex heavy chain promotes proper disulfide bond formation and stable peptide binding. J. Biol. Chem. 269, 22276–22281 (1994). [PubMed] [Google Scholar]
- 39.Fielding B. C., et al. , Characterization of a unique group-specific protein (U122) of the severe acute respiratory syndrome coronavirus. J. Virol. 78, 7311–7318 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson C. A., Pekosz A., Lee C. A., Diamond M. S., Fremont D. H., Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure 13, 75–85 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moriyama M. L., et al. , SARS-CoV-2 variants do not evolve to promote further escape from MHC-I recognition. bioRxiv (2022). 10.1101/2022.05.04.490614. [DOI]
- 42.Zheng S., The SARS-CoV-2 accessory factor ORF7a downregulates MHC class I surface expression. bioRxiv (2022). 10.1101/2022.05.29.493850. [DOI]
- 43.Cao Z., et al. , Ubiquitination of SARS-CoV-2 ORF7a promotes antagonism of interferon response. Cell Mol. Immunol. 18, 746–748 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Addetia A., et al. , Identification of multiple large deletions in ORF7a resulting in in-frame gene fusions in clinical SARS-CoV-2 isolates. J. Clin. Virol. 129, 104523 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nemudryi A., et al. , SARS-CoV-2 genomic surveillance identifies naturally occurring truncation of ORF7a that limits immune suppression. Cell Rep. 35, 109197 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lobiuc A., Sterbuleac D., Sturdza O., Dimian M., Covasa M., A conservative replacement in the transmembrane domain of SARS-CoV-2 ORF7a as a putative risk factor in COVID-19. Biology 10, 1276 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young B. E., et al. , Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: An observational cohort study. Lancet 396, 603–611 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie X., et al. , An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe 27, 841–848.e843 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu J. C., Laurent-Rolle M., Pawlak J. B., Wilen C. B., Cresswell P., Translational shutdown and evasion of the innate immune response by SARS-CoV-2 NSP14 protein. Proc. Natl. Acad. Sci. U.S.A. 118, e2101161118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weingarten-Gabbay S., et al. , Profiling SARS-CoV-2 HLA-I peptidome reveals T cell epitopes from out-of-frame ORFs. Cell 184, 3962–3980.e3917 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lei X., et al. , Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 11, 3810 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parham P., Brodsky F. M., Partial purification and some properties of BB7.2. A cytotoxic monoclonal antibody with specificity for HLA-A2 and a variant of HLA-A28. Hum. Immunol. 3, 277–299 (1981). [DOI] [PubMed] [Google Scholar]
- 53.Jacobsen C. N., Aasted B., Broe M. K., Petersen J. L., Reactivities of 20 anti-human monoclonal antibodies with leucocytes from ten different animal species. Vet. Immunol. Immunopathol. 39, 461–466 (1993). [DOI] [PubMed] [Google Scholar]
- 54.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kihara M., et al. , Conformation of amyloid fibrils of beta2-microglobulin probed by tryptophan mutagenesis. J. Biol. Chem. 281, 31061–31069 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Nelson C. A., Minasov G., Shuvalova L., Fremont D. H.; Center for Structural Genomics of Infectious Diseases (CSGID), Structure of the SARS-CoV-2 ORF7A encoded accessory protein (2020).DOI: 10.2210/pdb6W37/pdb [DOI]
- 57.Khan A. R., Baker B. M., Ghosh P., Biddison W. E., Wiley D. C., "Crystal structure of HLA-A*0201/Octameric tax peptide complex" (2000). 10.2210/pdb1duy/pdb [DOI] [PubMed]
- 58.Berman H. M., et al. , The protein data bank. Nucleic. Acids. Res. 28, 235–242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma J., Wang S., Algorithms, applications, and challenges of protein structure alignment. Adv. Protein. Chem. Struct. Biol. 94, 121–175 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Desta I. T., Porter K. A., Xia B., Kozakov D., Vajda S., Performance and its limits in rigid body protein-protein docking. Structure 28, 1071–1081.e1073 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vajda S., et al. , New additions to the ClusPro server motivated by CAPRI. Proteins 85, 435–444 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kozakov D., et al. , The ClusPro web server for protein-protein docking. Nat. Protoc. 12, 255–278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kozakov D., et al. , How good is automated protein docking? Proteins 81, 2159–2166 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vangone A., Bonvin A. M., Contacts-based prediction of binding affinity in protein-protein complexes. Elife 4, e07454 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xue L. C., Rodrigues J. P., Kastritis P. L., Bonvin A. M., Vangone A., PRODIGY: A web server for predicting the binding affinity of protein-protein complexes. Bioinformatics 32, 3676–3678 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Phillips J. C., et al. , Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Best R. B., et al. , Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory comput. 8, 3257 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Altamash T., Ahmed W., Rasool S., Biswas K. H., Intracellular ionic strength sensing using nanoluc. Int. J. Mol. Sci. 22, E677 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ahmed W., Phillip A. M., Biswas K. H., Stable interaction of the UK B. 1.1. 7 lineage SARS-CoV-2 S1 spike N501Y mutant with ACE2 revealed by molecular dynamics simulation. bioRxiv [Preprint] (2021). 10.1101/2021.01.07.425307. [DOI]
- 71.Geethakumari A., et al. , A Genetically encoded BRET-based SARS-CoV-2 Mpro protease activity sensor. bioRxiv [Preprint] (2022). 10.1101/2022.01.31.478460. [DOI] [PMC free article] [PubMed]
- 72.Ribeiro J. V., et al. , QwikMD–Integrative molecular dynamics toolkit for novices and experts. Sci. Rep. 6, 26536 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33 (1996). [DOI] [PubMed] [Google Scholar]
- 74.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.