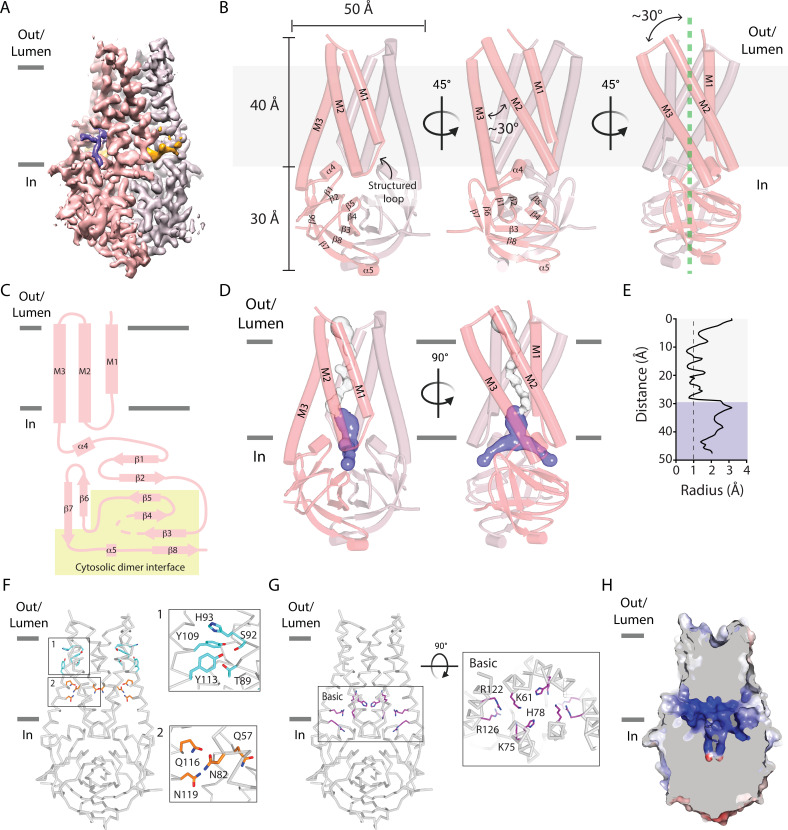
Figure 3. A narrow cavity detected in the SARS-CoV-2 Orf3a TM region is unlikely to conduct cations.
(A–C) Overall architecture of SARS-CoV-2 (CoV-2) Orf3a. (A) Cryo-EM map of dimeric CoV-2 Orf3a (dark and light pink), with density for lipids colored (orange, purple). (B) Three side views of CoV-2 Orf3a depicting dimeric architecture (dark and light pink) and key structural elements. (C) 2D topology of CoV-2 Orf3a. The region forming the cytosolic dimer interface is shown (yellow). (D) Inspection of the CoV-2 Orf3a TM region for a pore, depicted as the minimal radial distance from its center to the nearest van der Waals contact (HOLE program) (Smart et al., 1996). A region too narrow to conduct ions (white) and an aqueous vestibule (dark blue) are highlighted. (E) Radius of the pore (from D) as a function of the distance along the ion pathway. Dashed lines indicate the minimal radius that would permit a dehydrated ion. Blue and white colors follow (D). (F) Two layers of polar residues (1 and 2, cyan and orange) identified in the TM region, with a zoom in of each region. (G) Basic residues located in the aqueous vestibule (purple) with zoom in of the region. (H) Cutaway of the CoV-2 Orf3a molecular surface to view the aqueous vestibule is colored according to the electrostatic potential (APBS program) (Jurrus et al., 2018). Coloring: blue, positive (+10 kT/e) and red, negative (–10 kT/e).