Abstract
The domestication history of the avocado (Persea americana) remains unclear. We created a reference genome from the Gwen varietal, which is closely related to the economically dominant Hass varietal. Our genome assembly had an N50 of 3.37 megabases, a BUSCO score of 91%, and was scaffolded with a genetic map, producing 12 pseudo-chromosomes with 49,450 genes. We used the Gwen genome as a reference to investigate population genomics, based on a sample of 34 resequenced accessions that represented the 3 botanical groups of P. americana. Our analyses were consistent with 3 separate domestication events; we estimated that the Mexican group diverged from the Lowland (formerly known as “West Indian”) and Guatemalan groups >1 million years ago. We also identified putative targets of selective sweeps in domestication events; within the Guatemalan group, putative candidate genes were enriched for fruit development and ripening. We also investigated divergence between heterodichogamous flowering types, providing preliminary evidence for potential candidate genes involved in pollination and floral development.
Keywords: domestication, avocado, population genomics, heterodichogamy, reference genome, Plant Genetics and Genomics
Introduction
Avocado (Persea americana Mill.) is a perennial, subtropical crop that is in ever-increasing demand. In the United States, for example, per capita avocado consumption has tripled over the last 2 decades. Demand in the U.S. is met partly by domestic production but principally by imports from Mexico and elsewhere. Mexico is the largest producer, where the crop is worth an estimated $2.5 billion per year (Rendón-Anaya et al. 2019), but other major producers include the Dominican Republic, Peru, Brazil, Indonesia, Israel, and Kenya (http://www.fao.org/faostat/en/#data/QC/visualize). Although the popularity of avocados is primarily a 20th century phenomenon (Schaffer et al. 2013), they have quickly grown to be a global commodity.
Remarkably, avocado cultivation is dominated by a single variety (Hass) that represents ∼90% of cultivation worldwide (Rendón-Anaya et al. 2019). All Hass trees are derived clonally from a tree patented in 1935. Despite the shockingly narrow genetic base of agricultural production, avocado sensu lato is quite genetically diverse. Some of this diversity stems from the fact that there are 3 domesticated botanical groups (formerly called “races”): P. americana var. americana Miller [(which we will call the “Lowland” group in recognition that the previously accepted name of West Indian is inaccurate (Me and Arzate-Fernández 2010)], var. drymifolia (Schltdl. & Cham.) S.F. Blake (the Mexican group), and var. guatemalensis (L.O. Williams) Scora (the Guatemalan group) (Bergh and Ellstrand 1986). The strikingly different fruit morphologies among the groups suggest that they may have been domesticated separately, a conjecture supported by genetic data (Furnier et al. 1990; Ashworth and Clegg 2003; Rendón-Anaya et al. 2019). Further genetic and ethnobotanical data suggest that the 3 groups did not come into contact until the 16th century (Storey et al. 1986; Ashworth and Clegg 2003). One practical consequence of this history is that each group likely contains separate alleles and/or genes of interest for crop improvement, due to their different domestication histories. Another consequence is that hybridization between groups can produce unique allelic combinations, potentially leading to agronomically useful hybrid offspring. Hass may be, in fact, an example; although its precise breeding history is not known, genetic evidence has suggested that it is a hybrid between Guatemalan and Mexican groups (Chen et al. 2009; Schaffer et al. 2013; Rendón-Anaya et al. 2019).
The high demand for, and economic importance of, avocados motivates breeding efforts, but breeding remains challenging for at least 3 reasons. First, the avocado is a large tree that matures slowly [5–8 years before production (Lahav and Lavi 2009)] requiring substantial space, water, and finances (Ashworth et al. 2019). A second major obstacle is the reproductive system. A single tree typically produces more than 1 million flowers, of which only 0.1% or fewer yield mature fruit (Davenport 1986; Davis et al. 1998; Bender 2002), making controlled pollinations difficult (Chen et al. 2007). Finally, avocado is heterodichogamous, with 2 flowering types: A and B. Type A trees are female (receptive to pollen) in the morning of the first day and shed pollen as males in the afternoon of the following day. In contrast, type B trees are female in the afternoon of the first day and male in the morning of the next day. Although heterodichogamy is likely caused by a simple underlying genetic mechanism (Renner 2001), the system is complicated by the fact that there is some leakiness that depends on environmental cues (Davenport 1986). Heterodichogamy enhances outcrossing, but it does not ensure it. Selfing has been measured at rates from 2% to >90%, depending on the variety and environmental conditions (Torres and Bergh 1978; Degani and Gazit 1984; Davenport 1986). As a result of the complexities of the mating system, avocado breeding has historically relied on open-pollinated and inter-group hybridization, to the extent that most individual varieties lack accurate breeding records (Davis et al. 1998; Scora et al. 2002; Ashworth and Clegg 2003).
These complications argue that genomics and molecular breeding are central to the continued improvement of the avocado. Molecular markers for flowering types may be particularly useful because type B avocados are crucial for pollination but typically less productive than type A varieties (Davenport 1986). Recently, Rendón-Anaya et al. (2019) made an important contribution toward molecular breeding by producing draft genomes of Hass and a wild Mexican accession (P. americana ssp. drymifolia). They anchored the Hass assembly to a genetic map and ultimately produced a reference genome with 512 scaffolds and a genome size of 419 Megabases (Mb), which is less than half the expected 1C genome size of 896 Mb (Arumuganathan and Earle 1991). Using this reference, they explored the hybrid history of Hass and aspects of the evolutionary genomics of the avocado. Another Hass genome was published in 2021 (Sharma et al. 2021); this genome was 788 Mb, which was nearly 88% of the expected genome size, but it was not anchored into scaffolds. Still, more recently, a third study has further improved the assembly of the Hass genome, leading to resolved haplotypes (Nath et al. 2022). To date, however, Hass has remained the only avocado genome deciphered, and several important features of the evolutionary genomics of avocados still remain unexplored. These features include further unraveling its domestication history, using evolutionary genomic tools to identify chromosomal regions of potential agronomic interest, and focusing on genomic diversity in the context of interesting traits, like A vs B flowering types.
Here, we report the genome of the Gwen variety and use that genome as a reference for evolutionary analyses. Gwen is a grandchild of Hass with similar flavor characteristics (Witney and Martin 1995) but with higher yields and better fruit storage on the tree (Bergh and Whitsell 1982). Accordingly, Gwen has been the subject of intensive breeding efforts for 3 decades, and 1 motivation for generating a Gwen reference is to bolster these efforts. In addition, we utilized a whole-genome resequencing data set from 34 avocado accessions to address 4 sets of questions. First, what does the Gwen genome tell us about the patterns of genic hemizygosity within an avocado accession? Genomic analysis of grapevine (Vitis vinifera), another perennial clonally propagated crop, revealed that as many as 1 in 7 genes are hemizygous, perhaps due to structural mutations that have accrued during clonal propagation (Zhou et al. 2019). Is avocado similar? Second, we use the Gwen genome as a reference to explore genetic diversity within avocados, specifically to assess relationships among the 3 botanical groups and to assess the hybrid origin of well-known cultivars. This last question builds on several previous investigations of genetic diversity (Ashworth and Clegg 2003; Chen et al. 2008, 2009) but extends the work to a genomic scale. Third, we investigate features of avocado domestication, including demographic history, selective sweeps, and chromosomal regions of high genetic differentiation. Do the 3 groups share regions of selective sweeps, which may indicate parallel selection on genic regions associated with specific traits? Finally, we perform a preliminary investigation between the A and B flowering types, with the goal of identifying genomic regions that may contribute to heterodichogamy.
Materials and methods
Reference sequencing and genome assembly
Gwen (US Patent: USPP5298P 1983) is a grandchild of Hass and has been central to the University of California, Riverside breeding program. We sampled young leaf materials from a Gwen clone and isolated high molecular weight genomic DNA (gDNA) using the method of (Chin et al. 2016). We included a containing buffer (100 mM Tris-HCl pH 8.0, 0.35 M Sorbitol, 5 mM EDTA pH 8.0, 1% (v/v) PVP 40, 2% (v/v) of 2-mercaptoethanol) prior to cell lysis to avoid co-precipitation of polysaccharides and phenolics with DNA. DNA quality was assessed with a Nanodrop 2000 spectrophotometer (Thermo Scientific, IL, USA), by a Qubit 2.0 Fluorometer with the DNA High Sensitivity kit (Life Technologies, CA, USA), and by pulsed-field gel electrophoresis. From this, a 20 µg of high molecular weight genomic DNA was needle-sheared and used as input into the SMRTbell library preparation, using the SMRTbell Express Template Preparation V2 kit (Pacific Biosciences, CA, USA). Libraries were size selected to 22–80 Kb with the BluePippin (Sage Science, MA, USA) and sequenced on the Pacific Biosciences Sequel using P6-C4 chemistry (DNA Technology Core Facility, University of California, Davis).
We assembled SMRT reads with Canu version 2.1 (Koren et al. 2017), using default settings and including all reads. Once assembled, polishing was performed with 2 passes of PacBio GenomicConsensus v2.33, followed by 2 passes with Pilon v1.23 (Walker et al. 2014) using default parameters and 19 × coverage of Gwen short-read Illumina sequencing data. HapSolo v0.1 (Solares et al. 2021), which identifies and removes alternative haplotypes, was then run on the assembly with default parameters and 50,000 iterations, producing the Canu + Hapsolo (C + H) assembly. Scaffolding was based on a Gwen × Fuerte genetic map (Ashworth et al. 2019) by aligning the C + H assembly using NCBI BLAST v2.2.31+ (Altschul et al. 1990). The alignments were then ordered based on linkage group ID and cM distance, using only the alignments with the highest percent identity and evalue scores to identify contig order. When the orientation of a contig could not be determined, which was true for 112 contigs representing ∼274 Mb, it was placed in the “+” (or positive) direction and marked with an asterisk in the scaffolding annotation file (10.5281/zenodo.6392169). Where necessary, contigs were bridged using N's.
Gene and repeat annotation
Repeat annotation was based on RepeatModeler v2.0.1 in conjunction with RepeatMasker v4.1.1. RepeatModeler (Smit 2015) was run to generate a repeat database for avocado since a database for closely related species was not available. The repeat database was built using the option BuildDatabase on the Gwen C + H assembly and then used in RepeatModeler with the LTRStruct option. RepeatMasker was run using default parameters. The Gwen genome was subsequently soft masked for repeats using Bedtools v2.29.2 (Quinlan 2014) using the maskfasta run option.
For gene annotation, we mapped RNASeq reads from previous studies (Ibarra-Laclette et al. 2015; Xoca-Orozco et al. 2017; Barbier et al. 2019) onto the repeat masked genome using HiSat2 version 2.2.1. The resultant BAM files were merged and indexed using Samtools v1.10 (Li et al. 2009; Barnett et al. 2011) and fed to the BRAKER v2.1.6 (Hoff et al. 2019)/Augustus (Stanke et al. 2006) v3.4.0 pipeline. BRAKER was used in default mode for RNASeq data, with an additional option for soft masked reference assemblies (−softmasking). Finally, we excluded (as possible pseudogenes) any genes with exons that overlapped annotated repeats and filtered for a specific criterion related to gene integrity (Minio et al. 2021).
Functional annotations and Gene Ontology analyses were performed with Blast2GO v6.01 (Conesa and Götz 2008). Genes were extracted from assemblies, based on the gene annotation gff file, and then mapped to the NCBI nonredundant protein database, SwissProt, and uniref90 databases using NCBI's blastx, blastx-fast option. A protein family search was also performed using an InterPro scan. These results were then merged into a single annotation for GO mapping and Enrichment, which was performed using Blast2GO based on default options.
Genic hemizygosity was calculated following Zhou et al. (2019) by (1) remapping raw SMRT reads to the 12 pseudo-chromosomes using NGMLR v0.2.7, (2) inferring structural variants by feeding the alignment file to SNIFFLES (Sedlazeck et al. 2018), requiring a read-coverage of at least 4 to substantiate an SV, and (3) and quantitating insertion–deletion events that overlapped with 20% of the coding region.
Diversity samples, sequencing, and SNP calling
We collected leaf tissue for a total of 20 P. americana accessions and 3 outgroups, all of which were sampled from the South Coast Research and Extension Center in Irvine, CA (Table 1). For each sample, genomic DNA was extracted from leaf samples with the Qiagen DNeasy plant kit. Paired-end sequencing libraries were constructed with an insert size of 300 bp according to the Nextera Flex (Illumina, Inc) library preparation protocol. Libraries were sequenced on the Illumina NovaSeq with cycles to target 25 × coverage. We also used Illumina reads for 10 previously sequenced P. americana accessions and 1 putative outgroup (Rendón-Anaya et al. 2019), yielding a combined data set of 34 accessions that included 30 P. americana accessions and 4 putative outgroup species. Short reads from all accessions were mapped to the C + H and scaffolded assemblies using BWA, version 0.7.17 (Li and Durbin 2010), and realigned and recalibrated using GATK v3.7 (McKenna et al. 2010). Alignment filtering was done using BCFTools v1.10.2 (Danecek et al. 2021) using parameters -s LowQual -e “%QUAL < 20 || DP > 32”.
Table 1.
A list of the accessions used in this study; the historical ecotype assignment follows Rounds (1950) or later sources when indicated by a footnote.
| Accession, variety or species | Historical ecotypea | Admixed %a | Flowering typei | PopGen groupj |
Origin |
|---|---|---|---|---|---|
| 069-02 | Mb | G: 37.61 M: 62.39 L: 0.00 |
N/A | Unknown | |
| 263-C | Lb | G: 0.00 M: 0.00 L: 1.00 |
N/A | L | Hunucma, Yucatan, Mexico |
| Anaheimk | G | G: 93.06 M: 6.94 L: 0.00 |
A | G | Anaheim, California, 1910 |
| Baconk | Mc (GxMd) Ge | G: 37.69 M: 62.31 L: 0.00 |
B | Buena Park, California, 1928 | |
| Carlsbadk | G | G: 1.00 M: 0.00 L: 0.00 |
A | G | Mexico, budwood collected 1912 |
| CH-CR-25 | CRb | G: 50.01 M: 7.98 L: 42.01 |
N/A | W | Matapalo, Puntarenas, Costa Rica |
| CH-G-07 | Gb | G: 84.08 M: 0.00 L: 15.92 |
N/A | W | San Cristobal de las Casas, Chiapas, Mexico |
| CH-G-10 | Gb | G: 89.28 M: 0.00 L: 10.72 |
N/A | W | Olanca, Chiapas, Mexico |
| CH-G-11 | Gb | G: 89.96 M: 0.00 L: 10.04 |
N/A | W | Olanca, Chiapas, Mexico |
| Fairchildk | GxL | G: 0.00 M: 0.00 L: 1.00 |
A | L | Coconut Grove, Florida, seedling of Collin Red, 1925 |
| Fuertek | GxM (Me) | G: 58.48 M: 41.52 L: 0.00 |
B | Atlixco, Mexico, budwood collected 1911 | |
| Ganterk | M | G: 0.00 M: 1.00 L: 0.00 |
B | M | Whittier, California, 1905 |
| Gwenk | GxMc (Ge) | G: 1.00 M: 0.00 L: 0.00 |
A | SCREC, Irvine, California, late 1960s | |
| Hass | G (GxMb,c,e) | G: 1.00 M: 0.00 L: 0.00 |
A | G | La Habra Heights, California |
| Lindak | G | G: 1.00 M: 0.00 L: 0.00 |
B | G | Antigua, Guatemala, budwood collected 1914 |
| Lyonk | G (GxMc) (Me) | G: 83.78 M: 16.22 L: 0.00 |
B | G | Hollywood, California, 1908 |
| Mendez | GxMf | G: 1.00 M: 0.00 L: 0.00 |
A | La Habra Heights, California, 1926 | |
| Nabalk | G | G: 1.00 M: 0.00 L: 0.00 |
B | G | Antigua, Guatemala, budwood collected 1917 |
| Nimliohk | G | G: 1.00 M: 0.00 L: 0.00 |
B | G | Antigua, Guatemala, budwood collected 1917 |
| Ocotea botranthak | N/A | N/A | N/A | O | Unknown |
| Pequeno Charly | Mb | G: 0.00 M: 1.00 L: 0.00 |
N/A | M | Unknown |
| Persea donnell-smithiik | N/A | N/A | N/A | O | Unknown |
| Persea hintoniik | N/A | N/A | N/A | O | Unknown |
|
Persea schiedeana
(CH-GU-01) |
N/A | N/A | N/A | Mazatenango, Suchitepequez, Guatemala | |
| Pinkertonk | GxMc (Ge) | G: 81.00 M: 19.00 L: 0.00 |
A | Saticoy, California, late 1960s | |
| Reedk | Gc | G: 1.00 M: 0.00 L: 0.00 |
A | G | Carlsbad, California, putative Anaheim x Nabal progeny, 1960 |
| Simmondsk | L | G: 0.00 M: 0.00 L: 1.00 |
A | L | Miami, Florida, seedling of Pollock, 1908 |
| Taftk | G | G: 1.00 M: 0.00 L: 0.00 |
A | G | Seed from purchased fruit, likely Mexican origin, 1899 |
| Thillek | GxMc (Ge) | G: 1.00 M: 0.00 L: 0.00 |
B | G | Santa Paula, California, 1946 |
| TopaTopak | M | G: 0.00 M: 1.00 L: 0.00 |
A | M | Ojai, California, 1907 |
| VC26k | Lg | G: 0.00 M: 0.00 L: 1.00 |
A | L | Volcani Institute, Israel |
| Velvick | Lh | G: 59.18 M: 0.00 L: 40.82 |
N/A | University of Queensland, Australia | |
| Waldink | L | G: 0.00 M: 0.00 L: 1.00 |
A | L | Homestead, Florida, 1909 |
| Zutanok | M (GxMe) | G: 38.35 M: 61.65 L: 0.00 |
B | Fallbrook, California, 1926 |
G refers to Guatemalan, M to Mexican, and L to Lowland. The admixed percentage refers to our Admixture results with K = 3.
University of California Avocado website (http://ucavo.ucr.edu/avocadovarieties/varietyframe.html).
N/A – not available.
Denotes the designation for use in nonhybrid phylogenetic and population genetic analyses. L = Lowland (n = 5); M = Mexican (n = 3); G = Guatemalan (n = 10); W = wild (n = 3); O = Outgroup (n = 3). See text for details.
Collected from the UC ANR Research Station (Irvine, CA), with data generated for this study.
Phylogeny and population structure
We performed PCA using VCFTools v0.1.17 git commit 954e607 (Danecek et al. 2011) and PLINK v1.9 (Purcell et al. 2007). All samples had < 50% missing data for P. americana and <75% for outgroup taxa. We created a maximum likelihood phylogeny with a reduced number of sites from SNPhylo (Lee et al. 2014) using IQ-TREE v1.6.12 (Nguyen et al. 2015) and the “PMB + F + G4” substitution model, as chosen by Model Finder (Kalyaanamoorthy et al. 2017). We employed the ultrafast bootstrap with 1,000 replicates to obtain support values. Admixture plots and analyses were performed with NGSadmix from the ANGSD package version 0.930 (build 2020 January 6, 13:30:06). CLUMPAK (Kopelman et al. 2015) was used to identify the best number across K = 1–10 groups, each with 10 replicates. To apply TreeMix (Pickrell and Pritchard 2012), we first filtered the SNP data containing 33 samples (all avocado accessions and 3 of the 4 outgroups because P. schiedeana proved to be a poor outgroup; see below) to keep sites without missing data. We then pruned the SNPs based on linkage disequilibrium using PLINK v1.90b6.16 (Purcell et al. 2007) with a 20 kb window, 5 kb step, and an r2 threshold of 0.1. The pruned SNPs in VCF format were then converted using the “vcf2treemix.sh” script included in the TreeMix package to create the input format. We ran TreeMix v1.13 with migration edges ranging from 1 to 6 with 5 replicates each. Finally, we used the Evanno method from the package OptM v0.1.6 (Fitak 2021) to determine the optimal number of edges.
Population genetic analyses
To infer demographic histories, we used MSMC2 v2.1.1 (Mallick et al. 2016) with phased SNPs from the entire scaffolded assembly, which included the 12 pseudo-chromosomes and unplaced contigs. For each genetic group, we applied MSMC2 to the 3 accessions with the highest average sequencing coverage per genetic group (Guatemalan: Lyon, Carlsbad, Nimlioh; Lowland: Fairchild, Waldin, VC26; Mexican: Ganter, PequeñoCharly, TopaTopa). We created a mappability mask and a coverage mask (Supplementary Methods 1). Phase-informative SNPs (PIRs) were extracted for each sample as described in (Delaneau et al. 2013), which identifies reads that span at least 2 heterozygous SNPs; we then used the PIRs and the VCF files for each genetic group by chromosome as input to shapeit2 v2.r904 (Delaneau et al. 2013) for phasing. Finally, we used the “generate_multihetsep.py” script from the MSMC tools (https://github.com/stschiff/msmc-tools) to create MSMC2 input, taking into account mappability and coverage masks. To calculate the relative cross-coalescence rate (rCCR) across groups, we ran MSMC2 with all possible haplotype combinations between groups. To calculate split times, we used “plot_msmc.py” (Schiffels and Wang 2020) to estimate the time at rCCR = 0.5, based on a rate of 5.4e-9 mutations per site per year (Liang et al. 2019) and a generation time of 7 years.
To infer selective sweeps and to examine divergence between groups using Fst, we focused on a representative subset of accessions that (1) reduced nonrandom sampling in the Guatemalan group by removing 2 of the first- and second-degree relatives of Hass (Table 1), and (2) including only nonhybrid accessions, as inferred from Admixture analysis. For sweeps, we applied SweeD (Pavlidis et al. 2013) with default parameters on the scaffolded assembly, including unplaced contigs. To perform the analyses, we split the genome into nonoverlapping 10 kb windows created by bedtools v2.27.1 (Quinlan 2014), ignoring gaps, and focused on windows within the top 1% of the Composite Likelihood Ratio (CLR) statistic. For visualizing the results across chromosomes, we Loess smoothed across windows using the “geom_smooth” and “loess” methods with a span of 0.5 in ggplot (Wickham 2016), after assigning CLR values in gaps to zero. SweeD identifies the location of putative sweeps; to identify genes encompassed in that sweep, we included genes of 5 kb on either side of the location. Once genes were identified, we performed 2 types of analyses: GO enrichment, as described above, and the statistical significance of the number of shared sweep genes between groups. To evaluate significance, we permuted labels on genes (either sweep or nonsweep) within each race, recalculated the number of sweep genes in common between groups, and repeated the permutation 10,000 times. We used PLINK (Purcell et al. 2007) on 20 kb windows to calculate mean Fst for each window between samples, focusing on the top 1% windows.
Results
Gwen genome assembly and characterization
Gwen genome assembly
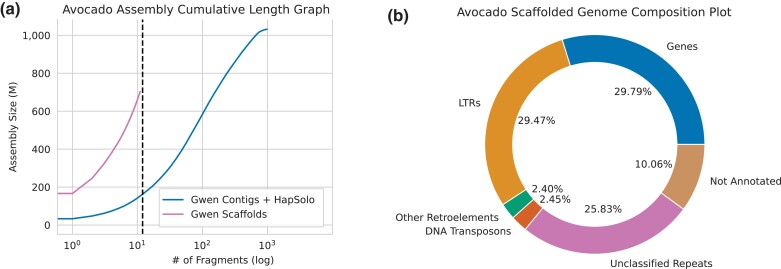
We generated 81.2 gigabases, equivalent to roughly 90 × coverage, based on the expected 1C genome size of 896 Mb (Arumuganathan and Earle 1991). We assembled PacBio SMRT reads using Canu (v 2.1), producing a genome of 1,456 Mb with 5,122 contigs, and then applied HapSolo (Solares et al. 2021) to remove putative secondary contigs (or haplotigs). The Canu + HapSolo (C + H) genome resulted in a primary assembly of 1,032 Mb, a longest contig of 17 Mb, a BUSCO score of 91%, and an N50 of 3.37 Mb (Supplementary Table 1). One useful measure of an assembly is the percentage of the assembly that is encompassed in the largest x contigs, where x represents the number of chromosomes (which is 12 for 1C P. americana). For the C + H assembly, this percentage was 17%—i.e. the 12 largest contigs represented 17% of the genome.
To improve contiguity, we anchored and scaffolded the C + H assembly using a published genetic map, based on a cross between Gwen and Fuerte (Ashworth et al. 2019). This exercise resulted in 12 scaffolds that were assigned to 12 linkage groups, along with unplaced contigs. Scaffold N50 improved ∼18-fold (to 61.9 Mb) over the 3.37 Mb contig N50. The 12 largest scaffolds (pseudo-chromosomes) represented 78% of the expected genome size of 896 Mb (Fig. 1a), which is superior to some previously published Hass genomes (Supplementary Table 1). Although each chromosome could be identified among the 12 largest scaffolds, we interpret the smaller-than-expected genome size to imply that the density of the genetic map limited resolution. The first Hass primary assembly also decreased substantially in size when it was anchored to a genetic map (Rendón-Anaya et al. 2019)—e.g. only 47% of the Hass assembly could be anchored, resulting in a scaffolded genome of 421.7Mb, while the second lacked any scaffolding (Sharma et al. 2021).
Fig. 1.
a) The cumulative sum assembly graph size (cdf) shows the size of the assemblies with the next largest consecutive contig (or scaffold) being added to the sum along the x-axis for the 2 Gwen assemblies (C + H and scaffolded). b) The annotation results for the Gwen C + H assembly, showing the percentage of the genome attributable to genes and different types of transposable elements, including DNA transposons, unclassified retroelements, and long terminal repeat (LTR) retroelements.
Genome annotation
We annotated the C + H assembly and scaffolded assemblies by first identifying the repetitive content (see Methods). We estimated that ∼61% of the Gwen genome consisted of repetitive elements (Fig. 1b), of which 52% were long terminal repeat (LTR) retroelements (with more than half of these being Gypsy elements) and another 40% were unknown repeats (Fig. 1b). We masked repeats before annotating genes (see Methods), inferring 36,993 genes on the 12 pseudo-chromosomes and 12,457 in unplaced contigs. We compared these numbers to the Hass genome annotation (Rendón-Anaya et al. 2019), which reported 33,378 genes. We filtered this set to remove potential duplicates, yielding 25,211 genes. Of the 25,211, 94% were present in our scaffolded assembly; thus, our annotation corroborates most previous genic inferences but also annotates ∼1.5-fold more genes.
Genic hemizygosity
Recent studies have suggested that diploid genomes may be replete with genic hemizygosity. We assessed genic hemizygosity by remapping raw PacBio reads to the 12 pseudo-chromosomes of the scaffolded assembly, by inferring structural variants, and then by filtering for variants that overlapped annotated genes (see Methods). With this approach, we estimated that 3.8% of genes were hemizygous in the Gwen genome. This estimate serves as a benchmark that can be used to compare other diploid systems (see Discussion).
Analysis of P. americana genetic structure
Classification of groups and hybrid accessions
Given the Gwen reference genome, we performed a preliminary study of genetic variation and evolutionary genomics across avocados. To do so, we amassed a whole-genome resequencing data set of 30 P. americana accessions and 4 putative outgroups, all with at least 14 × coverage (Table 1). The sample represented the 3 botanical groups of avocado, based on historical designations, historically important cultivars (Chen et al. 2009), and both A and B flowering types (Table 1). Given the resequencing data, we mapped reads to the scaffolded assembly (including unplaced contigs) and identified 23,938,386 biallelic SNPs within our entire P. americana sample and 32,235,263 biallelic SNPs when the 4 outgroups were analyzed with the P. americana samples.
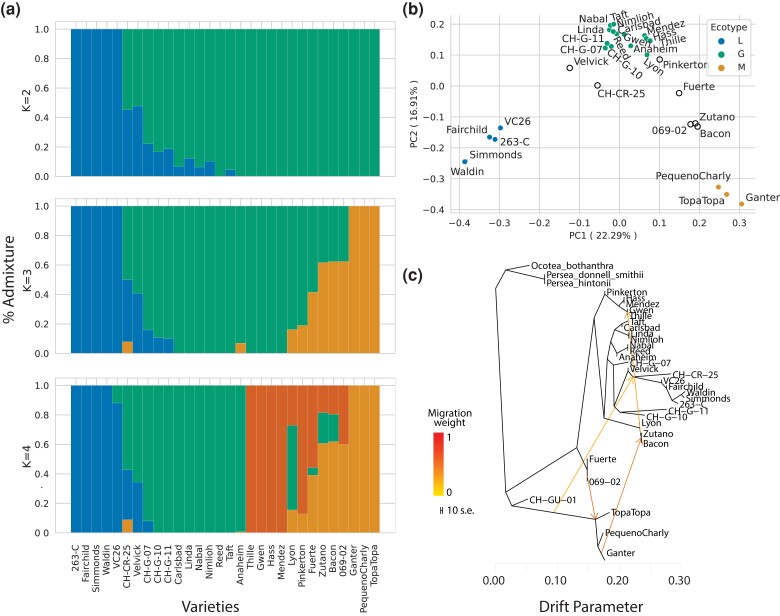
SNPs were then subjected to 3 types of clustering analysis—admixture mapping, principal component analysis (PCA), and phylogenetic inference—to define groups of accessions based on whole-genome data. We first investigated relationships among accessions using admixture mapping. The most highly supported analysis contained k = 4 groups: the 3 previously recognized botanical groups (Lowland, Mexican, Guatemalan) and a series of cultivars related to Gwen and Hass (Fig. 2a). This last group included Mendez, a somatic mutation of Hass (Illsley-Granich et al. 2011; Schaffer et al. 2013), and other close relatives like Thille. We suspect that this last group was defined by over-sampling accessions with first- or second-degree relationships to Hass. Indeed, the removal of 2 accessions from the “Hass group” yielded k = 3 as the most supported number of groups, with the 3 groups representing the previously recognized groups (Supplementary Fig. 1). Given this result and the historical designation of k = 3, we reported the proportion of each accession based on k = 3 groups (Table 1). These admixture maps reflect a hybrid history of some accessions, including Velvick, Fuerte, Bacon, Zutano, and also an accession (CH-CR-25) that was thought to represent a new racial ecotype—var. costaricensis (Ben-Ya’acov et al. 2003; Rendón-Anaya et al. 2019).
Fig. 2.
a) Admixture plots generated from the entire sample of 30 avocado accessions, using SNPs based on the Gwen scaffolded assembly. The plots show the inferred groups with K = 2, K = 3, and K = 4, which was inferred to be the most likely number of clusters with this data set. The K = 4 groups include the 3 historical groups (Mexican in orange on far right, Guatemalan in green in the center, and Lowland in blue on the left) and another group in red with accessions closely related to Hass. b) A PCA analysis of SNP diversity. Each dot represents an individual, and the color of the dot represents accessions with >80% assignment to a single group, including Guatemalan (G), Mexican (M), and Lowland (L). Open dots represent likely hybrid accessions (c) Results of TreeMix, with the optimal 4 edges. The accessions, including 3 outgroups are labeled.
Somewhat surprisingly, the admixture analyses also indicated that Hass is 100% Guatemalan, whereas previous work defined it as >50% Mexican (Rendón-Anaya et al. 2019) or 58% Guatemalan (Chen et al., 2009). We note that our results are consistent across Hass and its close relatives like Gwen, which is a grandchild of Hass, and Mendez. We also remark, however, that the reduced dataset suggested that Hass has a small genetic proportion (< 25%) attributable to the Mexican group (Supplementary Fig. 1), which is still far less than previous studies. Overall, these results suggest that Hass originated primarily from the guatemalensis group, with the complete dataset identifying a solely Guatemalan origin.
Second, we applied PCA to the P. americana data without outgroups. The results verified many of the admixture analyses, including clusters that represented previously identified Mexican, Guatemalan, and Lowland groups (Fig. 2a). The PCA consistently placed Hass and its relatives squarely within the Guatemalan group, but also clearly indicated that some accessions fell between groups on the PCA (e.g. Velvick), reinforcing hybrid origins. Furthermore, economically important accessions like Bacon and Zutano have been inconsistently inferred or assigned as hybrids (Chen et al. 2009), but our whole-genome analyses suggest likely hybrid origins (Fig. 2, a and b and Table 1). For completeness, we also applied PCA to a data set with the 4 putative outgroup taxa. The genetic placement of 3 species (P. donnell-smithii Mez, P. hintonii C.K. Allen, O. botrantha Rohwer) was consistent with their expected outgroup status, but P. schiedeana Nees clustered near the avocado ingroup (Supplementary Fig. 1). The P. schiedeana results are consistent with previous suggestions that it hybridizes with avocado (Ashworth and Clegg 2003), indicating it should not be used as an outgroup for evolutionary analyses as it has previously (Rendon-Ayala et al., 2019).
Finally, we investigated relationships among accessions and the 3 legitimate outgroups using 2 phylogeny-based methods. The first was TreeMix, which can reveal the potential presence of historical gene flow. Given that populations were difficult to define in our sample, due to apparently recent hybrid origins, we applied TreeMix to individuals and identified an optimum of 4 edge migration events (Fig. 2c). Unsurprisingly, TreeMix inferred migration events that likely reflect recent breeding history—e.g. directional migration between the pure Mexican accessions (represented by Topa Topa, PequenoCharly, and Ganter in Fig. 2c) and hybrid accessions like Zutano and Bacon. TreeMix also suggested ancient migration between the lineage leading to the Mexican group toward the base of the lineage representing the Costa Rican sample (CH-CR-25). This observation lends credence to the idea that var. costaricensis represents a distinct ecotype (Ben-Ya’acov et al. 2003), perhaps born of an ancient hybridization event.
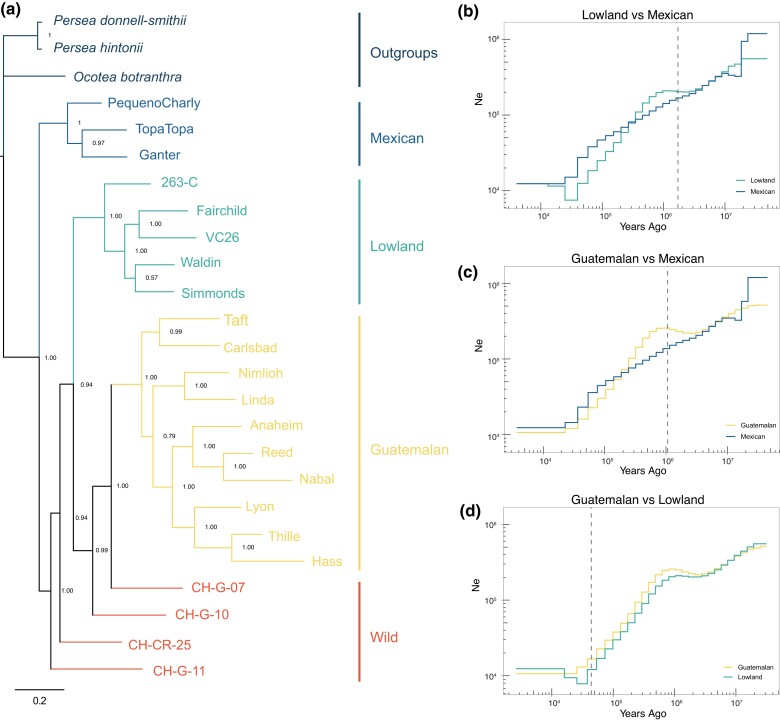
We also created a consensus phylogenetic tree to investigate relationships among the 3 historical groups. Because we intended to investigate phylogenetic history (and not the history of recent hybrid events due to breeding), we based this tree on a representative set of nonhybrid accessions that had high support (Qi > 80%) for inclusion in a single genetic group at K = 3 (Vigouroux et al. 2008; Castillo et al. 2010), reduced sampling of the near-relatives of Hass, and on a reduced number of SNPs to limit linkage disequilibrium (see Methods). The phylogeny had median bootstrap support of 88.5% for all nodes and strong support (> 76%) for nodes that separated the botanical groups (Fig. 3a; Table 1). Moreover, the accessions from each group formed well-supported monophyletic clades, justifying treating each named group as historically separate. An interesting feature of the rooted phylogeny is that the Mexican group is an early-diverging sister group, suggesting an early split from the Lowland and Guatemalan lineages and a separate domestication of the Mexican avocado. Another interesting feature is the placement of putatively wild accessions, CH-G-07 and CH-G-10. These accessions, which were from the study of Rendon-Ayala et al. (2019) and designated as wild in their analyses (Table 1), intercalate the Lowland and Guatemalan clades. Assuming these accessions are, in fact, wild, as opposed to feral escapees, their placement on the tree reinforces the idea that the Lowland and Guatemalan groups were domesticated separately (Furnier et al. 1990; Ashworth and Clegg 2003; Rendón-Anaya et al. 2019).
Fig. 3.
a) Genome-wide phylogeny of P. americana ingroup with 3 species outgroups. Accessions were chosen if they had 80% of higher assignments to a race, including putatively wild accessions (CH-G-07, CH-G-10, CH-G-11, and CH-CR-25). The numbers on the nodes represent bootstrap proportions. b) Demographic inference based on the Mexican and Lowland samples. The x-axis represents the time from present to past, and the y-axis is inferred as effective population size (Ne). The vertical dashed lines represented the estimated divergence time between the 2 groups. c) Like b, except for Mexican and Guatemalan samples. d) Like b, except for the Lowland and Guatemalan samples.
Population diversity analyses
Our sampling was designed to include representative samples from each botanical group. However, analyses of the genetic structure identified hybrids that were unlikely to be helpful for inferring the historical dynamics of specific groups. Accordingly, we focused population genetic analyses on representative samples with Qi > 80% (Fig. 3a). This resulted in different sample sizes for the 3 groups, with the Mexican sample being the smallest at n = 6 chromosomes from 3 diploid individuals (Table 1). These sample sizes likely limit inferential power for some analyses, but we nonetheless retained > 10 million SNPs within each sample, with nucleotide diversity (π) similar among groups, at ∼0.0035 per nucleotide site (Supplementary Table 2).
Demographic history
We applied the Sequentially Markovian Coalescent to infer 2 distinct aspects of the history of botanical groups. The first was to infer whether any experienced a domestication bottleneck or other dramatic demographic event, recognizing that perennial crops often lack a signature of such events (Gaut et al. 2018). After applying MSMC to the 3 individuals with the highest coverage from each group (to ensure equal sample sizes across groups; see Methods), the results indicated a consistent reduction of Ne over time but without any evidence of a particularly notable bottleneck or rapid post-bottleneck expansion (Fig. 3b). The second was to estimate the timing of the split of botanical groups; the divergence times complemented the phylogeny by indicating an early split (∼1.3 million years ago) of the Mexican group. In contrast, the Lowland and Guatemalan groups diverged more recently, at ∼44,000 years ago. These split times predate expected domestication times, and thus, likely reflect divergence times between ancestral wild progenitor populations (Fig. 3b).
Selective sweep mapping
For the 3 sets of samples representing potentially distinct domestication events, we investigated 2 additional features of their evolutionary genomics. The first was sweep mapping within each botanical group and the second was the divergence between groups, as measured by Fst. We performed these analyses to assess whether similar (or entirely different) sets of genes bear marks of selection across groups, to investigate whether highly differentiated chromosomal regions between groups overlapped with selected genes, and to generate a list of candidate-selected genes with potential functions.
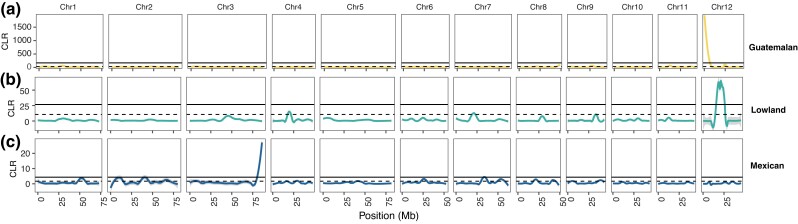
Sweep mapping relied on the CLR statistic and focused on 10 kb nonoverlapping genomic windows of the scaffolded assembly. Using an empirical cutoff of 1%, we identified 1,300 windows from each race, for which 638, 436, and 92 had genes in the Mexican, Lowland, and Guatemalan samples, respectively (Supplementary Table 3). Given these genes, we first hypothesized that separate domestication events may have targeted particularly important sets of genes in parallel. Visually, based on smoothed curves of the CLR statistic, this did not appear to be the case (Fig. 4), because putative sweep regions differed markedly among samples, although both the Lowland and Guatemalan samples had prominent sweep regions on chromosome 12. However, we also asked the question more formally by calculating the number of shared sweep genes between pairs of groups and then permuting labels (CLR, non-CLR) to test significance. We found no enrichment for shared CLR genes between the Guatemalan sample, and either the Mexican (P = 0.3692; 2 shared genes; Supplementary Table 4) or the Lowland samples (P = 1.00; 0 shared genes). However, the Lowland and Mexican groups had 18 CLR genes in common, a number significantly higher than random expectation (P < 0.0001) (Supplementary Table 5).
Fig. 4.
Loess smoothed composite likelihood values reflecting potential regions in samples from each of the 3 botanical groups: a) Mexican, b) Lowland, and (c) Guatemalan. Each panel represents the 12 scaffolded chromosomes, with chromosome number and location (in Mb) provided above and below each individual graph. In each graph, the dashed lines represent 1% and 5% cutoff values for significance; 95% Confidence Intervals are indicated by gray shades. Note the different scales of x-axes among the groups.
We evaluated sets of selected genes using GO enrichment. In the Guatemalan race, for example, the putative swept genes were enriched for functions related to fruit ripening, fruit development, anatomical structure maturation, and other functions (Supplementary Fig. 3). Since these functional enrichments reflect properties potentially associated with domestication, we consider this genic set to be noteworthy. In contrast, we did not identify the enrichment of gene function related to fruit maturation or suspected domestication traits for the Mexican and Lowland samples (Supplementary Figs. 4 and 5), perhaps reflecting statistical uncertainty due to their small sample sizes. We did, however, identify gene enrichments related to functions s stress response, terpene production, and metabolic processes. The set of shared 18 genes was also particularly interesting, due to the potential for parallel selection during separate domestication events. A subset of the genes had positive hits to functional annotation databases (Supplementary Table 5). However, with the possible exception of 2 genes that function in sugar transport, none have functions related to obvious domestication traits like fruit size or development.
Divergence mapping
We next examined divergence between groups using Fst based on 20 kb nonoverlapping windows along scaffolded chromosomes, focusing on regions that include the top 1% of Fst scores (Supplementary Fig. 6). For example, for the pairwise comparison between the Mexican and Lowland samples, we identified peaks containing 396 genes, with similar numbers for the other pairwise comparisons (Mexican–Guatemalan 387 genes; Lowland–Guatemalan 384 genes) (Supplementary Table 6). In contrast to the potential for parallel domestication pressures on some genes, we expected Fst results to be enriched for genes that contribute to agronomic differences between the 2 groups. Several genes in the top 1% were related to disease resistance and response, particularly drought and cold/heat response. We performed GO enrichment on these samples of genes (Supplementary Figs. 7–9), finding enrichment for light stimulus, pollen recognition, and several cellular and metabolic processes. We also assessed whether genes were shared between Fst and sweep analyses, hypothesizing that the selection of genes within 1 race could contribute to genetic divergence between groups. To perform this analysis, we identified genes shared between Fst and CLR analyses—e.g. we compared the set of 396 genes from Mexican–Lowland Fst analysis to the 436 CLR genes in the Lowland sample. We found 10 genes shared between the 2 lists, which was an enrichment relative to random expectation (permutation P < 0.0028). The number of shared genes between Fst and CLR analyses was higher than expected for 4 of 6 comparisons (Supplementary Table 4). The shared genes again constitute another set of credible domestication or improvement genes (Supplementary Table 7).
Contrasting A and B flowering types
To date, the genetic causes of heterodichogamy have not been identified in any system (Endress 2020). For that reason, we thought it worthwhile to explore genetic factors associated with type A and B flowers. Many of our resequenced accessions had known flowering types, with samples of n = 13 A types and n = 9 B types in total (Table 1). Importantly, within each flowering type, the samples traversed genetic groups—e.g. the A types included samples from each of the 3 groups (Mexican, Guatemalan, and Lowland), and the B types were distributed across the Mexican, Guatemalan and hybrid samples. Given the distribution of A and B types across groups, we thought that contrasting the 2 samples may provide preliminary insights into genomic regions that contribute to this interesting phenotype, perhaps without being overly confounded by population structure.
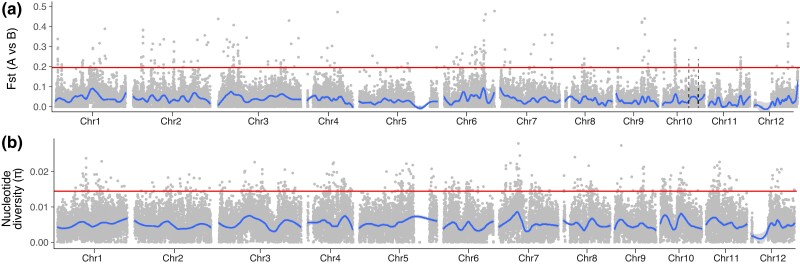
Therefore, we performed Fst analyses between the 2 groups, producing a plot with peaks of differentiation between types (Fig. 5a). The average value of Fst was low (at 0.038) compared to Fst differentiation between groups (average Fst: Guatemalan vs Mexican = 0.241, Guatemalan vs Lowland = 0.223; Lowland vs Mexican = 0.325) (Supplementary Fig. 6), reflecting again the fact that the A vs B samples do not represent highly differentiated samples. There were, nonetheless, regions of visually compelling Fst peaks between flowering types—e.g. evident peaks on chromosomes 6 and 10, among others. These peaks could be an artifact of the population histories of the samples, but they may also contain genes that differentiate the A and B morph. Consistent with the latter interpretation, the set of 466 genes within the top 1% of Fst windows (Supplementary Table 8) were enriched for functions—like pollination, floral development, and photoperiodism—that likely contribute to heterodichogamy (Supplementary Fig. 10). Given the functional enrichments, we explored the list of 466 genes to find genes related to floral development and timing, yielding several genes with homologs that affect floral development, circadian rhythm, photoperiodism, and the production of volatiles (Table 2). Reasoning that these results should be consistent for the subset of A vs B accessions within the Guatemalan group, we repeated the analyses in the Guatemalan sample, finding again an Fst signal for 4 of the candidates (Table 2). Finally, our results identified a prominent peak on Chromosome 10, but not in a location that overlapped with a previously identified QTL region for the flowering type (Chen et al. 2009) (Fig. 5a).
Fig. 5.
a) A genomic plot of Fst windows contrasting the A and B flowering type accessions. The horizontal line denotes the top 1% Fst peaks, which include 466 genes. The black dashed lines on chromosome 10 show the approximate location of a previously identified QTL for flowering type b) A plot of nucleotide diversity (π) across the 12 pseudo-chromosomes based on the “pure” Guatemalan (n = 20 chromosomes) sample. The horizontal lines indicate an empirical 1% cutoff.
Table 2.
Candidate genes found in Fst peaks between the samples of A- and B-type flowering accessions with apparent functions in flower development.
| Genea | Homologb | Homolog function |
|---|---|---|
| ChrU.ver1.g447950, ChrU.ver1.g387390 |
VOZ1
c
(Arabidopsis thaliana) |
Regulates Flowering Locus C (FLC) and Flowering Locus T (FT) |
| ChrU.ver1.g414850 |
CCR1
(Petunia hybrida) |
Biosynthesis of volatile compounds in flowers |
| Chr5.ver1.g224860, ChrU.ver1.g377230 |
EOBI
(Petunia hybrida) |
Transcription factor for volatile compounds in flowers |
| Chr10.ver1.g45990 | SPA1 (Arabidopsis thaliana) |
Controls normal photoperiodic flowering and regulates circadian rhythms |
| ChrU.ver1.g440620, ChrU.ver1.g440630 | KCS11c (Arabidopsis thaliana) |
Response to cold and light stimulus |
| Chr3.ver1.g159200 | AP2 (Arabidospis thaliana) |
Transcriptional activator that promotes early floral meristem identity |
| Chr5.ver1.g230020, Chr5.ver1.g230030 | MTERF2c (Arabidopsis thaliana) |
Transcription termination factor; knock-outs delay growth and flowering |
| Chr1.ver1.g24210 | NPR5c (Arabidopsis thaliana) |
Acts redundantly with BOP2. BOP1/2 promote leaf and floral meristem fate and determinacy in a pathway targeting AP1 and AGL24 |
| ChrU.ver1.g393700 | CSU2 (Arabidopsis thaliana) | Inhibits COP1, which is involved in seedling growth and photoperiodism in flowering |
Gene number in Gwen annotation file.
A homolog to the gene, as identified by functional analyses with SwissProt, with the species in which the homolog was identified. [Note: SPA1 is not present in the scaffolded assembly and was identified on Fst analyses of the C + H assembly.]
Also identified in Fst analyses based on the A vs B sample of only Guatemalan accessions.
We sought another piece of evidence to provide additional support for any of the genes in Table 2. We first wondered whether heterodichogamy could be caused by dosage effects and, therefore, investigated whether any of the 466 Fst-based genes were hemizygous in Gwen, an A-type flower morph. Of the 466 genes, 17 were hemizygous, a percentage (3.6%) nearly identical to the genome average (3.8%), suggesting hemizygosity was not a refining criterion. None of the hemizygous genes were related to floral function (Table 2). Second, we hypothesized that the A vs B polymorphism has been subject to balancing selection, given that heterodichogamy predates the diversification of P. americana (Renner 2001). If true, we expected causal genomic region(s) to have especially high levels of nucleotide diversity within a sample that contained both flowering types. We examined the Guatemalan sample to scan for 5 kb regions of high nucleotide diversity. While there were some weakly apparent peaks of diversity, their locations did not correspond with Fst peaks between the A and B flowering types (Fig. 5b); none of the genes in Table 2 were among the set of 1% most diverse genes. Overall, 3 genes were found in both Fst peaks and high diversity windows—a Leucine-Rich Repeat gene (Geneid Chr6.ver1.g275850) and 2 genes similar to the mitochondrial transcription termination factor MTERF2 (Geneids Chr5.ver1.g230020 and Chr5.ver1.g230030).
Discussion
We have assembled and annotated the genome of Gwen avocado. Annotation identified ∼65% of the genome as repetitive with 49,450 genes (Fig. 1b). The latter number is almost 2-fold higher than those predicted on the Hass genome (Rendón-Anaya et al. 2019), but approaches the 63,000 genes predicted in a transcriptome analysis (Chabikwa et al. 2020). Among the genes, 3.8% have been detected as hemizygous, due to structural variants that affect >20% or more of the coding region. This genic hemizygosity value is intermediate among a range that includes perennial grapevines (>12%) (Vondras et al. 2019; Zhou et al. 2019), an outcrossing annual relative of rice (8.9%) and selfing rice (<<1%) (Kou et al. 2020). Given that avocado is clonally propagated, we naively expected hemizygosity to be similar to the grapevine. The low value for Gwen probably reflects a low divergence between its parents and, perhaps, the fact that the sampled Gwen tree does not yet have an extensive history of clonality, which promotes the maintenance of hemizygous deleterious variants (Zhou et al., 2017). We do know that genic hemizygosity contributes to trait differences among grapevines (Carbonell-Bejerano et al. 2017); it will be interesting to monitor similar issues in avocados for accessions like Mendez, a somatic variant of Hass.
We generated the Gwen genome as a tool for breeding but used it here for investigating questions about the evolutionary history of P. americana—e.g. as a reference to address questions about the number of domestication events and the timing of divergence among botanical groups. Our analyses are consistent with 3 independent domestication events (Furnier et al. 1990; Ashworth and Clegg 2003; Rendón-Anaya et al. 2019), because admixture, PCA, and phylogenetic analyses clearly differentiated among the 3 botanical groups (Figs. 2 and 3). We also estimated divergence times (Fig. 3), which varied from ∼40,000 years between the Lowland and Guatemalan groups to >1.0 My between the Mexican and the 2 other groups. These estimates are much older than the expected domestication times for perennial crops (Miller and Gross 2011; Gaut et al. 2015), and hence, likely reflect divergence among wild lineages that eventually became the sources for domestication. The early divergence among groups may have been driven in part by ecological differences among regions, especially given the evidence that wild germplasm in Mexico is genetically subdivided by elevation (Chen et al. 2009).
Our analyses have also provided insights into the inter-racial hybrid origins of some cultivars, representing the first whole-genome insights for most of our samples. Many of our results confirmed findings based on microsatellites and other marker types (Ashworth and Clegg 2003; Chen et al. 2008, 2009)—e.g. accessions such as Zutano and Bacon were previously thought to be hybrids between botanical groups, which was confirmed in our analyses (Table 1)—but also offered surprises. The most notable was the genetic history of Hass, which was traditionally thought to be of Guatemalan origin (Ashworth and Clegg 2003), but had been inferred to be roughly 50% Guatemalan and 50% Mexican from genetic analyses (Chen et al. 2009; Rendón-Anaya et al. 2019). We find, however, that Hass falls predominantly into the Guatemalan group, as do the close relatives of Hass in our sample (Supplementary Fig. 2). Although we do not know the source of disagreement among studies, the genetic provenance of Hass—the most cultivated accession in the world—may not yet be fully resolved.
We removed hybrid accessions to define “pure” samples for population genetic analyses and to investigate the history of groups. The resulting sizes of samples varied and likely contributed to some of the variances in results across groups. For example, the smaller sample size of the Mexican sample likely led to false positives for sweep genes (n = 6 chromosomes with 638 CLR genes) compared to the Guatemalan sample (n = 20 chromosomes with 92 CLR genes). We have, nonetheless, found a few compelling patterns. For example, the set of high-CLR genes in the Guatemalan race was enriched for functions related to fruit development, likely a trait targeted for domestication. This gene set is, then, fitting for further study and has the potential to help disentangle the origin of some agronomic traits. In addition, some genes were shared as putative domestication genes more often than expected, including 18 genes between the Lowland and Mexican groups. In theory, these genes could represent genomic regions that are particularly prone to maintaining the history of sweeps (i.e. low recombination regions), but genome-wide patterns of the CLR statistic do not superficially suggest this as the cause (Fig. 4). We consider it more likely that these genes represent parallel selection pressures for independent domestication events, but we have few insights into how they may have contributed to domestication traits.
We used similar approaches to investigate regions of genomic divergence between samples defined either by race or flowering type. The latter yielded the most promising insights, representing (to our knowledge) the first attempt to define genomic regions that may contribute to heterodichogamy. Our inferences are at best preliminary, but they have yielded some interesting candidates, including homologs to VOZ1, which regulates FLC, a transcription factor that functions as a repressor of floral transition and contributes to temperature compensation of the circadian clock (Mitsuda et al. 2004; Yasui et al. 2012); SPA1, which contributes to the regulation of circadian rhythms of flowering processes in A. thaliana (Ishikawa et al. 2006); and APETALA2, which binds to thousands of loci in the developing flower and controls various aspects of floral development and organ identity (Yant et al. 2010). We have also documented a prominent peak on chromosome 10 (Fig. 5a), the same chromosome that housed a flowering type QTL (Ashworth et al. 2019). However, our peak does not overlap with the QTL region identified previously, and we also found no particularly noteworthy candidate genes under the peak identified in this study. It is unclear why the 2 studies do not correspond, but potential reasons include a low resolution for the QTL study, potential structural variants between the parents of the QTL study or between Gwen haplotypes, and potential nonincorporation of key contigs into the scaffolded assembly.
Interestingly, none of our candidate genes for heterodichogamy bear an obvious signal of balancing selection, which is something we hypothesize should be present for a long-lived genetic polymorphism that contributes to A vs B flowering types, perhaps similar to the S-locus of Arabidopsis species (Roux et al. 2013). The few genes that do overlap between diversity and divergence seem unlikely to affect heterodichogamy [although it should be noted that MTERF2 knock-outs affect flowering and plant growth (Lee et al. 2021)]. There are several reasons why it might be difficult to detect a balancing polymorphism if there is indeed a balanced polymorphism, but these candidate genes, nonetheless, suggest a way forward to study this interesting biological phenomenon. Future work will focus on additional genome sequencing and analyses of A vs B genomes, on characterizing segregation patterns of polymorphisms within candidate genes in a larger sample of A vs B avocado accessions, and also on searching for trans-specific polymorphisms (Charlesworth 2006) across species of the Lauraceae, since heterodichogamy is also found in other Lauraceae species (Renner 2001).
Supplementary Material
Acknowledgments
R. Gaut generated resequencing libraries, G. Gaut provided analysis insights, J. Ross-Ibarra provided feedback on ideas, and 4 reviewers provided helpful feedback.
Contributor Information
Edwin Solares, Deptartment of Ecology and Evolutionary Biology, University of California, Irvine, Irvine, CA 92697-2525, USA.
Abraham Morales-Cruz, Deptartment of Ecology and Evolutionary Biology, University of California, Irvine, Irvine, CA 92697-2525, USA.
Rosa Figueroa Balderas, Department of Viticulture and Enology, University of California, Davis, Davis, CA 95616, USA.
Eric Focht, Department of Botany and Plant Sciences, University of California, Riverside, Riverside, CA 92521, USA.
Vanessa E T M Ashworth, Department of Botany and Plant Sciences, University of California, Riverside, Riverside, CA 92521, USA.
Skylar Wyant, Deptartment of Ecology and Evolutionary Biology, University of California, Irvine, Irvine, CA 92697-2525, USA.
Andrea Minio, Department of Viticulture and Enology, University of California, Davis, Davis, CA 95616, USA.
Dario Cantu, Department of Viticulture and Enology, University of California, Davis, Davis, CA 95616, USA.
Mary Lu Arpaia, Department of Botany and Plant Sciences, University of California, Riverside, Riverside, CA 92521, USA.
Brandon S Gaut, Deptartment of Ecology and Evolutionary Biology, University of California, Irvine, Irvine, CA 92697-2525, USA.
Data availability
The data for this study were submitted to NCBI under BioProject: PRJNA758103, which contains all raw PacBio and Illumina sequencing data, as well as the scaffolded genome assembly. A gff file describing the genes and the scaffold annotation files are available at Zenodo: https://doi.org/10.5281/zenodo.6392169. Published resequencing data used in this study from Rendon-Ayala et al. (2019) were from NCBI numbers: SRR8295599, SRR8295600, SRR8295601, SRR8295602, SRR8295603, SRR8295604, SRR8295605, SRR8295607, SRR8295608, SRR8295609, SRR8295610, and SRR8295611. The RNAseq data used for gene annotation were downloaded from NCBI with accession numbers SRR6116327, SRR6116328, SRR6116329, SRR6116330, and SRR2000042. The scripts used for analyses are available from https://github.com/GautLab/avo_ref_paper
Supplemental material is available at G3 online.
Funding
E. Solares was supported by the NSF graduate research fellowship program, the University of California President's Pre-Professoriate Fellowship, and a UC Presidential Postdoctoral Fellowship. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1548562. Specifically, it used the Bridges system, which is supported by NSF award number ACI-1445606, at the Pittsburgh Supercomputing Center (PSC). The XSEDE grants were MCB190050 and MCB180035 to ES. This work was supported in part by a USDA grant #2020-04640 to M.L. Arpaia.
Author contributions
ES, AM-C, SW, VA, and AM performed analyses; RFB and DC optimized DNA extraction and supervised long-read sequencing; EF and MLA aided sampling. ES, AM-C, EF, MLA, and BSG designed the project. ES, AM-C, and BSG co-wrote the manuscript with feedback from all authors.
Communicating editor: M. Hufford
Literature cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anderson G. The Australian avocado industry. Calif. Avocado Soc. Yearbook. 2004–2005;87:55–58. [Google Scholar]
- Arumuganathan K, Earle ED. 1991. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 9(3):208–218. doi: 10.1007/BF02672069. [DOI] [Google Scholar]
- Ashworth VETM, Chen H, Calderón-Vázquez CL, Arpaia ML, Kuhn DN, Durbin ML, Tommasini L, Deyett E, Jia Z, Clegg MT, et al. Quantitative trait locus analysis in avocado: the challenge of a slow-maturing horticultural tree crop. J Am Soc Hortic Sci. 2019;144(5):352–362. doi: 10.21273/JASHS04729-19. [DOI] [Google Scholar]
- Ashworth VETM, Clegg MT. Microsatellite markers in avocado (Persea americana Mill.): genealogical relationships among cultivated avocado genotypes. J Hered. 2003;94(5):407–415. doi: 10.1093/jhered/esg076. [DOI] [PubMed] [Google Scholar]
- Barbier FF, Chabikwa TG, Ahsan MU, Cook SE, Powell R, Tanurdzic M, Beveridge CA. A phenol/chloroform-free method to extract nucleic acids from recalcitrant, woody tropical species for gene expression and sequencing. Plant Methods. 2019;15(1):62. doi: 10.1186/s13007-019-0447-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett DW, Garrison EK, Quinlan AR, Strömberg MP, Marth GT. Bamtools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011;27(12):1691–1692. doi: 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ya’acov A, Michelson E. Avocado rootstocks In: J. Janick, editor. Horticultural Reviews. Vol. 17. New York (NY): John Wiley and Sons, Inc.; 1995. p. 381–429. [Google Scholar]
- Ben-Ya’acov A, Solis-Molina A, Bufler G. The mountain avocado of Costa Rica, a new sub-species. In: Proceedings V World Avocado Congress. 2003. p. 27–33. [Google Scholar]
- Bender GS. Avocado flowering and pollination. Avocado Prod Calif. 2002;1:39–49. [Google Scholar]
- Bergh B, Ellstrand N. Taxonomy of the avocado. Calif Avocado Soc Yearbook. 1986;70:135–146. [Google Scholar]
- Bergh BO, Whitsell RH. Three new patented avocados. In: California Avocado Society Yearbook. Vol. 66. 1982. p. 51–56. [Google Scholar]
- Carbonell-Bejerano P, Royo C, Torres-Pérez R, Grimplet J, Fernandez L, Franco-Zorrilla JM, Lijavetzky D, Baroja E, Martínez J, García-Escudero E, et al. Catastrophic unbalanced genome rearrangements cause somatic loss of berry color in grapevine. Plant Physiol. 2017;175(2):786–801. doi: 10.1104/pp.17.00715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo A, Dorado G, Feuillet C, Sourdille P, Hernandez P. Genetic structure and ecogeographical adaptation in wild barley (Hordeum chilense Roemer et Schultes) as revealed by microsatellite markers. BMC Plant Biol. 2010;10(1):266. doi: 10.1186/1471-2229-10-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabikwa TG, Barbier FF, Tanurdzic M, Beveridge CA. De novo transcriptome assembly and annotation for gene discovery in avocado, macadamia and mango. Sci Data. 2020;7(1):9. doi: 10.1038/s41597-019-0350-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2006;2(4):e64. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Ashworth VE, Xu S, Clegg MT. Quantitative genetic analysis of growth rate in avocado. J Am Soc Hortic Sci. 2007;132(5):691–696. doi: 10.21273/JASHS.132.5.691. [DOI] [Google Scholar]
- Chen H, Morrell PL, Ashworth VE, De La Cruz M, Clegg MT. Tracing the geographic origins of major avocado cultivars. J Hered. 2009;100(1):56–65. doi: 10.1093/jhered/esn068. [DOI] [PubMed] [Google Scholar]
- Chen H, Morrell PL, de la Cruz M, Clegg MT. Nucleotide diversity and linkage disequilibrium in wild avocado (Persea americana Mill.). J Hered. 2008;99(4):382–389. doi: 10.1093/jhered/esn016. [DOI] [PubMed] [Google Scholar]
- Chin C-S, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O’Malley R, Figueroa-Balderas R, Morales-Cruz A, et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 2016;13(12):1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Götz S. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. 2008;2008:1–12. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10(2):giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport TL. Avocado flowering. In: J. Janick, editor. Horticultural Reviews. Vol. 8. New York (NY): John Wiley and Sons, Inc.; 1986. p. 257–289. [Google Scholar]
- Davis J, Henderson D, Kobayashi M, Clegg MT. Genealogical relationships among cultivated avocado as revealed through RFLP analyses. J Hered. 1998;89(4):319–323. doi: 10.1093/jhered/89.4.319. [DOI] [Google Scholar]
- Degani C, Gazit S. Selfed and crossed proportions of avocado progenies produced by caged pairs of complementary cultivars. HortScience. 1984;19(2):258–260. doi: 10.21273/HORTSCI.19.2.258. [DOI] [Google Scholar]
- Delaneau O, Howie B, Cox AJ, Zagury J-F, Marchini J. Haplotype estimation using sequencing reads. Am J Hum Genet. 2013;93(4):687–696. doi: 10.1016/j.ajhg.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress PK. Structural and temporal modes of heterodichogamy and similar patterns across angiosperms. Bot J Linn Soc. 2020;193(1):5–18. doi: 10.1093/botlinnean/boaa001. [DOI] [Google Scholar]
- Fitak RR. Optm: estimating the optimal number of migration edges on population trees using treemix. Biol Methods Protoc. 2021;6(1):bpab017. doi: 10.1093/biomethods/bpab017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnier GR, Cummings MP, Clegg MT. Evolution of the avocados as revealed by DNA restriction fragment variation. J Hered. 1990;81(3):183–188. doi: 10.1093/oxfordjournals.jhered.a110963. [DOI] [Google Scholar]
- Gaut BS, Díez CM, Morrell PL. Genomics and the contrasting dynamics of annual and perennial domestication. Trends Genet. 2015;31(12):709–719. doi: 10.1016/j.tig.2015.10.002. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Seymour DK, Liu Q, Zhou Y. Demography and its effects on genomic variation in crop domestication. Nat Plants. 2018;4(8):512–520. doi: 10.1038/s41477-018-0210-1. [DOI] [PubMed] [Google Scholar]
- Hoff KJ, Lomsadze A, Borodovsky M, Stanke M. Whole-genome annotation with BRAKER. Methods Mol Biol. 2019;1962:65–95. doi: 10.1007/978-1-4939-9173-0_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra-Laclette E, Méndez-Bravo A, Pérez-Torres CA, Albert VA, Mockaitis K, Kilaru A, López-Gómez R, Cervantes-Luevano JI, Herrera-Estrella L. Deep sequencing of the Mexican avocado transcriptome, an ancient angiosperm with a high content of fatty acids. BMC Genomics. 2015;16(1):599. doi: 10.1186/s12864-015-1775-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illsley-Granich C, Brokaw R, Ochoa-Ascencio S, Bruwer T. Hass carmen®, a precocious flowering avocado tree. In: Proceedings VII World Avocado Congress. 2011. p. 5–9. [Google Scholar]
- Ishikawa M, Kiba T, Chua N-H. The Arabidopsis SPA1 gene is required for circadian clock function and photoperiodic flowering. Plant J. 2006;46(5):736–746. doi: 10.1111/j.1365-313X.2006.02737.x. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. Modelfinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14(6):587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour. 2015;15(5):1179–1191. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27(5):722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou Y, Liao Y, Toivainen T, Lv Y, Tian X, Emerson JJ, Gaut BS, Zhou Y. Evolutionary genomics of structural variation in Asian rice (Oryza sativa) domestication. Mol Biol Evol. 2020;37(12):3507–3524. doi: 10.1093/molbev/msaa185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahav E, Lavi U. Avocado Genetics and Breeding. Breeding Plantation Tree Crops: Tropical Species. New York (NY): Springer; 2009. p. 247–285. [Google Scholar]
- Lee TH, Guo H, Wang X, Kim C, Paterson AH. SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics. 2014;15(1):162. doi: 10.1186/1471-2164-15-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Leister D, Kleine T. Arabidopsis mitochondrial transcription termination factor mTERF2 promotes splicing of group IIB introns. Cells. 2021;10(2):315. doi: 10.3390/cells10020315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Duan S, Sheng J, Zhu S, Ni X, Shao J, Liu C, Nick P, Du F, Fan P, et al. Whole-genome resequencing of 472 Vitis accessions for grapevine diversity and demographic history analyses. Nat Commun. 2019;10(1):1–12. doi: 10.1038/s41467-019-09135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick S, Li H, Lipson M, Mathieson I, Gymrek M, Racimo F, Zhao M, Chennagiri N, Nordenfelt S, Tandon A, et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016;538(7624):201–206. doi: 10.1038/nature18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Me G-T, Arzate-Fernández AM. West Indian avocado: where did it originate? Phyton (B Aires). 2010;79(1):203. doi: 10.32604/phyton.2010.79.203. [DOI] [Google Scholar]
- Miller AJ, Gross BL. From forest to field: perennial fruit crop domestication. Am J Bot. 2011;98(9):1389–1414. doi: 10.3732/ajb.1000522. [DOI] [PubMed] [Google Scholar]
- Minio A, Cochetel N, Vondras A, Massonnet M, Cantu D. Assembly of complete diploid phased chromosomes from draft genome sequences. G3 (Bethesda). 2021;12(8):jkac143. doi: 10.1093/g3journal/jkac143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Hisabori T, Takeyasu K, Sato MH. VOZ; isolation and characterization of novel vascular plant transcription factors with a one-zinc finger from Arabidopsis thaliana. Plant Cell Physiol. 2004;45(7):845–854. doi: 10.1093/pcp/pch101. [DOI] [PubMed] [Google Scholar]
- Nath O, Fletcher SJ, Hayward A, Shaw LM, Masouleh AK, Furtado A, Henry RJ, Mitter N. A haplotype resolved chromosomal level avocado genome allows analysis of novel avocado genes. Hortic Res. 2022;9:uhac157. doi: 10.1093/hr/uhac157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Živković D, Stamatakis A, Alachiotis N. Sweed: likelihood-based detection of selective sweeps in thousands of genomes. Mol Biol Evol. 2013;30(9):2224–2234. doi: 10.1093/molbev/mst112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8(11):e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR. BEDTools: the Swiss-army tool for genome feature analysis. Curr Protoc Bioinformatics. 2014;47(1):11.12.1–11.12.34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendón-Anaya M, Ibarra-Laclette E, Méndez-Bravo A, Lan T, Zheng C, Carretero-Paulet L, Perez-Torres CA, Chacón-López A, Hernandez-Guzmán G, Chang T-H, et al. The avocado genome informs deep angiosperm phylogeny, highlights introgressive hybridization, and reveals pathogen-influenced gene space adaptation. Proc Natl Acad Sci U S A. 2019;116(34):17081–17089. doi: 10.1073/pnas.1822129116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner SS. How common is heterodichogamy? Trends Ecol Evol. 2001;16(11):595–597. doi: 10.1016/S0169-5347(01)02280-7. [DOI] [Google Scholar]
- Rounds MB. Check list of avocado varieties. Calif. Avocado Soc. Yearbook. 1950;35:178–205. [Google Scholar]
- Roux C, Pauwels M, Ruggiero M-V, Charlesworth D, Castric V, Vekemans X. Recent and ancient signature of balancing selection around the S-locus in Arabidopsis halleri and A. lyrata. Mol Biol Evol. 2013;30(2):435–447. doi: 10.1093/molbev/mss246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer BA, Wolstenholme BN, Whiley AW. The Avocado: Botany, Production and Uses. CABI; 2013. [Google Scholar]
- Schiffels S, Wang K. MSMC And MSMC2: the multiple sequentially markovian coalescent. Methods Mol Biol. 2020;2090:147–166. doi: 10.1007/978-1-0716-0199-0_7. [DOI] [PubMed] [Google Scholar]
- Scora RW, Wolstenholme BN, Lavi U. Taxonomy and botany. In: The Avocado: Botany, Production and Uses. Wallingford, UK: CABI Publishing; 2002. p. 15–37. [Google Scholar]
- Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, von Haeseler A, Schatz MC. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods. 2018;15(6):461–468. doi: 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Al-Dossary O, Alsubaie B, Al-Mssallem I, Nath O, Mitter N, Rodrigues Alves Margarido G, Topp B, Murigneux V, Kharabian Masouleh A, et al. Improvements in the sequencing and assembly of plant genomes. Gigabyte. 2021;2021:1–10. doi: 10.46471/gigabyte.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0; 2015.
- Solares EA, Tao Y, Long AD, Gaut BS. Hapsolo: an optimization approach for removing secondary haplotigs during diploid genome assembly and scaffolding. BMC Bioinformatics. 2021;22(1):9. doi: 10.1186/s12859-020-03939-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34(Web Server):W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey WB, Bergh B, Zentmyer GA. The origin, indigenous range and dissemination of the avocado. Calif Avocado Soc Yearbook. 1986;70:127–133. [Google Scholar]
- Torres AM, Bergh BO. Isozymes as indicators of outcrossing among “Pinkerton”seedlings. Calif Avocado Soc Yearbook. 1978;62:103–110. [Google Scholar]
- Vigouroux Y, Glaubitz JC, Matsuoka Y, Goodman MM, Sánchez GJ, Doebley J. Population structure and genetic diversity of new world maize races assessed by DNA microsatellites. Am J Bot. 2008;95(10):1240–1253. doi: 10.3732/ajb.0800097. [DOI] [PubMed] [Google Scholar]
- Vondras AM, Minio A, Blanco-Ulate B, Figueroa-Balderas R, Penn MA, Zhou Y, Seymour D, Ye Z, Liang D, Espinoza LK, et al. The genomic diversification of grapevine clones. BMC Genomics. 2019;20(1):972. doi: 10.1186/s12864-019-6211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B J, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9(11):e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag; 2016. ISBN 978-3-319-24277-4. [Google Scholar]
- Witney G, Martin G. Taking the California avocado breeding program into the next century. In: Proceedings of The World Avocado Congress III. 1995. p. 118. [Google Scholar]
- Xoca-Orozco L-Á, Cuellar-Torres EA, González-Morales S, Gutiérrez-Martínez P, López-García U, Herrera-Estrella L, Vega-Arreguín J, Chacón-López A. Transcriptomic analysis of avocado hass (Persea americana Mill) in the interaction system fruit-chitosan-colletotrichum. Front Plant Sci. 2017;8:956. doi: 10.3389/fpls.2017.00956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, Chen X, Schmid M. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell. 2010;22(7):2156–2170. doi: 10.1105/tpc.110.075606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui Y, Mukougawa K, Uemoto M, Yokofuji A, Suzuri R, Nishitani A, Kohchi T. The phytochrome-interacting vascular plant one-zinc finger1 and VOZ2 redundantly regulate flowering in Arabidopsis. Plant Cell. 2012;24(8):3248–3263. doi: 10.1105/tpc.112.101915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- California Avocado Society , 1951Yearbook.
- Zhou Y, Massonnet M, Sanjak JS, Cantu D, Gaut BS. Evolutionary genomics of grape (Vitis vinifera ssp vinifera) domestication. Proc Natl Acad U S A. 2017;114(44):11715–11720. doi: 10.1073/pnas.1709257114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Minio A, Massonnet M, Solares E, Lv Y, et al. The population genetics of structural variants in grapevine domestication. Nat Plants. 2019;5(9):965–979. doi: 10.1038/s41477-019-0507-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data for this study were submitted to NCBI under BioProject: PRJNA758103, which contains all raw PacBio and Illumina sequencing data, as well as the scaffolded genome assembly. A gff file describing the genes and the scaffold annotation files are available at Zenodo: https://doi.org/10.5281/zenodo.6392169. Published resequencing data used in this study from Rendon-Ayala et al. (2019) were from NCBI numbers: SRR8295599, SRR8295600, SRR8295601, SRR8295602, SRR8295603, SRR8295604, SRR8295605, SRR8295607, SRR8295608, SRR8295609, SRR8295610, and SRR8295611. The RNAseq data used for gene annotation were downloaded from NCBI with accession numbers SRR6116327, SRR6116328, SRR6116329, SRR6116330, and SRR2000042. The scripts used for analyses are available from https://github.com/GautLab/avo_ref_paper
Supplemental material is available at G3 online.