Abstract
Flavonoids are rich in tea plants (Camellia sinensis), and responsible for the flavor and healthful benefits of tea beverage. The anthocyanin levels in the purple tender shoots are higher than in the general green leaves of tea plant, which provide special materials to search metabolic mechanisms of flavonoid enrichment in plant. In this work, flavonoid differences between purple and green shoots from tea cultivars “Zijuan” (ZJ) and “Yunkang10” (YK-10) were investigated through metabolomic analysis, and mechanisms for their difference were surveyed by comparative transcriptomic and proteomic analysis. Levels of 34 flavonoids were different between ZJ and YK-10 shoots. Among them, 8 and 6 were marker metabolites in ZJ and YK-10, respectively. The differentially expressed genes (DEGs), differentially expressed proteins (DEPs), and different-level metabolites (DLMs) between ZJ and YK-10 were researched, respectively; and interactions including DEG-DLM, DEP-DLM, DEG-DEP, and DEG-DEP-DLM were analyzed; the contents of 18 characteristic flavonoids in tea leaves and expressions of 34 flavonoid metabolic genes were measured to verify the omics results. Integrated above analyses, a proposed model of flavonoids biosynthesis in tea shoots were established. The differential expression of the leucoanthocyanidin reductase (LAR), anthocyanidin synthase (ANS), anthocyanidin reductase (ANR), UDPG-flavonoid glucosyltransferase (UGT) 75L12 and 94P1 at gene level, and the ANS, ANR, and UGT78A15 at protein level, were closely associated with differences in flavonoids between ZJ and YK-10 shoot. Together, this study provides new information on the flavonoid accumulation mechanism in tea plant.
Keywords: flavonoids, metabolome, transcriptome, proteome, tea plant
Introduction
Flavonoids are important plant secondary metabolites that exist broadly in fruit, vegetables, spices, herbs, and beverage plants (Zhao et al. 2020). They produce multiple human health benefits, such as antioxidability, anti-inflammatory, anticancer, antiallergic, immunomodulatory, antihypertensive, antibacterial, and antiviral properties (Montenegro-Landívar et al. 2021), and their presence is associated with the bitter and astringent flavors of plant-based foods (Drewnowski 2001; McShea et al. 2008). Plant flavonoids are synthetized through phenylpropanoid, flavonoid, anthocyanin, flavone, and flavonol biosynthesis, and further modification of glycosylation, acylation and methylation (Chen, Shi et al. 2020). Many pivotal and functional enzymes are involved in the biosynthesis or modification of flavonoids, including phenylalanine ammonia-lyase (PAL), cinnamic 4-hydroxy-lase (C4H), 4-coumarate CoA ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone-3-hydroxylase (F3H), flavonoid synthase (FLS), flavonoid 3′-hydroxylase (F3′H), flavonoid 3′5′-hydroxylase (F3′5′H), dihydroflavonol-4-reductase (DFR), leucoanthocyanidin reductase (LAR), anthocyanidin synthase (ANS), anthocyanidin reductase (ANR), and UDPG-flavonoid glucosyltransferase (UGT) (Liu et al. 2021; Ma et al. 2021). Because flavonoids are responsible for the flavor and healthful benefits of plant foods and herbs, their biosynthesis and modifications have both scientific and commercial importance.
Tea is the most consumed beverage after water, and it is the dried product of tender tea shoots of the tea plant [Camellia sinensis (L.) O. Ktze]. Flavonoids, especially catechins, such as (–)-epigallocatechin-3-gallate (EGCG) and (–)-epigallocatechin (EGC), are important bioactive and flavorful substances in tea (Kim et al. 2014; Reygaert 2018; Davis and Running 2021). Interestingly, in many countries and regions, like China, India, Kenya, and Japan, there are some cultivars have red or purple tender shoots, such as “Zijuan” (ZJ), “Ziyan,” and “TRFK 306” (Jiang et al. 2013; Kerio et al. 2013; Joshi et al. 2015; Lai, Li et al. 2016; Maritim, Korir et al. 2021). They are color mutants of the tea plant with green shoot and can be inherited stably through asexual reproduction. The percentage of anthocyanins in the red or purple tender shoots is 1%–3% of dry weight, which is higher than that in green shoots (Saito et al. 2011; Jiang et al. 2013). In addition, the phenolic acids, amino acids, alkaloids and flavonoids like flavan-3-ols, proanthocyanins, flavonol, and flavone glycosides were also differentially abundant metabolites between purple and green tea leaves. The fold change values of flavonoids were obviously higher than that of other compounds (Li et al. 2021). Therefore, tea germplasms with purple shoots can be used for studying chemical compositions and physiological functions of anthocyanins, as well as the mechanism of flavonoid biosynthesis.
The glycosylated anthocyanins, pelargonidin 3-rhamnoside 5-glucoside, quercetin 3-rutinoside-4′-glucoside, cyanidin 3-diglucoside 5-glucoside, pelargonidin 3-sophoroside 5-glucoside, delphinidin/cyanidin-3-O-galactoside, and delphinidin/cyanidin 3-O-(6-O-p-coumaroyl) galactoside, have been identified in ZJ (Jiang et al. 2013; Mei et al. 2020; Li et al. 2021). Additionally, the levels of CHS, CHI, DFR, ANS, and UGT enzymes may be responsible for anthocyanin accumulation (Wang et al. 2017). The significantly increased genes expression levels of enzymes including UGT, ANS, anthocyanidin 3-O-glucosyltransferase, and anthocyanidin 3-O-glucoside 6″-O-acyltransferase, play important roles in increasing anthocyanin levels and their transformation (Wang, Liu et al. 2019; Maritim, Masand et al. 2021). In addition, the transcription factors MYB, bHLH, WD40, and WRKY influence anthocyanin accumulation by regulating the target genes in purple tea (Zhao et al. 2013; Lai, Du et al. 2016; Wei et al. 2019; Mei et al. 2020; Peng et al. 2020; Premathilake et al. 2020). However, the differences in the compositions and contents of flavonoids between purple and green tea leaves are not well-studied. The reasons for these differences also require further study.
Here, we explored the differences in flavonoids between the ZJ purple shoots and Yunkang-10 (YK-10) green shoots using metabolomics and high-performance liquid chromatography (HPLC) analyses. To improve the accuracy and specially explore the flavonoids biosynthesis mechanism, the targeted metabolome was chosen to analyze the flavonoids difference, both the primary and secondary spectrum data were qualitatively analyzed. The differences in expression levels of genes and enzymes involved in flavonoid biosynthesis were investigated using transcriptomics and proteomics analyses. The correlations between flavonoids and biosynthesized genes of enzymes were investigated and further verified by real-time quantitative polymerase chain reaction (RT-qPCR) and HPLC measurement. Our research increases our understanding of flavonoids and their formation mechanism in the tea plant.
Materials and methods
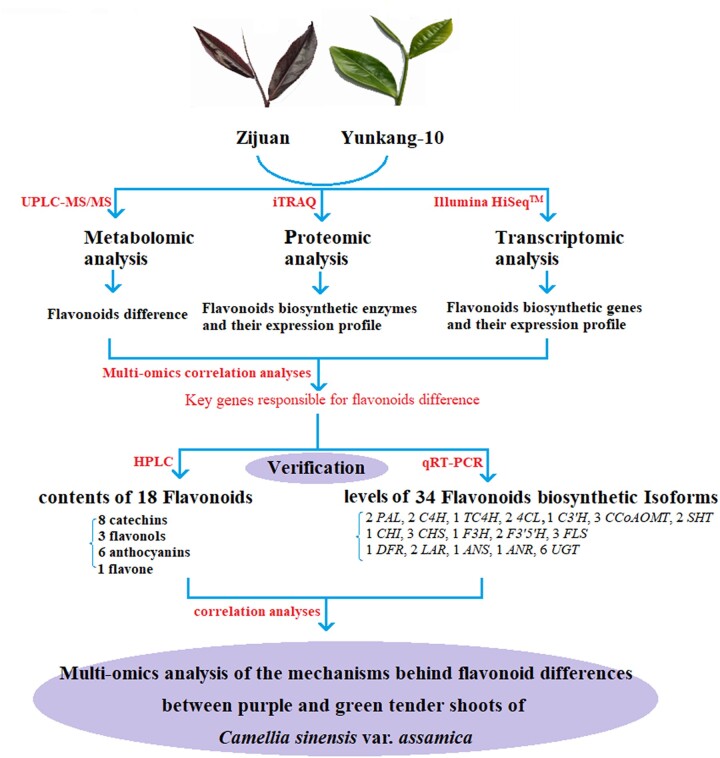
Plant materials and samples preparation
All the plant materials were collected from the Pu’er City Institute of Tea Science (N 22°02′-24°50′, E 99°09′–102°19′), Yunnan Province, China, on September 30, 2018. Under the same picking standard (1 bud and 2 leaves), all replicate samples picked from healthy tea plants with similar growth status to ensure highly consistent. One bud and 2 leaves of purple shoots from ZJ and green shoots from YK-10 were collected for metabolomics, transcriptomics, and proteomics analyses, each with 3 biological duplications. The expression levels of 34 flavonoid biosynthetic genes were determined using RT-qPCR, and the contents of 18 tea characteristic flavonoids were measured using by HPLC (Fig. 1). The samples used for HPLC and metabolomics analyses were fixed and dried by microwave, and the samples used for transcriptomics, proteomics and RT-qPCR analyses were immediately frozen in liquid nitrogen and stored at −80°C.
Fig. 1.
Overview of the multiomics analyses in this study.
Transcriptomic analysis
Total RNA was extracted using the TRIzol method and evaluated using a NanoDrop 2000 Spectrophotometers (Thermo Fisher Scientific, Wilmington, USA) and Agilent 2100 Bioanalyzer (Agilent, California, USA). A cDNA library was constructed by amplifying full-length cDNA to generate full-length transcriptome through PacBio Isoform sequencing, as described in our previous report (Chen, Shi et al. 2020). Another cDNA library was constructed with amplified cDNA of greater than 300 bp to generate the 2nd generation transcriptome using Illumina HiSeq2000 sequencing. High-quality isoforms or clean reads were obtained after filtration, and then were mapped to the YK-10 genome (http://www.plantkingdomgdb.com/tea_tree/) (Xia et al. 2017). The mapped reads were assembled using Cufflinks software and Reference Annotation Based Transcripts method (Trapnell et al. 2010; Roberts et al. 2011). Then, the assembled results of different replicate samples were merged for further analysis.
The gene expression levels were calculated using the Fragments Per Kb per Million reads (FPKM) method, and the differentially expressed genes (DEGs) were filtered using edgeR (https://bioconductor.org/packages/release/bioc/html/edgeR.html) with the criteria False Discovery Rate < 0.05 and |log2[Fold Change (FC)]| > 1. Based on the genes annotation description of reference genome, the DEGs were used for cluster and Gene Ontology (GO) (http://www.geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) significant enrichment analyses (Kanehisa et al. 2008). The new genes (length ≥ 200 bp and exon number ≥ 2) were annotated by blasting into non-redundant proteins sequences database in NCBI (http://www.ncbi.nlm.nih.gov/) and KEGG (E-value < 1 × 10−5) database.
Proteomic analysis
Proteins were extracted from tea shoots using the fractional precipitation method with organic solvents, and protein concentrations were measured using a Bicinchoninic Acid detection kit (P0012, Biyuntian, Shanghai, China). Extracted proteins were labeled using an iTRAQ label kit (AB SCIEX, Framingham, USA). The labeled peptides were pre-separated using high-pH reversed-phase liquid chromatography and analyzed by low-pH nano-HPLC-MS/MS (Orbitrap Fusion Tribrid, Thermo Fisher Scientific, Wilmington, USA) (Wang et al. 2018). Then, the MS data were analyzed using the Mascot search engine 2.3.2 (Matrix Science, London, UK). The Mascot database was established for protein identification using the tea plant genome.
According to the GO and KEGG annotations, enrichment analyses of the target proteins were fulfilled under Fisher’s exact test. The medians of relative expression levels after all “Unique Peptides” normalization were taken as the protein expression level. The differentially expressed proteins (DEPs) were filtered using the criteria |FC| >1.2 and P <0.05. On the basis of gene/protein expression profiles, correlations among the quantity and the GO and KEGG enrichments between the proteome and transcriptome were analyzed. The 4/9-quadrant figures were used for separating the correlated genes and proteins. To better identify the key genes/proteins, the GO and KEGG annotations of the DEPs and DEGs were further analyzed.
Metabolomic analysis
The freeze-dried tea shoot samples were crushed into powders and extracted overnight at 4°C with 1.0 ml 70% aqueous methanol. Then, the extracts were analyzed using an LC-ESI-MS/MS system (UPLC, Shim-pack UFLC SHIMADZU CBM30A system, http://www.shimadzu.com.cn/; MS, Applied Biosystems 4500 Q TRAP, http://www.appliedbiosystems.com.cn/). The effluent was alternatively connected into an ESI-triple quadrupole-linear ion trap (Q TRAP)-MS (Chen et al. 2013). On the basis of the Metware database established by Metware Biotechnology Co., Ltd. (Wuhan, China), the obtained data were further processed using the software Analyst 1.6.1 (AB SCIEX). The different-level metabolites (DLMs) were filtered using the criteria VIP >1, P <0.05 and |log2(FC)| ≥1, and then, they were used for the KEGG annotation and enrichment analysis by MBRole (http://csbg.cnb.csic.cs/mbrole), cluster analysis by Hierarchy Cluster and integrated analysis by iPath2.0 (http://pathways.embl.de).
According to metabolites abundance and gene/protein expression levels, the correlations between metabolome and transcriptome/proteome were analyzed. Three models were established to analyze the correlations, the KEGG Pathways, Bidirectional orthogonal projections to latent structures and Pearson’s correlation coefficients. Furthermore, the correlations among the metabolome, proteome, and transcriptome were analyzed on the basis of the DLMs, DEPs, and DEGs involved in the flavonoid synthetic pathways.
HPLC determination of the 18 flavonoids
The presence of 18 flavonoids, including 8 catechins [catechin (C), epicatechin (EC), catechin gallate (CG), gallocatechin (GC), EGC, epicatechin-3-gallate (ECG), gallocatechin gallate (GCG), and EGCG], 6 anthocyanins [delphinidin (Dp), cyanine (Cy), pelargonidin (Pg), peonidin (Pn), petunidin (Pt), and malvidin (Mv)], 3 flavonols [myricetin (My), quercetin (Qu), kaempferol (Kf)], and luteolin (Lu), in ZJ and YK-10 shoots were determined by HPLC (Agilent 1200, USA) with a TSKgel ODS-80TM column (4.6 mm Φ × 250 mm, 5 µm, TOSOH, Japan). In total, 8 ml of chromatographic methanol was added to 0.20 g tea-leaf powder and refluxed for 1.5 h. Then, the 10 mL diluted extract was filtered with a 0.45 µm nylon membrane. Each sample was extracted twice, and each extraction was detected 3 times.
The set conditions of the HPLC were as follow: mobile phase A was 5% acetonitrile and 0.261% phosphoric acid, phase B was 80% methanol; flow rate was 0.8 ml/min; column temperature was 35°C; injection volume was 2 μl; and the detection wavelengths were 280 nm for catechins, 530 nm for anthocyanins, and 360 nm for flavonols. The elution gradient program was 0–16 min, and B increased from 10% to 45% linearly; 16–22 min, B from 45% to 65%; 25–25.9 min, B from 65% to 100%; 25.9–29.0 min, B maintained 100%; 30 min, B decreased from 100% to 10% linearly and was then maintained at 10% for 6 min.
RT-qPCR analysis of genes expression
The total RNA was extracted using a total RNA Extraction Kit for polysaccharide polyphenolic plants (TIANGEN, Beijing, China) and reverse transcribed to cDNA using a PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China). Using the TB Green Premix Ex Taq II Kit with Tli RNaseH Plus (TaKaRa), the 20-μl reaction system for RT-qPCR was established as follows: 10 μl TB Green Premix Ex Taq II (2×), 0.8 μl forward/reverse primers (10 μM), 0.4 μl ROX Reference Dye II (50×), 2 μl cDNA and 6 μl ddH2O.
Using the software Primer 3 Plus, the primers for RT-qPCR were designed (Supplementary Table 1). The reaction program for the RT-qPCR was as follows: 95°C pre-denaturation for 30 s, 40 cycles of 95°C denaturation for 5 s and 60°C extension for 30 s; followed by 60–95°C for a melting curve analysis. The relative expression levels of genes were calculated using the 2–ΔΔCT method (Livak and Schmittgen 2001).
Statistical analyses
The means and standard deviation represent the average values of 3 determination results, and P <0.05 was considered as significant difference after t-tests among independent samples. The SPSS 22 software (Cronk 2016) (IBM SPSS Statistics, Chicago, USA) and TBtool (Chen, Chen et al. 2020) were used for statistical analyses, and SIMCA14.1 software was used for PCA and OPLS-DA analysis. The heatmap, network diagram and scatter plot were constructed using Heml, Cytoscape and Excel software, respectively.
Results
Difference in flavonoids between shoots of ZJ and YK-10
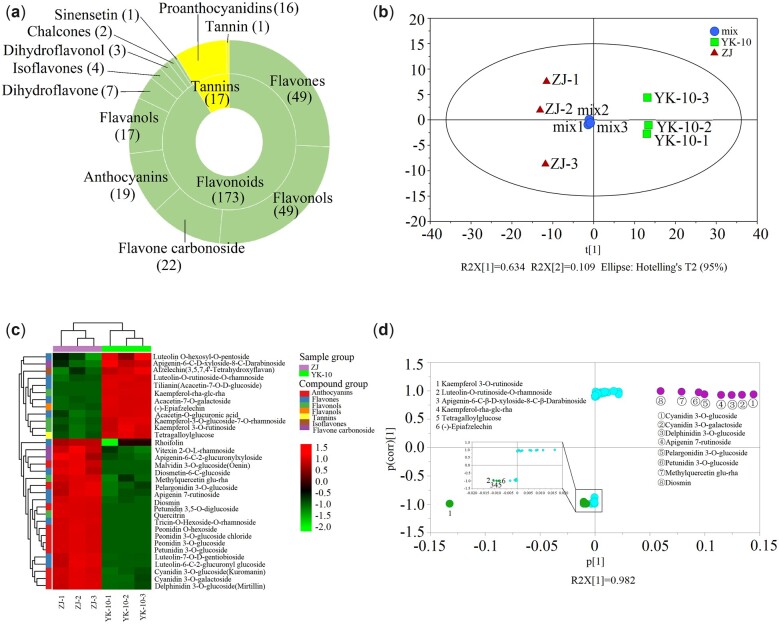
Through the UPLC-MS/MS metabolomic analysis, 187 and 178 flavonoids were identified in ZJ and YK-10 shoots, respectively. A total of 190 flavonoids and related compounds were identified, including 71 flavones and their carbon glycoside derivatives, 49 flavonols, 19 anthocyanins, 17 flavanols (catechins), 10 procyanidins, and 4 isoflavones. In the PCA, ZJ and YK-10 were grouped into different clusters, indicating that the compositions and contents of the flavonoids in ZJ and YK-10 differed (Fig. 2, a and b). In total, 20 and 14 metabolite levels were significantly higher (VIP > 1, P < 0.05 and FC ≥ 2) and lower (VIP > 1, P < 0.05 and FC < 0.5), respectively, in ZJ than in YK-10. Those flavonoids were further classified into anthocyanins, flavones, flavone oxyglycosides, flavonols, flavanols, isoflavones, and tannins. In ZJ shoots, the DLMs that were anthocyanins showed significantly higher contents, but the isoflavone (afzelechin), flavanol ((–)-epiafzelechin), and tannin (tetragalloylglucose) contents were lower than those in YK-10 (Fig. 2c; Table 1).
Fig. 2.
Results of metabolomics analysis. Classification of total identified flavonoids (a), PCA scores (b), heatmap (c) and S-plot (d) of 34 DLMs between ZJ and YK-10 shoots. Note: a: the inside circle of concentric circles represent Flavonoids and Tannins, the outside circle represent the classified compounds of Flavonoids and Tannins.
Table 1.
The list of DLMs between ZJ and YK-10 shoots.
| Compounds | Class II | VIP | FC | LogFC |
|---|---|---|---|---|
| Petunidin 3-O-glucoside | Anthocyanins | 3.09 | a | — |
| Peonidin 3-O-glucoside chloride | Anthocyanins | 2.84 | a | — |
| Peonidin 3-O-glucoside | Anthocyanins | 2.84 | a | — |
| Peonidin O-hexoside | Anthocyanins | 2.82 | a | — |
| Malvidin 3-O-glucoside (Oenin) | Anthocyanins | 2.60 | a | — |
| Petunidin 3,5-O-diglucoside | Anthocyanins | 2.51 | a | — |
| Cyanidin 3-O-galactoside | Anthocyanins | 1.66 | 68.47 | 6.10 |
| Cyanidin 3-O-glucoside (Kuromanin) | Anthocyanins | 1.65 | 64.96 | 6.02 |
| Delphinidin 3-O-glucoside (Mirtillin) | Anthocyanins | 1.64 | 62.69 | 5.97 |
| Pelargonidin 3-O-glucoside | Anthocyanins | 1.59 | 49.81 | 5.64 |
| (-)-Epiafzelechin | Flavanols | 1.26 | 0.09 | −3.53 |
| Tricin-O-Hexoside-O-rhamnoside | Flavones | 2.94 | a | |
| Tilianin (Acacetin-7-O-β-D-glucoside) | Flavones | 2.40 | b | — |
| Diosmetin-6-C-glucoside | Flavones | 2.16 | a | — |
| Luteolin-6-C-2-glucuronylglucoside | Flavones | 2.14 | a | — |
| Diosmin | Flavones | 2.12 | a | — |
| Rhoifolin | Flavones | 1.83 | 77.83 | 6.28 |
| Acacetin-O-glucuronic acid | Flavones | 1.72 | b | — |
| Apigenin 7-rutinoside (Isorhoifolin) | Flavones | 1.64 | 65.25 | 6.03 |
| Luteolin-7-O-β-D-gentiobioside | Flavones | 1.59 | 49.56 | 5.63 |
| Acacetin-7-O-galactoside | Flavones | 1.46 | 0.04 | −4.75 |
| Luteolin-O-rutinoside-O-rhamnoside | Flavones | 1.27 | 0.08 | −3.59 |
| Quercetin coumarin glucoside | Flavones | 1.17 | 9.04 | 3.18 |
| Luteolin O-hexosyl-O-pentoside | Flavones | 1.09 | 0.14 | −2.79 |
| Apigenin-6-C-2-glucuronylxyloside | Flavones oxyglycoside | 1.12 | 7.36 | 2.88 |
| Apigenin-6-C-β-D-xyloside-8-C-β-darabinoside | Flavones oxyglycoside | 1.11 | 0.15 | −2.78 |
| Vitexin 2′′-O-β-L-rhamnoside | Flavones oxyglycoside | 1.08 | 6.08 | 2.60 |
| Kaempferol rha-glc-rha | Flavonols | 2.86 | b | — |
| Quercitrin | Flavonols | 2.79 | a | — |
| methylquercetin glu-rha | Flavonols | 2.15 | 976.10 | 9.93 |
| Kaempferol-3-O-glucoside-7-O-rhamnoside | Flavonols | 1.07 | 0.17 | −2.57 |
| Kaempferol 3-O-rutinoside (Nicotiflorin) | Flavonols | 1.02 | 0.20 | −2.31 |
| Afzelechin (3,5,7,4′-Tetrahydroxyflavan) | Isoflavones | 1.33 | 0.06 | −3.95 |
| Tetragalloylglucose | Tannin | 1.07 | 0.17 | −2.53 |
: Compounds only detected in ZJ;
: Compounds only detected in YK-10.
Through S-plot analyses, 8 and 6 compounds were designated as marker metabolites in ZJ and YK-10, respectively (Fig. 2d). In the former, they were cyanidin 3-O-galactoside, cyanidin 3-O-glucoside (kuromanin), delphinidin 3-O-glucoside (mirtillin), petunidin 3-O-glucoside, pelargonidin 3-O-glucoside, apigenin 7-rutinoside (isorhoifolin), methylquercetin glu-rha, and diosmetin, and their levels were 49.81 times greater than in YK-10. In the latter, they were kaempferol 3-O-rutinoside (nicotiflorin), kaempferol rha-glc-rha, luteolin-O-rutinoside-O-rhamnoside, apigenin-6-C-D-xyloside-8-C-darabinoside, (–)-Epiafzelechin, and tetragalloylglucose, and their levels were 4.96 times greater than in ZJ.
Flavonoid biosynthetic gene expression differences between ZJ and YK-10 shoots
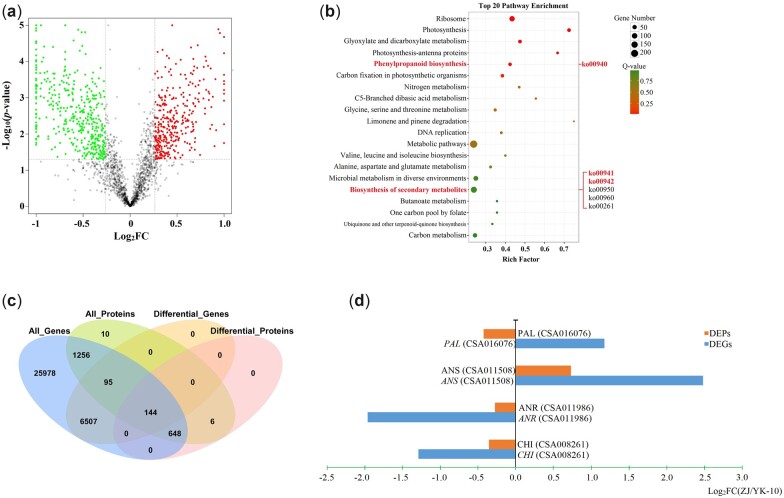
Using Illumina HiSeq2000 sequencing and data analysis, 3,531 and 3,215 genes were identified as having higher and lower expression levels in ZJ compared with in YK-10, respectively. Additionally, 64, 22, 12, and 3 DEGs were assigned into the KEGG pathways of phenylpropanoid biosynthesis (ko00940), flavonoid biosynthesis (ko00941), anthocyanin biosynthesis (ko00942), and flavone & flavonol biosynthesis (ko00944), respectively, which are related to the biosynthesis of flavonoid.
The metabolic pathways of flavonoids biosynthesis were showed in Supplementary Fig. 1. Structural genes were annotated into these pathways, and among them, expression of 34 genes were different significantly between ZJ and YK-10. Compared with in YK-10, expression of 17 genes in ZJ were increased significantly, included 3 PALs (CSA016076, 022024, 022025), 2 DFRs (CSA003949, XLOC_010242), 1 ANS (CSA011508), 1 CHS (CSA029775), 1 SHT (CSA023956), 1 CCoAOMT (XLOC_018193), 6 UGT75L12/13 (CSA005544, 005545, 010001, 036671, 036672, 029026), and 2 UGT94P1 (CSA007394, 008750). In addition, expression of other 17 genes decreased in ZJ significantly, included 1 4CL (CSA001434), 1 DFR (CSA035727), 1 CHI (CSA008261), 1 FLS (CSA008358), 1 ANS (CSA035767), 1 ANR (CSA011986), 1 CHS (CSA029773), 2 LARs (CSA014943, XLOC_016774), 1 F3′5′H (CSA031792), 1 SHT (XLOC_018541), 2 CCoAOMT (CSA009706, 030460), 2 UGT75L12s (CSA008693, 028873), and 2 UGT94P1s (CSA005965, 026000) (Supplementary Fig. 1 and Table 2).
In a previous work, we obtained full-length transcriptomes of ZJ and YK-10 using PacBio Iso-sequencing (Chen, Shi et al. 2020). In total, 84, 46, 6, and 7 isoforms, respectively, in ZJ and 133, 37, 4, and 2 isoforms, respectively, in YK-10 were annotated to the ko00940, ko00941, ko00942, and ko00944 KEGG pathways (Supplementary Table 3). After an integrated analysis of transcriptomes, 34 isoforms (2 PALs, 2 C4Hs, 1 TC4H, 2 4CLs, 1 CHI, 3 CHSs, 1 F3H, 2 F3′5′Hs, 3 FLSs, 1 DFR, 2 LARs, 1 ANS, 1 ANR, 6 UGTs, 1 C3′H, 3 CCoAOMTs, and 2 SHTs) were finally chosen for further RT-qPCR analysis (Supplementary Table 4).
Flavonoids biosynthetic enzyme expression differences between ZJ and YK-10 shoots
Using a proteomics analysis, 4,186 proteins were identified. In comparison to YK-10, 415 and 383 proteins showed increased and decreased expression levels in ZJ (Fig. 3a). Among them, 25, 3, and 1 DEP were enriched int phenylpropanoid, flavonoid and anthocyanin biosynthesis pathways, respectively, and these pathways were among the 20 top enriched pathways (Fig. 3b). Among the 29 DEPs, 3 PALs, 1 CHI, 1 ANS, 1 ANR, and 1 UGT78A15 were directly involved in the biosynthesis of catechins and anthocyanins (Supplementary Table 5).
Fig. 3.
Results of the proteomics analyses. A volcano plot (a) and the top 20 KEGG enriched pathways (b) of DEPs; a venn diagram of all the proteins/genes and DEPs/DEGs (c); the expression profiles of DEP-DEG pairs involved in flavonoid biosynthesis (d).
To further understand the functions of the DEPs in flavonoid biosynthesis, the correlations between DEPs and DEGs of ZJ and YK-10 were analyzed. The expression abundances of 144 DEPs were associated with the corresponding coding DEG levels, including 130 positively and 14 negatively correlated DEP-DEG pairs (Fig. 3c). Among them, 13 and 3 DEP-DEG pairs were enriched in the phenylpropanoid and flavonoid biosynthetic pathways, respectively. Only 4 DEP-DEG pairs were related to flavonoid biosynthesis. Among them, CHI (CSA008261), ANS (CSA011508), and ANR (CSA011986) showed the same expression trends as their corresponding DEGs, but PAL (CSA016076) showed a different expression trend (Fig. 3d; Supplementary Table 6a).
Correlation analyses of DLMs and DEGs/DEPs
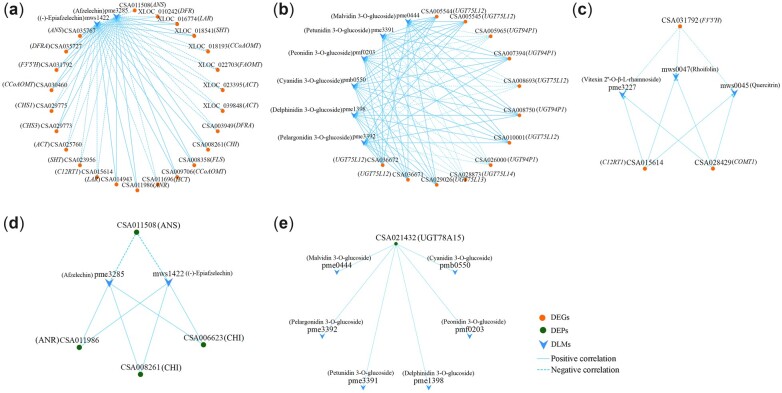
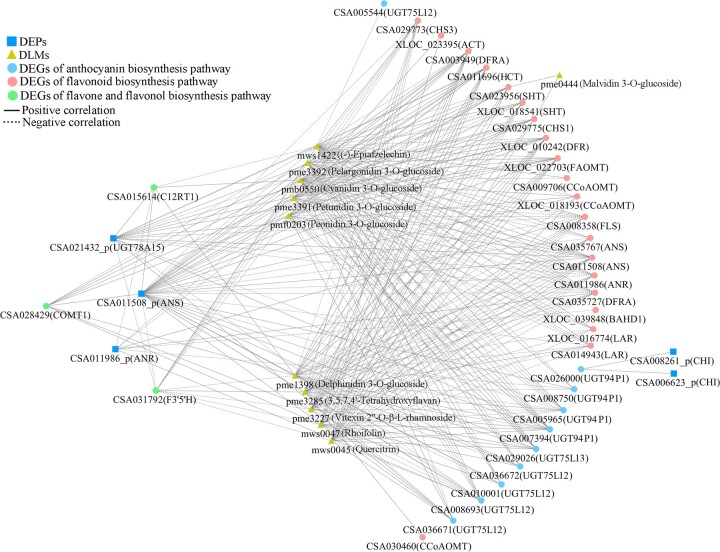
The correlation analyses of DLMs and DEGs revealed 3 shared KEGG pathways related to flavonoid biosynthesis. They were the flavonoid (2 DLMs and 24 DEGs), anthocyanin (6 DLMs and 12 DEGs), and flavone and flavonol (3 DLMs and 3 DEGs) biosynthetic pathways. Positive correlations were shown between the levels of some DEGs and DLMs, including 12 DEGs (1 SHT, 2 CCoAOMTs, 1 CHI, 1 CHS, 1 F3′5′H, 1 FLS, 1 DFR, 2 LARs, 1 ANS, 1 ANR) and 2 flavanols [(−)-epiafzelechin, afzelechin], 8 DEGs (5 UGT75L12, 1 UGT75L13, 2 UGT94P1), and 6 anthocyanin 3-O-glucosides (pelargonidin/delphinidin/cyanidin/peonidin/petunidin/malvidin 3-O-glucosides), and 2 DEGs (1 C12RT1, 1 COMT1) and 3 flavone glycosides (quercitrin, rhoifolin, vitexin 2′′-O-β-L-rhamnoside) (Fig. 4, a–c; Supplementary Table 6b).
Fig. 4.
The correlation networks of DLMs and DEGs/DEPs in flavonoid (a, d), anthocyanin (b, e), flavone and flavonol (c) biosynthesis.
Flavonoid biosynthetic (2 DLMs and 4 DEPs) and anthocyanin biosynthetic (6 DLMs and 1 DEP) pathways were both annotated in the DLM and DEP correlation analysis. The expression of UGT (CSA021432) was positively correlated with the levels of 6 anthocyanin 3-O-glucosides. The DEPs expression of CHI (CSA008261, 006623) and ANR (CSA011986) were positively correlated with the flavanol accumulations of (−)-epiafzelechin and afzelechin, whereas the expression of ANS (CSA011508) was negatively correlated with their levels (Fig. 4, d and e; Supplementary Table 6c). Interestingly, the expression of CHI (CSA008261) and ANR (CSA011986) showed positive correlation, while expression of ANS (CSA011508) showed negative correlation with the abundances of (−)-epiafzelechin and afzelechin, respectively.
Integrated analysis of transcriptomic, proteomic and metabolomic data
To gain a deeper understanding of the flavonoid metabolic mechanisms, the transcriptome-proteome-metabolome correlations were analyzed on the bases of DEGs-DEPs-DLMs involved in the flavonoid pathway (Fig. 5; Supplementary Table 7). In the flavonoid biosynthetic pathway, the expression of most DEGs showed opposite correlations with the expression of ANS (CSA011508) and accumulations of (−)-epiafzelechin and afzelechin. The accumulations of (−)-epiafzelechin and afzelechin presented negative correlations with the expression of ANS (CSA011508) but had no direct correlation with that of ANR (CSA011986). The DEGs expression of DFR (CSA035727) and FLS (CSA008358) showed negative associations with the DEP level of ANS, while they showed positive association with DEP level of ANR and DLMs accumulations of (−)-epiafzelechin and afzelechin. However, the expression of DFR (CSA003949) and HCT (CSA011696) positively correlated with level of ANS but negatively correlated with level of ANR, (−)-epiafzelechin, and afzelechin.
Fig. 5.
The correlation network of DEGs, DEPs and DLMs related to flavonoids biosynthesis.
In the anthocyanin biosynthetic pathway, the expression of 8 UGT genes (UGT94P1 and UGT75L12/13) correlated with the accumulation of 5 anthocyanin 3-O-glucosides. Only the expression of 4 DEGs (2 UGT94P1 and 2 UGT75L12) directly correlated with the levels of DEP UGT78A15 (CSA021432) and anthocyanin 3-O-glucosides at the same time. The levels of UGT94P1 (CSA005965) and UGT75L12 (CSA008693) were negatively correlated with the levels of UGT78A15 and anthocyanin 3-O-glucosides, but those of the UGT94P1 (CSA007394) and UGT75L12 (CSA036671) showed opposite correlations. In addition, the expression of UGT94P1 (CSA026000) was positively correlated with the level of the DEP CHI (CSA008261, 006623), but it showed no correlation with the abundances of anthocyanin 3-O-glucosides. In the flavone and flavonol biosynthetic pathway, the expression of the DEGs F3′5′H (CSA031792) and CCoAOMT1 (CSA028429) presented negative and positive correlations, respectively, with the flavone glycosides levels of rhoifolin, quercitrin, and vitexin 2′′-O-β-L-rhamnoside. There was no DEP level in this pathway was associated with DEG and DLM levels. However, the DEG expression of C12RT1 (CSA015614) showed correlations with the DEP and DLM levels in other pathways. Generally, the connections among the DEG, DEP, and DLM levels were not limited to the same pathway.
Verification of the correlations between gene expression and flavonoid contents
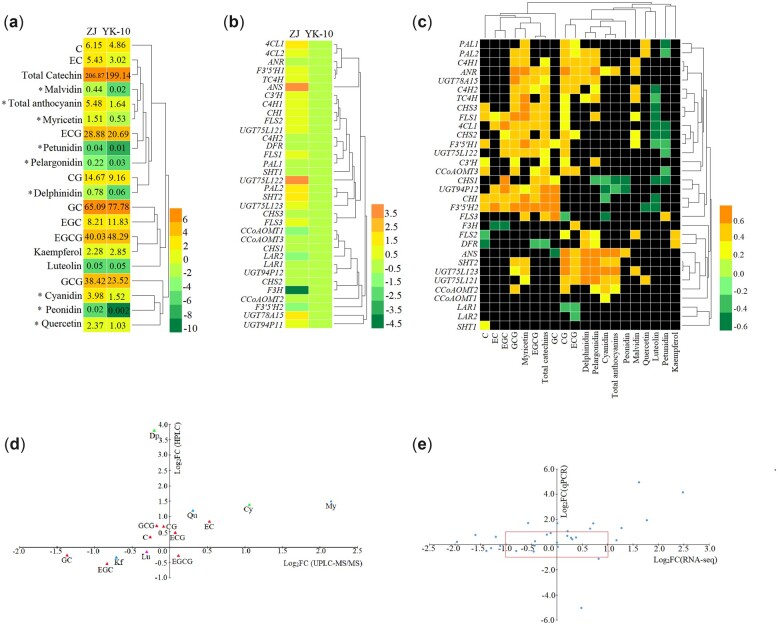
To verify the UPLC-MS/MS and RNA-seq results, 18 flavonoids and 34 biosynthetic genes in ZJ and YK-10 shoots were detected using HPLC and RT-qPCR, respectively (Fig. 6, a and b). Among them, 14 flavonoids and 30 genes had been determined using omics technology. In ZJ/YK-10, the contents of 9 flavonoids showed similar trends in the metabolomics analysis and HPLC measurements. Additionally, the expression patterns of 20 genes showed similar trends in the RNA-seq analysis and RT-qPCR. Additionally, the differences in flavonoid and genes levels were mostly not significant, as indicated by the absolute log2FC values being less than one in the omics data (Fig. 6, c and d). The HPLC results showed that the contents of Cy and 6 anthocyanins in ZJ were significantly greater than in YK-10, but the contents of 8 catechins showed no significant differences. Interestingly, the expression levels of most genes were higher in ZJ than in YK-10.
Fig. 6.
The HPLC analyses of the contents of 18 flavonoids (a, c), RT-qPCR analyses of 34 related biosynthetic genes (b, d) and correlation analyses between them (e). Note: a: the numbers represent contents of compounds (mg/g); * represents significant differences (log2FC > 1 or log2FC < −1).
Correlation analyses showed that the most significant correlations were positive, and the expression of most early genes played positive roles in catechin accumulation. The expression of ANS, which had a 17 times greater level in ZJ compared with in YK-10, positively correlated with the Cy and total anthocyanin accumulations. The expression of ANR, UGT75L121/123, SHT, and CCoAOMT2 also promoted anthocyanin biosynthesis. Additionally, the expression of DFR and the contents of EGCG showed negative correlations. The expression of ANS and the content of GC, as well as the expression of CHS1, CHI, and FLS3 and the level of Cy, showed negative correlations (Fig. 6e).
Discussion
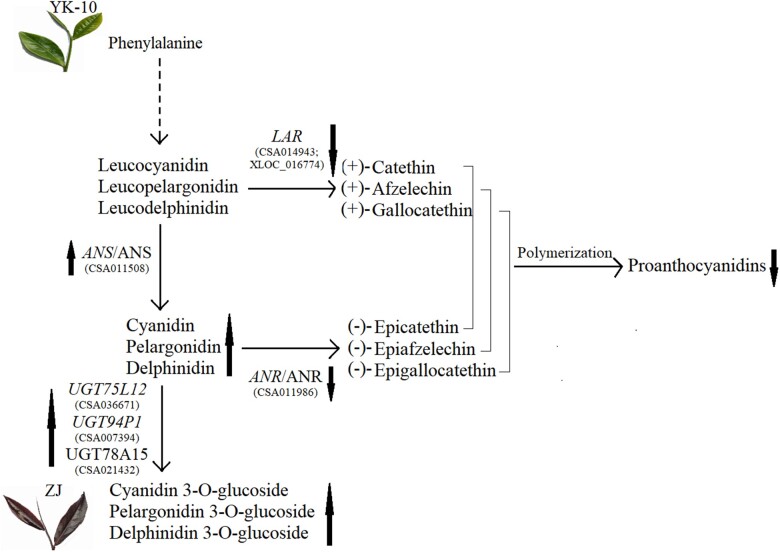
Using the 2nd transcriptome, 101 DEGs of ZJ/YK-10 encoding flavonoid biosynthetic enzymes were identified. The expression levels of 2 DFRs, 1 ANS (CSA011508), and 8 UGTs in ZJ were significantly higher than in YK-10. However, the expression of ANR (CSA011986) presented the opposite profile. Furthermore, the expression of ANS (CSA011508) and ANR (CSA011986) showed consistent trends with their encoding genes, and negatively and positively correlated with the DLMs of flavanols. Therefore, we speculate that the DEGs with high FPKM values, especially ANS (CSA011508) and ANR (CSA011986), play key roles in the accumulation and distribution of catechins and anthocyanins in ZJ. The high expression of ANS (CSA011508) and low expression of ANR (CSA011986) stimulate the accumulation of anthocyanins by reducing the conversion to catechins (Seitz et al. 2003; Mei et al. 2020).
In the DLM-DEG and DLM-DEP integration analyses, there were more correlated DLM-DEG pairs than DLM-DEP pairs involved in the flavonoids pathways. There were no interacting DLM-DEP pairs in the flavone and flavonol biosynthetic pathway, which may indicate that flavonoid accumulation is regulated mainly at the gene level but not at the protein level. The contents of 6 anthocyanidin 3-O-glucosides, 3 flavone glycosides and 2 flavanols were correlated with DEG/DEP expression. The 2 flavanols, (−)-epiafzelechin and afzelechin, are dimer and trimer units of propelargonidin, which is a proanthocyanidin (Omar et al. 2011). The expression of LAR (CSA014943, XLOC_016774) showed positive correlations with the contents of (−)-epiafzelechin and afzelechin. In our research, the lower level of LAR in ZJ was consistent with the reported results (Hossain et al. 2018; Mei et al. 2020), but the displayed correlations with (−)-epiafzelechin and afzelechin were noted for the first time. Therefore, we speculated that the lower expression of LAR resulted in the lower accumulations of proanthocyanidins in ZJ than in YK-10.
In addition, the importance of the gene UGT in anthocyanin biosynthesis was demonstrated once again by a multi-omics correlation analysis (Fang et al. 2021). UGT75L12 and UGT78A15 are responsible for flavonol 7-O-glucosides and flavonol 3-O-galactosides biosynthesis (Cui et al. 2016). UGT94P1 converts volatile organic compounds into β-primeverosides by sequential xylosylation (Ohgami et al. 2015). In our research, the expression of UGT94P1 (CSA005965, 007394) and UGT75L12 (CSA008693, 036671) showed direct correlations with the expression of UGT78A15 (CSA021432) and accumulation of anthocyanin 3-O-glucosides. Furthermore, the expression levels of UGT75L12 (CSA036671), UGT94P1 (CSA007394), and the UGT78A15 (CSA021432) enzyme were positively correlated with the contents of anthocyanin 3-O-glucosides. Thus, the higher expression levels of genes UGT75L12 (CSA036671), UGT94P1 (CSA007394), and enzyme UGT78A15 (CSA021432) promoted a greater accumulation of anthocyanin 3-O-glucosides in ZJ/YK-10.
These special DEGs [ANS (CSA011508), ANR (CSA011986), LAR (XLOC_016774), UGT75L12 (CSA036671), UGT94P1 (CSA007394)] and DEPs [ANS (CSA011508), ANR (CSA011986), UGT78A15 (CSA021432)] may play indispensable roles in the molecular mechanisms associated with the purple color in ZJ shoots. The competition for reaction substrates may responsible for the opposite expression profiles between ANS and LAR, ANR genes, and ANS and ANR enzymes (Mei et al. 2020). Under the collective effects of these DEGs and DEPs, the ZJ shoots present as purple with the decrease in proanthocyanidins and increase in anthocyanins. The anthocyanins were further converted into anthocyanins glucosides, which are more stable structures (Liu et al. 2018) (Fig. 7).
Fig. 7.
The proposed model of molecular mechanisms of purple-color formation in tea shoots. Note: the orange and green ellipse represents DEG and DEP; the red arrows represent the DEG and DEP trends; the green arrows represent the DLM trends.
In ZJ shoots, the total catechins, and especially anthocyanin, concentrations were higher than in YK-10 shoots, which corresponds to the quality characteristics of ZJ as described previously (Yang et al. 2009). Among the catechins, the content of EGCG analyzed by HPLC in ZJ was less than in YK-10, which was opposite of the UPLC-MS/MS data. This may be because the heating during the extraction process promotes the isomerization and reduction of EGCG (Jiang et al. 2021). The higher concentration of GCG in our experiment, which is a transisomer of EGCG, was also probably related to the extraction method.
The RT-qPCR results confirmed that the higher expression levels of most biosynthetic genes were important reasons for higher catechin and anthocyanin levels in ZJ/YK-10(Aza-González et al. 2013). The roles of early genes in anthocyanin accumulation have been shown in other purple plants (Wang, Luo et al. 2019), and the correlations between Dp and Pg abundance and early gene expression levels were almost positive in our study. The high gene expression level and catalytic activity of UGT promotes flavonoid accumulation (He et al. 2018), and 5 UGT isoforms with higher values in ZJ were verified by HPLC. The differential expression levels of ANS (CSA011508), LAR (CSA014943), and UGT (CSA036672) identified by RT-qPCR were consistent with the transcriptome predictions. Furthermore, the ANS level correlated negatively with GC content but positively with Cy, Dp, Pg and total 6 anthocyanins content in our study. This was consistent with ANS expression promoting the anthocyanin accumulation in Solanaceous vegetables (Liu et al. 2018). This indicates that the proposed model of purple-color formation in tea shoots is credible to some extent.
In general, we attempted to understand more clearly the molecular mechanisms behind the flavonoid differences that exist in tea shoots showing color variations using omics integrated analyses. The key ANS, ANR, LAR, UGT75L12, and UGT94P1 genes and the key ANS, ANR, UGT78A15 enzymes were predicted to play essential roles in anthocyanin accumulation. This demonstrated that the anthocyanin accumulation was positively associated with ANS and UGT75L12 expression as determined by HPLC and RT-qPCR combined analyses. However, the results of the RT-qPCR and HPLC experiments were not totally consistent with those of the omics analyses. Furthermore, the expression profiles of flavonoid biosynthetic genes are not only influenced by many environment factors, such as light (Li et al. 2020) and abiotic stresses (Gai et al. 2020), they are also regulated at the transcriptional and translational levels (Wu et al. 2020). The regulator genes of transcription factors like MYB, bHLH, WD40 and complex of them involving in the flavonoid biosynthesis, and their regulation mechanism through target genes are complex but worth to study. It is the most important work in progress for our team to explore and verify the regulator genes using transgenic and other biotechnologies.
Supplementary Material
Acknowledgments
We sincerely thank Pu’er City Institute of Tea Science, Yunnan Province, China, for providing us the samples of Zijuan and Yunkang10 tea plants. We sincerely thank the Gene Denovo Biotechnology Co. (Guangzhou, China) for technical assistance in the RNA-seq and proteomics, and the MetWare Biotechnology Co. (Wuhan, China) for technical assistance in the metabolomics.
Funding
The research was supported by the National Natural Science Foundation of China (31560221, 31760225, and 32160728), the earmarked fund for China Agriculture Research System (CARS-19) and the scientific research fund for teacher project of Yunnan Provincial Department of Education (2021J0111).
Author contributions
Conceived and designed the experiments: M.Z. and Y.M. Performed the experiments: S.-M.D., X.-Y.S. and B.N. Analyzed the data: Z.-W.L., S.-M.D., X.-Y.S. and B.N. Contributed experiments/analysis tools and funds: M.Z., L.-J.C., G.-H.Z. and C.-Y.L. Wrote the paper: Z.-W.L. Revised the paper: M.Z. and Z.-W.L. All authors reviewed and approved the final manuscript.
Contributor Information
Zhi-Wei Liu, College of Tea Science, State Key Laboratory of Conservation and Utilization of Bio-resources in Yunnan, and National & Local Joint Engineering Research Center on Germplasm Innovation & Utilization of Chinese Medicinal Materials in Southwest China, Yunnan Agricultural University, Kunming, Yunnan 650201, China.
Xing-Yun Shi, College of Tea Science, State Key Laboratory of Conservation and Utilization of Bio-resources in Yunnan, and National & Local Joint Engineering Research Center on Germplasm Innovation & Utilization of Chinese Medicinal Materials in Southwest China, Yunnan Agricultural University, Kunming, Yunnan 650201, China.
Shuang-Mei Duan, Tea Technique Extension Station of the Agriculture Bureau of Dehong, Yunnan 678400, China.
Bo Nian, College of Tea Science, State Key Laboratory of Conservation and Utilization of Bio-resources in Yunnan, and National & Local Joint Engineering Research Center on Germplasm Innovation & Utilization of Chinese Medicinal Materials in Southwest China, Yunnan Agricultural University, Kunming, Yunnan 650201, China.
Li-Jiao Chen, College of Tea Science, State Key Laboratory of Conservation and Utilization of Bio-resources in Yunnan, and National & Local Joint Engineering Research Center on Germplasm Innovation & Utilization of Chinese Medicinal Materials in Southwest China, Yunnan Agricultural University, Kunming, Yunnan 650201, China.
Guang-Hui Zhang, College of Tea Science, State Key Laboratory of Conservation and Utilization of Bio-resources in Yunnan, and National & Local Joint Engineering Research Center on Germplasm Innovation & Utilization of Chinese Medicinal Materials in Southwest China, Yunnan Agricultural University, Kunming, Yunnan 650201, China.
Cai-You Lv, College of Tea Science, State Key Laboratory of Conservation and Utilization of Bio-resources in Yunnan, and National & Local Joint Engineering Research Center on Germplasm Innovation & Utilization of Chinese Medicinal Materials in Southwest China, Yunnan Agricultural University, Kunming, Yunnan 650201, China.
Yan Ma, College of Tea Science, State Key Laboratory of Conservation and Utilization of Bio-resources in Yunnan, and National & Local Joint Engineering Research Center on Germplasm Innovation & Utilization of Chinese Medicinal Materials in Southwest China, Yunnan Agricultural University, Kunming, Yunnan 650201, China.
Ming Zhao, College of Tea Science, State Key Laboratory of Conservation and Utilization of Bio-resources in Yunnan, and National & Local Joint Engineering Research Center on Germplasm Innovation & Utilization of Chinese Medicinal Materials in Southwest China, Yunnan Agricultural University, Kunming, Yunnan 650201, China.
Data Availability
All data generated or analyzed during this study are included in this published article and its Supplementary Materials. The raw data of transcriptome and proteome have been respectively deposited to Sequence Read Archive database (https://submit.ncbi.nlm.nih.gov/about/sra/) (PRJNA524304) and iProX database (https://www.iprox.org/) (PXD036754). The qualitative and quantitative analysis result of all flavonoids identified by target metabolome was uploaded as Supplementary Table 8.
Supplemental material is available at G3 online.
Literature cited
- Aza-González C, Herrera-Isidrón L, Núñez-Palenius HG, De La Vega OM, Ochoa-Alejo N.. Anthocyanin accumulation and expression analysis of biosynthesis-related genes during chili pepper fruit development. Biol Plant. 2013;57(1):49–55. [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R.. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–1202. [DOI] [PubMed] [Google Scholar]
- Chen W, Gong L, Guo Z, Wang W, Zhang H, Liu X, Yu S, Xiong L, Luo J.. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol Plant. 2013;6(6):1769–1780. [DOI] [PubMed] [Google Scholar]
- Chen L, Shi X, Nian B, Duan S, Jiang B, Wang X, Lv C, Zhang G, Ma Y, Zhao M.. Alternative splicing regulation of anthocyanin biosynthesis in Camellia sinensis var. assamica unveiled by PacBio Iso-Seq. G3 (Bethesda). 2020;10(8):2713–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronk B. How to Use IBM SPSS Statistics: A Step-by-Step Guide to Analysis and Interpretation. Routledge; 2016. [Google Scholar]
- Cui L, Yao S, Dai X, Yin Q, Liu Y, Jiang X, Wu Y, Qian Y, Pang Y, Gao L, et al. Identification of UDP-glycosyltransferases involved in the biosynthesis of astringent taste compounds in tea (Camellia sinensis). J Exp Bot. 2016;67(8):2285–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Running D.. Repeated exposure to epigallocatechin gallate solution or water alters bitterness intensity and salivary protein profile. Physiol Behav. 2021;242:113624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewnowski A. The science and complexity of bitter taste. Nutr Rev. 2001;59(6):163–169. [DOI] [PubMed] [Google Scholar]
- Fang K, Xia Z, Li H, Jiang X, Qin D, Wang Q, Wang Q, Pan C, Li B, Wu H, et al. Genome-wide association analysis identified molecular markers associated with important tea flavor-related metabolites. Hortic Res. 2021;8(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai Z, Wang Y, Ding Y, Qian W, Qiu C, Xie H, Sun L, Jiang Z, Ma Q, Wang L, et al. Exogenous abscisic acid induces the lipid and flavonoid metabolism of tea plants under drought stress. Sci Rep. 2020;10(1):12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhao X, Gao L, Shi X, Dai X, Liu Y, Xia T, Wang Y.. Isolation and characterization of key genes that promote flavonoid accumulation in purple-leaf tea (Camellia sinensis L.). Sci Rep. 2018;8(1):130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M, Kim H-T, Shanmugam A, Nath U, Goswami G, Song J-Y, Park J-I, Nou I-S.. Expression profiling of regulatory and biosynthetic genes in contrastingly anthocyanin rich strawberry (Fragaria × ananassa) cultivars reveals key genetic determinants of fruit color. Int J Mol Sci. 2018;19(3):656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Han Z, Qin C, Lai G, Wen M, Ho C-T, Zhang L, Wan X.. Model studies on the reaction products formed at roasting temperatures from either catechin or tea powder in the presence of glucose. J Agric Food Chem. 2021;69(38):11417–11426. [DOI] [PubMed] [Google Scholar]
- Jiang L, Shen X, Shoji T, Kanda T, Zhou J, Zhao L.. Characterization and activity of anthocyanins in Zijuan tea (Camellia sinensis var. kitamura). J Agric Food Chem. 2013;61(13):3306–3310. [DOI] [PubMed] [Google Scholar]
- Joshi R, Rana A, Gulati A.. Studies on quality of orthodox teas made from anthocyanin-rich tea clones growing in Kangra valley, India. Food Chem. 2015;176:357–366. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(suppl_1):D480–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerio L, Wachira F, Wanyoko J, Rotich M.. Total polyphenols, catechin profiles and antioxidant activity of tea products from purple leaf coloured tea cultivars. Food Chem. 2013;136(3–4):1405–1413. [DOI] [PubMed] [Google Scholar]
- Kim H, Quon M, Kim J.. New insights into the mechanisms of polyphenols beyond antioxidant properties; lessons from the green tea polyphenol, epigallocatechin 3-gallate. Redox Biol. 2014;2:187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai B, Du L-N, Liu R, Hu B, Su W-B, Qin Y-H, Zhao J-T, Wang H-C, Hu G-B.. Two LcbHLH transcription factors interacting with LcMYB1 in regulating late structural genes of anthocyanin biosynthesis in nicotiana and litchi chinensis during anthocyanin accumulation. Front Plant Sci. 2016;7:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y-S, Li S, Tang Q, Li H-X, Chen S-X, Li P-W, Xu J-Y, Xu Y, Guo X.. The dark-purple tea cultivar ‘ziyan’ accumulates a large amount of delphinidin-related anthocyanins. J Agric Food Chem. 2016;64(13):2719–2726. [DOI] [PubMed] [Google Scholar]
- Li M, Shen Y, Ling T, Ho C-T, Li D, Guo H, Xie Z.. Analysis of differentiated chemical components between zijuan purple tea and yunkang green tea by UHPLC-orbitrap-MS/MS combined with chemometrics. Foods. 2021;10(5):1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Tan L, Zou Y, Tan X, Huang J, Chen W, Tang Q.. The effects of ultraviolet A/B treatments on anthocyanin accumulation and gene expression in dark-purple tea cultivar ‘Ziyan’ (Camellia sinensis). Molecules. 2020;25(2):354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Liu Z, Wu Y, Zheng L, Zhang G.. Regulatory mechanisms of anthocyanin biosynthesis in apple and pear. Int J Mol Sci. 2021;22(16):8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Tikunov Y, Schouten RE, Marcelis LFM, Visser RGF, Bovy A.. Anthocyanin biosynthesis and degradation mechanisms in vegetables: a review. Front Chem. 2018;6:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD.. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- Ma Y, Ma X, Gao X, Wu W, Zhou B.. Light induced regulation pathway of anthocyanin biosynthesis in plants. Int J Mol Sci. 2021;22(20):11116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maritim TK, Korir RK, Nyabundi KW, Wachira FN, Kamunya SM, Muoki RC.. Molecular regulation of anthocyanin discoloration under water stress and high solar irradiance in pluckable shoots of purple tea cultivar. Planta. 2021;254(5):85. [DOI] [PubMed] [Google Scholar]
- Maritim TK, Masand M, Seth R, Sharma RK.. Transcriptional analysis reveals key insights into seasonal induced anthocyanin degradation and leaf color transition in purple tea (Camellia sinensis (L.) O. Kuntze). Sci Rep. 2021;11(1):1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShea A, Ramiro-Puig E, Munro SB, Casadesus G, Castell M, Smith MA.. Clinical benefit and preservation of flavonols in dark chocolate manufacturing. Nutr Rev. 2008;66(11):630–641. [DOI] [PubMed] [Google Scholar]
- Mei Y, Xie H, Liu S, Zhu J, Zhao S, Wei C.. Metabolites and transcriptional profiling analysis reveal the molecular mechanisms of the anthocyanin metabolism in the “Zijuan” tea plant (Camellia sinensis var. assamica). J Agric Food Chem. 2020;69(1):414–427. [DOI] [PubMed] [Google Scholar]
- Montenegro-Landívar MF, Tapia-Quirós P, Vecino X, Reig M, Valderrama C, Granados M, Cortina JL, Saurina J.. Polyphenols and their potential role to fight viral diseases: an overview. Sci Total Environ. 2021;801:149719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgami S, Ono E, Horikawa M, Murata J, Totsuka K, Toyonaga H, Ohba Y, Dohra H, Asai T, Matsui K, et al. Volatile glycosylation in tea plants: sequential glycosylations for the biosynthesis of aroma beta-primeverosides are catalyzed by two Camellia sinensis glycosyltransferases. Plant Physiol. 2015;168(2):464–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omar MH, Mullen W, Crozier A.. Identification of proanthocyanidin dimers and trimers, flavone C-Glycosides, and antioxidants in Ficus deltoidea, a Malaysian herbal tea. J Agric Food Chem. 2011;59(4):1363–1369. [DOI] [PubMed] [Google Scholar]
- Peng Y, Thrimawithana AH, Cooney JM, Jensen DJ, Espley RV, Allan AC.. The proanthocyanin-related transcription factors MYBC1 and WRKY44 regulate branch points in the kiwifruit anthocyanin pathway. Sci Rep. 2020;10(1):14161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premathilake AT, Ni J, Bai S, Tao R, Ahmad M, Teng Y.. R2R3-MYB transcription factor PpMYB17 positively regulates flavonoid biosynthesis in pear fruit. Planta. 2020;252(4):59. [DOI] [PubMed] [Google Scholar]
- Reygaert W. Green tea catechins: their use in treating and preventing infectious diseases. BioMed Res Int. 2018;2018:9105261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pimentel H, Trapnell C, Pachter L.. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27(17):2325–2329. [DOI] [PubMed] [Google Scholar]
- Saito T, Honma D, Tagashira M, Kanda T, Nesumi A, Maeda-Yamamoto M.. Anthocyanins from new red leaf tea ‘Sunrouge’. J Agric Food Chem. 2011;59(9):4779–4782. [DOI] [PubMed] [Google Scholar]
- Seitz C, Oswald N, Börstling D, Forkmann G, Martens S.. Being acyanic: an unavoidable fate for many white flowers? Acta Hortic. 2003;612(612):83–88. [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L.. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu B-y, Zhao Q, Sun X, Li Y, Duan Z, Miao X, Luo S, Li J.. Genomic variance and transcriptional comparisons reveal the mechanisms of leaf color affecting palatability and stressed defense in tea plant. Genes (Basel). 2019;10(11):929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Luo Z, Lu C, Zhou R, Zhang H, Zhao L, Yu W, Zhao T.. Transcriptome profiles reveal new regulatory factors of anthocyanin accumulation in a novel purple-colored cherry tomato cultivar Jinling Moyu. Plant Growth Regul. 2019;87(1):9–18. [Google Scholar]
- Wang Z, Ma H, Smith K, Wu S.. Two-dimensional separation using high-pH and low-pH reversed phase liquid chromatography for top-down proteomics. Int J Mass Spectrom. 2018;427:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Pan D, Liang M, Abubakar YS, Li J, et al. Regulation of anthocyanin biosynthesis in purple leaves of zijuan tea (Camellia sinensis var. kitamura). Int J Mol Sci. 2017;18(4):833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K, Wang L, Zhang Y, Ruan L, Li H, Wu L, Xu L, Zhang C, Zhou X, Cheng H, et al. A coupled role for CsMYB75 and CsGSTF1 in anthocyanin hyperaccumulation in purple tea. Plant J. 2019;97(5):825–840. [DOI] [PubMed] [Google Scholar]
- Wu L-Y, Lv Y-Q, Ye Y, Liang Y-R, Ye J-H.. Transcriptomic and translatomic analyses reveal insights into the developmental regulation of secondary metabolism in the young shoots of tea plants (Camellia sinensis L.). J Agric Food Chem. 2020;68(39):10750–10762. [DOI] [PubMed] [Google Scholar]
- Xia E-H, Zhang H-B, Sheng J, Li K, Zhang Q-J, Kim C, Zhang Y, Liu Y, Zhu T, Li W, et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol Plant. 2017;10(6):866–877. [DOI] [PubMed] [Google Scholar]
- Yang XR, Bao YX, Huang M.. The botanical and quality characteristics of the tea cultivar “zi-Juan” in Yunnan province. J Tea. 2009;17–18. [Google Scholar]
- Zhao L, Gao L, Wang H, Chen X, Wang Y, Yang H, Wei C, Wan X, Xia T.. The R2R3-MYB, bHLH, WD40, and related transcription factors in flavonoid biosynthesis. Funct Integr Genomics. 2013;13(1):75–98. [DOI] [PubMed] [Google Scholar]
- Zhao C, Wang F, Lian Y, Xiao H, Zheng J.. Biosynthesis of citrus flavonoids and their health effects. Crit Rev Food Sci Nutr. 2020;60(4):566–583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Supplementary Materials. The raw data of transcriptome and proteome have been respectively deposited to Sequence Read Archive database (https://submit.ncbi.nlm.nih.gov/about/sra/) (PRJNA524304) and iProX database (https://www.iprox.org/) (PXD036754). The qualitative and quantitative analysis result of all flavonoids identified by target metabolome was uploaded as Supplementary Table 8.
Supplemental material is available at G3 online.