Abstract
The European database on SSU rRNA can be consulted via the World WideWeb at http://rrna.uia.ac.be/ssu/ and compiles all complete or nearly complete small subunit ribosomal RNA sequences. Sequences are provided in aligned format. The alignment takes into account the secondary structure information derived by comparative sequence analysis of thousands of sequences. Additional information such as literature references, taxonomy, secondary structure models and nucleotide variability maps, is also available.
SMALL SUBUNIT RIBOSOMAL RNA (SSU rRNA)
Many thousands of SSU rRNA sequences, coded by the genomes of Bacteria, Archaea, Eucarya, plastids and mitochondria, have been determined since the first complete primary structure, that of the 16S rRNA of the bacterium Escherichia coli, was published (1). As for the secondary structure of SSU rRNA, this has been derived gradually, mainly by examination of sequence alignments and searches for compensating substitutions that reveal the existence of base-pairing. Recently, the tertiary structures of the SSU rRNA of the bacterium Thermus thermophilus was obtained with atomic resolution by X-ray diffraction crystallography of the ribosomal subunit (2). The exactness of the theoretically derived secondary structure model for bacterial SSU rRNA (3,4) was thus confirmed.
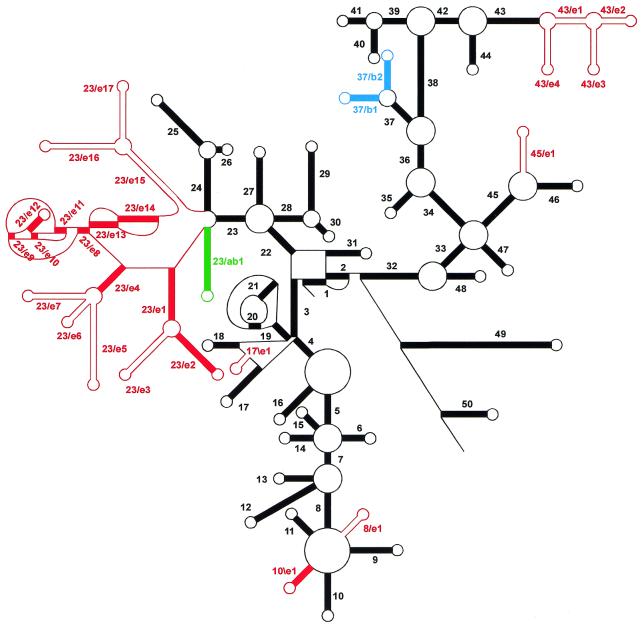
Figure 1 shows a schematic comparison of the secondary structure of SSU rRNAs of Archaea, Bacteria and Eucarya. A common core structure consisting of 50 helices can be distinguished. Archaea and Bacteria SSU rRNAs show only minor structural differences with this core structure. Eucarya SSU rRNAs, however, can possess insertions, or more rarely deletions, at several points of the common core, accounting for a larger average chain length of these RNAs as compared to Bacteria and Archaea. Some insertions are shared by most Eucarya SSU rRNAs, such as the large structure branching from helix 23 of the common core structure, for which a detailed folding model has been proposed (5). Other insertions are taxon, or species, specific and account for the extreme variability in length of these RNAs. As for the SSU rRNAs coded by organelle genomes, those of the plastids conform to the bacterial secondary structure model. Mitochondrial SSU rRNAs, on the other hand, are extremely variable in length. Some of them, such as those of the Fungi, have nearly the same secondary structure as bacterial SSU rRNAs. Others, such as those of the plants, possess insertions with respect to the common core structure, yet others, such as those of the animals and many protists possess a reduced structure missing several helices of the common core.
Figure 1.
Scheme of the secondary structure of SSU rRNA. The core of the structure common to SSU rRNAs of Archaea, Bacteria and most Eucarya is drawn in black. Helices are numbered in the order of occurrence of their 5′-strand when following the chain from 5′- to 3′-terminus. They bear a different number when separated by a multibranched loop, a pseudoknot loop or a single-stranded area not forming a loop. Bulge loops and internal loops are not shown. Coloured helices are present in Archaea and Bacteria (green), in Bacteria only (blue) or in Eucarya only (red). Those drawn as solid red bars are present in all Eucarya with the exception of the protist taxa Microsporidia, Diplomonadida and Parabasalidea, where some of these helices and even some core helices can be absent. Those drawn as parallel red lines are present only in certain eukaryotic taxa. A helix is numbered N/en if it is the nth Eucarya-specific (red) helix following the 5′-strand of the Nth common core (black) helix. It is numbered N\en if it follows the 3′-strand of the Nth helix. Analogous nomenclature, N/bn and N\bn, is used for Bacteria-specific helices, N/abn and N\abn for those common to Archaea and Bacteria.
CONTENTS OF THE DATABASE
The European database on SSU rRNA is regularly updated by scanning the EMBL nucleotide sequence database (6) for corrected or newly determined ribosomal RNA genes. In general, only complete or nearly complete sequences are compiled. Partial sequences are included only if the length of the aligned sequence amounts to 70% or more of the estimated complete chain length. All sequences are stored in aligned format with annotation of secondary structure. In September 2001, the SSU rRNA database contained 20 851 aligned sequences, of which 597 are from Archaea, 12 467 from Bacteria, 6561 from eukaryotes, 139 from plastids and 1087 from mitochondria. This represents an increase of 56% with respect to the previous release (7).
ACCESSIBILITY
The SSU rRNA database is available at http://rrna.uia.ac.be/ssu/. In order to simplify and speed up access to the data via the World Wide Web, each SSU rRNA sequence is stored in a separate file. Each of these files contains the sequence as well as secondary structure information, and annotations such as accession number, literature reference and detailed taxonomic specifications. Three interfaces are available to select and download the desired sequences: the list interface, where individual sequences can be selected; the forms interface, which allows the selection of groups of sequences; and the query interface, which allows the search for sequences by species name, accession number and literature data. Using the query interface, it is also possible to perform searches on the entire database or to limit the search to certain taxa.
Additional material available online on our SSU rRNA server includes:
secondary structure models of prokaryotic, eukaryotic, plastidial and mitochondrial rRNAs, updated with respect to those published previously (7) where necessary;
secondary structure variability maps of bacterial and eukaryotic SSU rRNA, based on substitution rate calibration (8);
tertiary structure variability maps of bacterial SSU rRNAs (J.Wuyts, Y.Van de Peer and R.De Wachter, manuscript submitted for publication);
information about primers for sequencing SSU rRNA sequences;
software available for sequence alignment (9), tree construction (10) and sequence alignment format conversion (11);
links to other relevant databases and resources.
If problems occur in connecting to the server or in retrieving data, the authors can be contacted by email: jan.wuyts@ua.ac.be, yvdp@gengenp.rug.ac.be or rupert.dewachter@ua.ac.be. Users publishing results based on data retrieved from our database are requested to cite this paper.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
J.W. holds a scholarship of the Special Research Fund of the University of Antwerp. Y.V.d.P. is a research fellow of the Fund for Scientific Research, Flanders.
REFERENCES
- 1.Brosius J., Dull,T.J., Sleeter,D.D. and Noller,H.F. (1981) Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol., 148, 107–127. [DOI] [PubMed] [Google Scholar]
- 2.Wimberly B.T., Brodersen,D.E., Clemons,W.M., Morgan-Warren,R.J., Carter,A.P., Vonrhein,C., Hartsch,T. and Ramakrishnan,V. (2000) Structure of the 30S ribosomal subunit. Nature, 407, 327–339. [DOI] [PubMed] [Google Scholar]
- 3.Gutell R.R. (1993) Collection of small subunit (16S- and 16S-like) ribosomal RNA structures. Nucleic Acids Res., 21, 3051–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neefs J.-M., Van de Peer,Y., De Rijk,P., Chapelle,S. and De Wachter,R. (1993) Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res., 21, 3025–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wuyts J., De Rijk,P., Van de Peer,Y., Pison,G., Rousseeuw,P. and De Wachter,R. (2000) Comparative analysis of more than 3000 sequences reveals the existence of two pseudoknots in area V4 of eukaryotic small subunit ribosomal RNA. Nucleic Acids Res., 28, 4698–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoesser G., Baker,W., van den Broek,A., Camon,E., Garcia-Pastor,M., Kanz,C., Kulikova,T., Lombard,V., Lopez,R., Parkinson,H. et al. (2001) The EMBL nucleotide sequence database. Nucleic Acids Res., 29, 17–21. Updated article in this issue: Nucleic Acids Res. (2002), 30, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van de Peer Y., De Rijk,P., Wuyts,J., Winkelmans,T. and De Wachter,R. (2000) The European small subunit ribosomal RNA database. Nucleic Acids Res., 28, 175–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van de Peer Y., Van der Auwera,G. and De Wachter,R. (1996) The evolution of stramenopiles and alveolates as derived by “substitution rate calibration” of small subunit ribosomal RNA. J. Mol. Evol., 42, 201–210. [DOI] [PubMed] [Google Scholar]
- 9.De Rijk P. and De Wachter,R. (1993) DCSE, an interactive tool for sequence alignment and secondary structure research. Comput. Appl. Biosci., 9, 735–740. [DOI] [PubMed] [Google Scholar]
- 10.Van de Peer Y. and De Wachter,R. (1994) TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci., 10, 569–570. [DOI] [PubMed] [Google Scholar]
- 11.Raes J. and Van de Peer,Y. (1999) ForCon: a software tool for the conversion of sequence alignments. EMBnet.news, 6, (http://www.hgmp.mrc.ac.uk/embnet.news/vol6_1/ForCon/forcon.html).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.