Abstract
The Yeast Protein Localization database (YPL.db) contains information about the localization patterns of yeast proteins resulting from microscopic analyses. The data and parameters of the experiments to obtain the localization information, together with images from confocal or video microscopy, are stored in a relational database, building an archive of, and the documentation for, all experiments. The database can be queried based on gene name, protein localization, growth conditions and a number of additional parameters. All experiment parameters are selectable from predefined lists to ensure database integrity and conformity across different investigators. The database provides a structure reference resource to allow for better characterization of unknown or ambiguous localization patterns. Links to MIPS, YPD and SGD databases are provided to allow fast access to further information not contained in the localization database itself. YPL.db is available at http://ypl.tugraz.at.
INTRODUCTION
The yeast Saccharomyces cerevisiae (baker’s yeast) is an important model system for cell biological research. The completion of the genomic DNA sequence in 1996 [yeast was the first eukaryote whose genome was completely sequenced (1)] has laid the basis for whole-genome analysis of an organism and has made yeast the paradigm for genomic research. The fact that multiple human disease genes have homologs in yeast (2) has supported the great biomedical interest in yeast research. Microarray technology for the analysis of the transcriptome (3) and proteome (4) was developed using yeast DNA and protein arrays, and multinational concerted projects focus on genome-wide analyses of gene function [e.g. European Function Analysis Network (EUROFAN; 5)]. Cell biological research on protein function and biochemical genomics (6) is complemented by genome-wide studies on two-hybrid protein–protein interaction (7) as well as protein localization (8,9). Knowledge about the precise subcellular localization of a protein, or its dynamics, is of great importance to understand protein function and its interaction with other factors in a metabolic pathway. The increased application of green fluorescent protein (GFP) as a tag for microscopic analysis in living yeast cells and immunological methods for subcellular localization studies have provided a wealth of localization data in the literature (10,11). Numerous methods for protein tagging with GFP or immuno-tags, either C- or N-terminally, as chromosomally integrated versions or on episomal plasmids have been described (11). In order to provide a reference of microscopic localization patterns of yeast proteins and, subsequently, subcellular structures, we have created a relational database to accommodate protein localization data in yeast, together with information on experimental design, strain background, growth conditions and other relevant information, in a readily web-accessible form. The database supports TIFF image files as well as JPEG images, and provides direct links to the relevant genomic and proteomic information, at MIPS (12), YPD (13) and SGD (14) yeast genome and protein databases. The implemented search functions allow the user to specify experimental design, type of construct, gene or protein name or localization patterns.
DESIGN
The Yeast Protein Localization database (YPL.db) was designed to support yeast investigators in their daily microscopy work and to serve as the central archive for all microscopy experiments, stored in the database in a consistent and reproducible manner. A number of features have been implemented to allow the addition of new experiments to the database, the modification of experiments, the definition of data and the definition of constructs. Additional features cover querying the database, processing the images using external image processing programs and the definition of user accounts.
Experiments, data and constructs
The core entity of YPL.db is an experiment. Experiments help to determine the localization of a certain protein in the yeast cell. Each experiment is primarily described by the gene investigated and the construct used to introduce the fluorescent (protein) tag into the cell (Fig. 1). The strain background, the growth medium, the growth time and temperature and the growth phase of the cell have to be specified. Additional experimental data are: the name of the investigator, the date and time the experiment was performed and an optional comment. Based on the fluorescence image(s) the investigator determines the protein localization(s) of the protein in the cell.
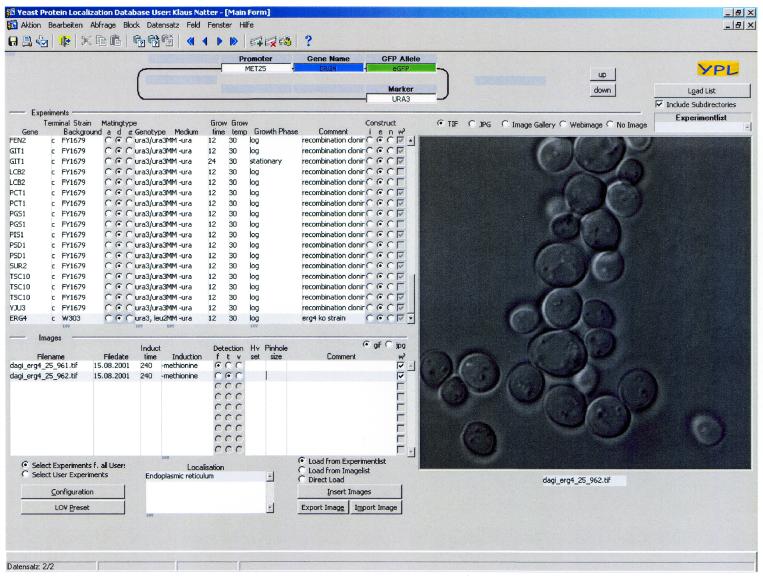
Figure 1.
A view of the Oracle forms interface for the YPL.db. The construct is shown in the upper panel and includes the investigated gene (ERG4), the FP-tag (eGFP), selection marker to introduce the construct (URA3) and promoter to drive the expression (MET25). The list on the left panel describing the experiments includes the name of the gene, type of fusion (C- or N-terminal), strain background, mating type (a, d or α), genotype, medium, growth time, growth temperature, growth phase, comment and construct (integral, episomal or ‘vital stain’). The selected image files are shown in the lower left panel with the appropriate file name, date, time, induction, detection and localization (endoplasmic reticulum). The image itself is shown on the right (transmission image of ERG4-GFP fluorescence).
An experiment is created in the database by importing one or more TIFF images obtained from the video or confocal microscope. It is assigned the default experimental parameters, which are defined in the user profile of the current user. The parameters for the current experiment may be modified to reflect the actual experimental conditions. The cellular localizations of the protein based on the microscopic observations have to be set in order to query the database based on this parameter. The nomenclature for localization patterns is according to YPD (13). If the observed pattern is ambiguous, the internal structure reference database can be accessed to correlate the observed pattern to known structures. Multiple localizations in the cell (e.g. ‘endoplasmic reticulum’ and ‘cytosol’) can be defined and searched for using the Oracle interface. A comment can be entered for the experiment for further detailed description or to reflect experimental conditions not covered by the database.
Collaborating investigators often use different terms to describe the same feature or parameter. To keep the experimental database consistent and to get meaningful and complete results from database queries, all parameter values have to be selected from predefined yet easily extensible lists. These menu lists are maintained by users, who have administrative access rights to the database, through easy to use dialogs. There are lists for each of the experimental parameters, including growth conditions, growth media, growth phase, growth temperature, cellular localization, components of constructs, strain backgrounds and ORFs, and synonyms (Fig. 1). An important feature is the definition of the constructs used to attach an immuno-tag or fluorescent protein (FP) to specific reading frames. A construct consists of the investigated gene, an FP or immuno-tag, a selection marker used to introduce the construct into yeast, promoters to drive expression and 3′ and 5′ primers, used for construction of the gene fusion. There are three different types of ‘constructs’, the episomal type and the integral type, which express fusion proteins, and the ‘vital stain’ type, where organelles are stained by organelle-specific fluorescent dyes. A ‘construct’ is composed by selecting the required components, their relative location and then connecting them together. Components may be added, deleted and repositioned and each modification is instantly visible through the graphical representation of the construct.
Specific proteins and/or constructs or vital dye labeling patterns are defined as references in the database to describe particular yeast subcellular structures as they occur under various growth conditions. This ‘structure reference database’ allows visual comparison of ‘undefined’ localization patterns obtained in a microscopic experiment with known representative localization patterns, to better enable identification of the respective protein localization.
Queries, external processing, user management and data display
YPL.db features extensive and flexible queries. A mask dedicated for that task allows the formulation of queries with respect to every single experiment attribute. Each of the query parameter values is selected from the same value lists used to define the experiment, ensuring meaningful and complete results. The experiments matching the query are displayed in tabular form containing all relevant parameters and the user can navigate through the results using the navigation buttons or the corresponding keyboard shortcuts. Once an image has been imported into the database, it may be processed further to add annotations directly to the image or to extract an interesting area of the image, which will be displayed when the database is accessed via the World Wide Web (Fig. 2). The program that is to be used for image processing can be defined in the user profile, and hence every investigator can use the tools of her/his choice. When invoking the image processing function, the image is exported from the database to the file system, and the previously defined program is called. Upon exiting the program, the image is imported under the same or a new name into the database. A new image is assigned to the same experiment to which the original image belonged.
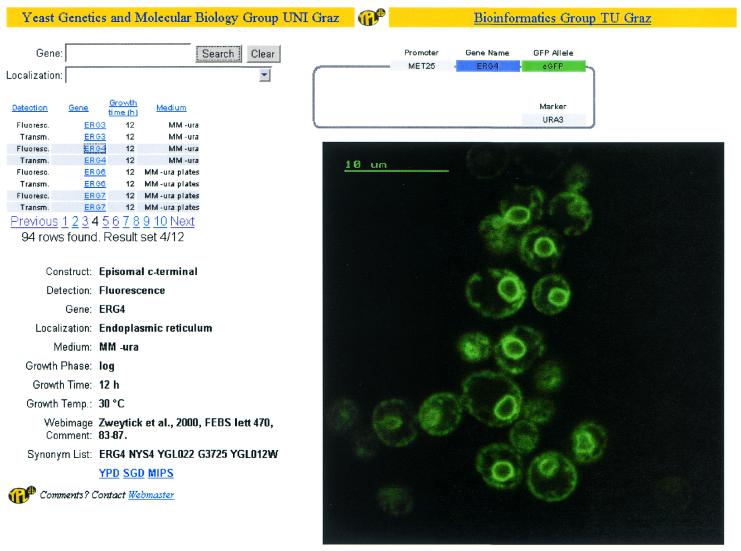
Figure 2.
A view of the result of a database search for ERG4 using the web interface. An image can be searched using gene name or localization and then selected from this list in the upper-left panel. The construct is shown in the upper-right panel and includes the investigated gene (ERG4), the FP-tag (eGFP), selection marker (URA3) and promoter (MET25). The screen describing an experiment is in the lower-left panel and shows: the used construct (episomal C-terminal), detection (fluorescence), the gene (ERG4), localization (endoplasmic reticulum), medium (MM-ura), growth phase (log), growth time (12 h), growth temperature (30°C), comment (reference for the image), synonym list and links to YPD, SGD and MIPS. The image itself (ERG4-GFP fluorescence image) is shown on the right.
Each investigator accessing YPL.db to enter data has to have a database account, to prohibit unauthorized access and modification of the database. An account is associated with a certain access level, defining the actions a user may perform, and a set of user preferences, which contain the default experiment parameters used during experiment creation and the name of the image processing software. Currently, three access levels are defined; the level of the super user, who can modify data and experiments of other investigators. The second level is assigned to users who can create experiments and modify them, but have read-only access to experiments of other users. The third level is the read-only access through the World Wide Web to selected image data contained in the database, and their links to experimental setup, or other databases.
IMPLEMENTATION
The database is based on Oracle 8.0.5 and is maintained and modified by the Oracle Forms and PL/SQL-based application. A two-tier client server architecture is utilized with the Oracle database as the server and the Forms application as a client. The content of the database is converted into a format suitable for standard web browser resulting in a three-tier architecture. The database was designed in normalized form to avoid redundant data storage.
The experiment table is the central table in the database, which links to all other tables. All experiment parameters are represented by their own table, which contains valid values for a certain parameter. The entries of these tables are presented in selectable lists of values, from which data have to be selected.
Images generated in our laboratory typically have a size of 512 × 512 pixels (8 bit dynamic range) at a pixel size of 60 nm, resulting in a 256 kB entry per image. Although the individual image size is not restricted, we strongly encourage submission of image files that are not larger than the given number, which may require cropping of original image data obtained from video or confocal systems. The fully populated database covering the entire yeast proteome (approximately 6200 proteins) will be in the range of 15–20 GB, depending on the number of images per experiment.
YPL.db features a web interface to access selected areas of the database. The database is queried through the web interface in the same way as when accessed directly. All of the experimental parameters can be specified, but not all data is accessible, unless the investigator who supplied the image information explicitly grants access to his experimental data. Investigator-defined images of an experiment are displayed in an image gallery, representing interesting areas derived from the raw microscopic images, and stored in compressed JPEG format for faster access times. Access to other relevant yeast genomic information is provided by direct links to the MIPS (12), YPD (13) and SGD (14) databases.
FUTURE DIRECTIONS
YPL.db is constantly increasing in size by adding new experiments. Our goal is to include data for the localization of all of the approximately 6200 yeast proteins and we encourage contributions of data sets from the yeast community to reach this goal. The next version of the database will support image sequences in Quicktime format and 3D volume models obtained from images taken from different planes of a cell. It will allow investigators from other research facilities to upload their experimental data and images via the World Wide Web. Additional new features will allow queries about metabolic pathways and will include data from protein–protein interactions (7).
CITING AND ACCESSING YPL.db
YPL.db should be cited with the present publication as a reference. Images should also be cited with the present publication unless the original authors are specified in the comment field. Access to YPL.db is possible through the World Wide Web at http://ypl.tugraz.at. The Oracle SQL scripts are available free of charge to academic, government and other non-profit institutions.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by grants from the Jubiläumsfonds der Oesterreichischen Nationalbank (7273, 7465 and 8773), the Austrian Science Fund (P14298, F706 and F718) and the Austrian Ministry for Education and Research (projects Biocomputing, AUSTROFAN and Thermogenesis).
REFERENCES
- 1.Goffeau A., Barrell,B.G., Bussey,H., Davis,R.W., Dujon,B., Feldmann,H., Galibert,F., Hoheisel,J.D., Jacq,C., Johnston,M. et al. (1996) Life with 6000 genes. Science, 274, 546, 563–547. [DOI] [PubMed] [Google Scholar]
- 2.Ploger R., Zhang,J., Bassett,D., Reeves,R., Hieter,P., Boguski,M. and Spencer,F. (2000) XREFdb: cross-referencing the genetics and genes of mammals and model organisms. Nucleic Acids Res., 28, 120–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown P.O. and Botstein,D. (1999) Exploring the new world of the genome with DNA microarrays. Nature Genet., 21, 33–37. [DOI] [PubMed] [Google Scholar]
- 4.Zhu H., Klemic,J.F., Chang,S., Bertone,P., Casamayor,A., Klemic,K.G., Smith,D., Gerstein,M., Reed,M.A. and Snyder,M. (2000) Analysis of yeast protein kinases using protein chips. Nature Genet., 26, 283–289. [DOI] [PubMed] [Google Scholar]
- 5.Oliver S. (1996) A network approach to the systematic analysis of yeast gene function. Trends Genet., 12, 241–242. [DOI] [PubMed] [Google Scholar]
- 6.Martzen M.R., McCraith,S.M., Spinelli,S.L., Torres,F.M., Fields,S., Grayhack,E.J. and Phizicky,E.M. (1999) A biochemical genomics approach for identifying genes by the activity of their products. Science, 286, 1153–1155. [DOI] [PubMed] [Google Scholar]
- 7.Uetz P., Giot,L., Cagney,G., Mansfield,T.A., Judson,R.S., Knight,J.R., Lockshon,D., Narayan,V., Srinivasan,M., Pochart,P. et al. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- 8.Ding D.Q., Tomita,Y., Yamamoto,A., Chikashige,Y., Haraguchi,T. and Hiraoka,Y. (2000) Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes Cells, 5, 169–190. [DOI] [PubMed] [Google Scholar]
- 9.Ross-Macdonald P., Coelho,P.S., Roemer,T., Agarwal,S., Kumar,A., Jansen,R., Cheung,K.H., Sheehan,A., Symoniatis,D., Umansky,L. et al. (1999) Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature, 402, 413–418. [DOI] [PubMed] [Google Scholar]
- 10.Ross-Macdonald P., Sheehan,A., Roeder,G.S. and Snyder,M. (1997) A multipurpose transposon system for analyzing protein production, localization, and function in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 94, 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohlwein S.D. (2000) The beauty of the yeast. Live cell microscopy at the limits of optical resolution. Microsc. Res. Technol., 51, 511–529. [DOI] [PubMed] [Google Scholar]
- 12.Mewes H.W., Frishman,D., Gruber,C., Geier,B., Haase,D., Kaps,A., Lemcke,K., Mannhaupt,G., Pfeiffer,F., Schüller,C., Stocker,S. and Weil,B. (2000) MIPS: a database for genomes and protein sequences. Nucleic Acids Res., 28, 37–40. Updated article in this issue: Nucleic Acids Res. (2002), 30, 31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costanzo M.C., Hogan,J.D., Cusick,M.E., Davis,B.P., Fancher,A.M., Hodges,P.E., Kondu,P., Lengieza,C., Lew-Smith,J.E., Lingner,C. et al. (2000) The Yeast Proteome Database (YPD) and Caenorhabditis elegans Proteome Database (WormPD): comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res., 28, 73–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ball C.A., Dolinski,K., Dwight,S.S., Harris,M.A., Issel-Tarver,L., Kasarskis,A., Scafe,C.R., Sherlock,G., Binkley,G., Jin,H. et al. (2000) Integrating functional genomic information into the Saccharomyces Genome Database. Nucleic Acids Res., 28, 77–80. Updated article in this issue: Nucleic Acids Res. (2002), 30, 69–72. [DOI] [PMC free article] [PubMed] [Google Scholar]