Abstract
Although many human genes have been associated with genetic diseases, knowing which mutations result in disease phenotypes often does not explain the etiology of a specific disease. Drosophila melanogaster provides a powerful system in which to use genetic and molecular approaches to investigate human genetic diseases. Homophila is an intergenomic resource linking the human and fly genomes in order to stimulate functional genomic investigations in Drosophila that address questions about genetic disease in humans. Homophila provides a comprehensive linkage between the disease genes compiled in Online Mendelian Inheritance in Man (OMIM) and the complete Drosophila genomic sequence. Homophila is a relational database that allows searching based on human disease descriptions, OMIM number, human or fly gene names, and sequence similarity, and can be accessed at http://homophila.sdsc.edu.
INTRODUCTION
The continuing progress in the sequencing of the human genome will accelerate the identification of many genes involved in human diseases. Although a map location, nucleotide sequence and even the identity of the protein involved in a specific disease may be known, it is often difficult to decipher the etiology of the disease without employing an experimental organism. One approach to deciphering the role of these genes in specific diseases is to investigate the function of cognate genes in model organisms. A number of groups (1–4) have examined various sets of genes for cognates in Drosophila, and it is clear that other groups will employ this powerful genetic model organism in the future.
Online Mendelian Inheritance in Man (OMIM) (5) is a catalog of human genes and genetic disorders. The OMIM Morbid Map describes those disease genes with known cytogenetic positions. Additional disease-related genes can be found in OMIM entries as allelic variants of a given gene. The combination of these two types of OMIM entries gives a relatively complete view of known genes involved in human diseases.
Homophila is a systematic examination of these human disease-related genes and their Drosophila cognates. This cross-genomic analysis bridges the gap between the human disease and the Drosophila genome databases (6). Furthermore, this information is available online in a searchable format supported by a relational database management system (RDBMS).
DATABASE CONTENT
Homophila integrates information from four main sources: human disease gene information from OMIM, information relating OMIM entries to specific sequences from LocusLink (7), Drosophila nucleotide and protein sequence data (8), and annotation of Drosophila genes from FlyBase (9).
Construction of Homophila began with a list of OMIM disease entries (ones that either appear in the Morbid Map or contain an allelic variant notation). Because of the narrative nature of the OMIM database, which often discusses entirely unrelated proteins that may have been excluded as the causes of the disease, it is not possible to simply look up the sequences related to each disease in OMIM. A more involved procedure relying on the NCBI LocusLink database was required. Each OMIM disease entry was looked up in the LocusLink mim2loc table, which relates OMIM entries to NCBI locus records. Each locus record was then used to locate the correct protein sequence records using the LocusLink loc2UG, loc2acc and loc2ref tables, which specify entries in the NCBI UniGene, protein and RefSeq databases, respectively.
Each of the protein sequence entries was compared to the complete Drosophila genome sequence using the BLASTP program (10). BLAST comparisons were performed using BLAST v.2.09 with the standard BLOSUM 62 and expect = 1 × 10–10 settings. The result of this procedure was a list of 5283 protein sequence entries associated with 911 OMIM disease loci and 666 matching Drosophila genes (Table 1).
Table 1. Statistical summary of the information contained in the Homophila database.
| Number of OMIM entries analyzed | 1858 |
| Number of OMIM entries with protein sequence | 1224 |
| Number of human protein sequences BLASTed | 5283 |
| Number of high hita OMIM entries | 911 |
| Number of high hita Drosophila genes | 666 |
| Number of Drosophila genes with P elements | 133 |
aHigh hit indicates a BLAST E-value of 1 × 10–10 or lower.
A relational database has been implemented to allow queries on these results and is available online (http://homophila.sdsc.edu) using the MySQL RDBMS (11). PERL scripts using the DBI package are used to convert queries entered on the Homophila web pages to SQL queries to the actual RDBMS.
A complete list of P-element locations in the Drosophila genomic sequence was kindly provided by FlyBase (9). This information is added to the results of the database searches in order to identify cognate genes for which null mutants in genes already exist (e.g. the P-element falls within the protein coding sequence of a gene) or for which it would be straightforward to generate null deletion mutations (e.g. by imprecise P-element excision).
ACCESS
Homophila is available for both browsing and searching online at http://homophila.sdsc.edu. The database content is also available in a relational version or as flat files upon request.
Many OMIM disease entries have multiple protein sequences linked to the disease through LocusLink. The BLAST search results for each of the probe sequences are merged and used to create a list of best matching sequences (Fig. 1).
Figure 1.
Precompiled list of best human–Drosophila gene matches (the Clear-Hit List). For each OMIM entry with known sequences, the MIM number, brief description of the disease (as given in OMIM), lowest E-value for a match to any Drosophila sequence, the ID number and description of the best Drosophila match, the probe sequence or sequences used in the search and the P-element insertions in or near the best matching Drosophila sequence are shown. The MIM number, best matching Drosophila sequence, and probe sequences, shown underlined, are hyperlinked to further information.
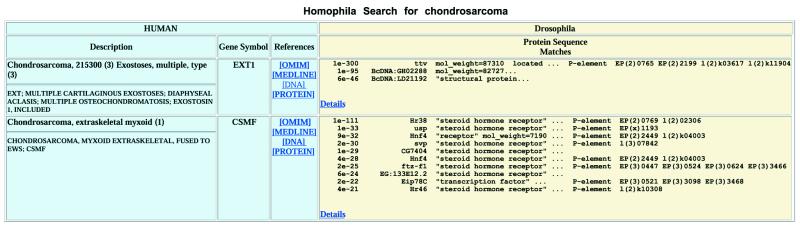
The precompiled list of best matches obviously gives an incomplete view of the correspondence between the gene probes for a specific disease and their Drosophila cognates. More complete information is available by directly searching the database. Searches based on OMIM entry number, human and Drosophila gene names and symbols, human disease description and text keywords are available. All entries matching the search query are displayed in a summary output (Fig. 2).
Figure 2.
Example of a Homophila Search. A search was made with the keyword query ‘chondrosarcoma’. Information related to the human disease is shown in blue on the left. Hyperlinks in the references column provide direct access to the OMIM entry, MEDLINE literature references, and the DNA and protein query sequences. Information related to Drosophila is shown in beige on the right. For each entry, a merged summary of the BLAST searches for all query sequences is shown. The ‘Details’ hyperlink gives complete access to the individual BLAST searches.
The information stored in Homophila is changing rapidly as new disease loci are sequenced. Homophila is updated approximately every 2 months using a semi-automated process to import source data and perform the requisite analyses.
FUTURE DEVELOPMENT PLANS
1. Complete automation of data update and analysis.
2. Extension of analyses to other genomes: Dictyostelium, Caenorhabditis elegans, Saccharomyces cerevisiae and Mus musculus.
3. Inclusion/linkage of more complete information about human diseases and Drosophila genes so that searches based on known human disease phenotypes and Drosophila mutant phenotypes can be used to identify potentially novel functional groupings of human and fly genes.
Acknowledgments
ACKNOWLEDGEMENT
This work is supported in part by the National Institutes of Health through a National Center for Research Resources program grant (P 41 RR08605-06) to the National Biomedical Computation Resource at the San Diego Supercomputer Center.
REFERENCES
- 1.Fortini M.E., Skupski,M.P., Boguski,M.S. and Hariharan,I.K. (2000) A survey of human disease gene counterparts in the Drosophila genome. J. Cell Biol., 150, F23–F30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Littleton J.T. and Ganetzky,B. (2000) Ion channels and synaptic organization: analysis of the Drosophila genome. Neuron, 26, 35–43. [DOI] [PubMed] [Google Scholar]
- 3.Potter C.J., Turenchalk,G.S. and Xu,T. (2000) Drosophila in cancer research: an expanding role. Trends Genet., 16, 33–39. [DOI] [PubMed] [Google Scholar]
- 4.Rubin G.M., Yandell,M.D., Wortman,J.R., Gabor Miklos,G.L., Nelson,C.R., Hariharan,I.K., Fortini,M.E., Li,P.W., Apweiler,R., Fleischmann,W. et al. (2000) Comparative genomics of the eukaryotes. Science, 287, 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyadijiev S.A. and Jabs,E.W. (2000) Online Mendelian Inheritance in Man (OMIM) as a knowledgebase for human developmental disorders. Clin. Genet., 57, 253–266. [DOI] [PubMed] [Google Scholar]
- 6.Reiter L.T., Potocki,L., Chien,S., Gribskov,M. and Bier,E. (2001) A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res., 11, 1114–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pruitt K.D., Katz,K.S., Sicotte,H. and Maglott,D.R. (2000) Introducing RefSeq and LocusLink: curated human genome resources at the NCBI. Trends Genet., 16, 44–47. [DOI] [PubMed] [Google Scholar]
- 8.Adams M.D., Celniker,S.E., Holt,R.A., Evans,C.A., Gocayne,J.D., Amanatides,P.G., Scherer,S.E., Li,P.W., Hoskins,R.A., Galle,R.F. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 9.The FlyBase Consortium (1999) The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res., 27, 85–88. Updated article in this issue: Nucleic Acids Res. (2002), 30, 106–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubois P. (2000) MySQL. New Riders, IN.