Abstract
The ImmunoDeficiency Resource (IDR), freely available at http://www.uta.fi/imt/bioinfo/idr/, is a comprehensive knowledge base on immunodeficiencies. It is designed for different user groups such as researchers, physicians and nurses as well as patients and their families and the general public. Information on immunodeficiencies is stored as fact files, which are disease- and gene-based information resources. We have developed an inherited disease markup language (IDML) data model, which is designed for storing disease- and gene-specific data in extensible markup language (XML) format. The fact files written by the IDML can be used to present data in different contexts and platforms. All the information in the IDR is validated by expert curators.
INTRODUCTION
Primary immunodeficiencies (PIDs) are a group of over 80 genetic disorders of the immune system (1). Information about genetic disorders such as PIDs is increasing rapidly due to the impact of the Human Genome Project (2–5). Patients with these intrinsic defects have increased susceptibility to recurrent and persistent infections. It is clear that validated and curated information about PIDs would be useful in the diagnosis and treatment of these diseases and for patients and their families, as well as for the general public, to learn about the disorders and how to live with them. The ImmunoDeficiency Resource (IDR; http://www.uta.fi/imt/bioinfo/idr/) has been developed to facilitate the need for readily accessible information on immunodeficiencies (6).
Over the last decade in particular, major advances have been made both in the study of normal biological processes as well as the basic molecular mechanisms underlying several PIDs. Information obtained from cellular, molecular and genetic studies has enabled the development of strategies for the modification, prevention and potential cure of human diseases. This includes gene therapy, which was first applied successfully to a PID, namely severe combined immunodeficiency (7). The amount of information about the human genome, gene expression and protein function is increasing rapidly. PIDs are relatively rare diseases, with an estimated rate of incidence of one per 10 000 births worldwide (8). Due to the rarity and the wide distribution of these diseases, information on them has been scattered around the Internet. Therefore, it is important that the information is now collected in one place offering relevant and valid information on PIDs.
The IDR is used here as a model to illustrate how molecular studies have not only redefined the standards for diagnosis of immunodeficiencies, but have also influenced management approaches, increased our understanding of fundamental disease-causing mechanisms and identified potential targets for therapeutic intervention.
The IDR is a freely accessible information resource which is continuously updated. New features, such as XML linking language (Xlink), will be added in order to create and describe more sophisticated links between resources.
EXPERT VALIDATION
The Internet contains a large amount of pages, but only a few contain information about data validation. Most of the search engines give thousands of links making it almost impossible to trawl through all this information. The most difficult task is usually not to find data but to differentiate useful and reliable data from other less important search results. In the IDR, the experts check all data, especially links to external information sources, and approve only those sites with solid scientific and medical information. There is at least one expert for each immunodeficiency from around the world. Some of them act also as regional experts, who check all data coming from their country. This is important because health care systems and research methods vary between countries. Nurse and patient societies are also involved in the data validation process for their own interest groups.
FACT FILES
We have developed inherited disease markup language (IDML) which is based on the extensible markup language (XML) standard, developed by the World Wide Web Consortium (W3C). The IDML provides a standard method for exchanging genetic and clinical data along with general disease description, diagnostic information and links to other related resources. Separation of data from the presentation enables the seamless integration of data from diverse sources. The IDML format is a published, documented open format, offered especially for the purpose of data interchange between platforms and databases in the Internet. The data from the IDML format to different systems, e.g. to hypertext markup language (HTML), is tranformed by using extensible stylesheet language transformations (XSLT) stylesheets. The IDML specification and document type definition (DTD) follow XML standards of W3C.
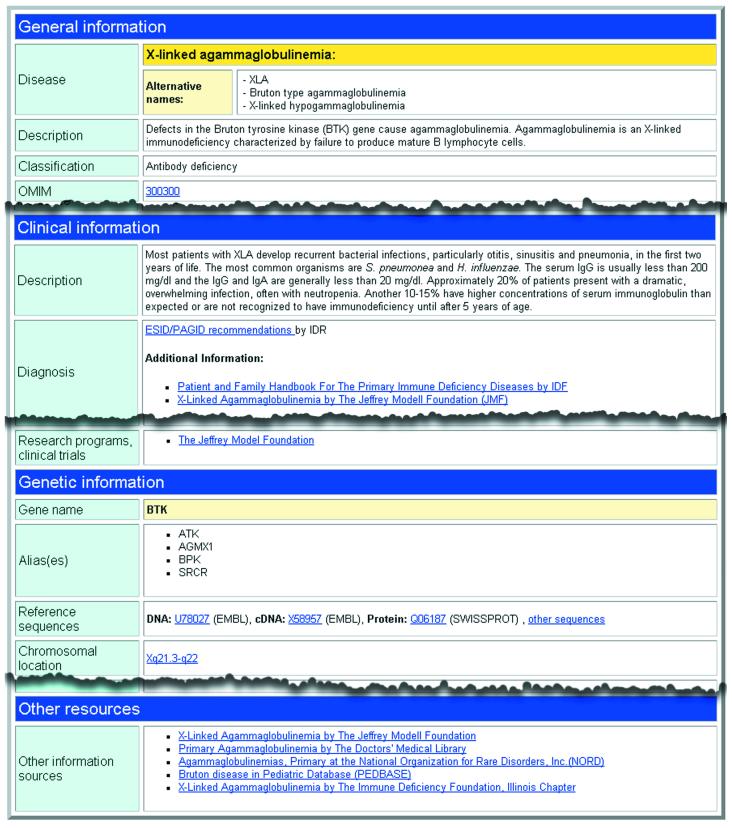
The basic information on PIDs is stored as fact files by using the IDML format (Fig. 1). The fact files are XML-based, validated data sources on PID-related information. Each fact file provides basic information on diseases and affected genes. The fact files also contain HTML hyperlinks to other Internet resources that are accepted to be reliable by the experts. The fact files act as a quick portal to further information on diseases.
Figure 1.
A transformed XML fact file for X-linked agammaglobulinemia presented in HTML-format in Internet browser.
CONTENTS
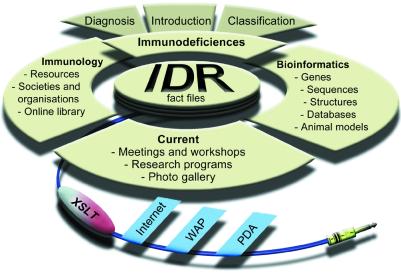
The main data categories of the IDR are shown in Figure 2. The IDR contains extensive cross-referencing and links to other services. The IDR integrates numerous web-based services e.g. sequence databases (EMBL, GenBank, SWISS-PROT), genome information (Ensembl, GDB, UniGene, GeneCard, GenAtlas, LocusLink, euGene), protein structural database (PDB), diseases (OMIM), references (PubMed), patient information (ESID registry), symptoms and diagnosis (ESID/PAGID recommendations), mutation data (IDbases), animal models (MGD, FlyBase, SacchDB) and information produced by the IDR team.
Figure 2.
The contents, links and distribution of data in the IDR. The major information categories are shown as well as transformation of the XML pages by XSLT for different platforms such as PCs, wireless application protocol (WAP)-compliant devices (e.g. mobile phones) and personal digital assistants (PDAs).
The immunodeficiencies category includes, for example, an introduction to, and classification of, immunodeficiencies. Information about the affected genes and loci are linked to corresponding servers. The recent ESID and PAGID recommendations for diagnostics criteria for immunodeficiences (9) are also distributed.
The immunology section includes immunology related data sources such as lectures on immunology and immunodeficiencies.
The IDbases section contains links to some 20 mutation registries. Most of them are maintained by IMT bioinformatics. The pages for animal models list links to knock-outs of immunodeficiency related genes in mice (MGD), Drosophila melanogaster (FlyBase) and Saccharomyces cerevisiae (SacchDB).
Interest groups for immunologist, nurses and patients are listed. Several societies are related to immunodeficiency research, care and patients.
The immunology laboratories contain a list of home pages of laboratories which are active in the many fields of immunodeficiency research including diagnosis, treatment and basic research on such areas as genetic analyses, protein structure determination and signal transduction. It is also possible to read about immunodeficiency related research programs.
Links to immunodeficiency related DNA and amino acid sequences are available as well as to three-dimensional structures of proteins. Furthermore, a picture gallery and a list of meetings and workshops are available.
NAVIGATION
The IDR is easy to navigate. The IDR pages are colour coded for different interest groups: researchers, physicians, nurses, patients and families. By selecting the group of interest, the user can get specific pages such as introduction written for this particular group. This makes it easier for the user to find interesting and useful information for her/his own area. The IDR also provides advanced text search facility, that can utilize boolean logic searches with multiple keywords.
CITING IDR
Authors who make use of the information provided by the IDR should cite this article as a general reference for the access to the content of the IDR, and quote the IDR home page address, http://www.uta.fi/imt/bioinfo/.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Jukka Lehtiniemi for implementation of the IDR. This project is funded by The European Union Commission from EU program on rare diseases, the Finnish Academy, the Medical Research Fund of Tampere University Hospital,and the Sigrid Juselius Foundation.
REFERENCES
- 1.Vihinen M., Arredondo-Vega,F.X., Casanova,J.L., Etzioni,A., Giliani,S., Hammarström,L., Hershfield,M.S., Heyworth,P.G., Hsu,A.P., Lähdesmäki,A. et al. (2001) Primary immunodeficiency mutation databases. Adv. Genet., 43, 103–188. [DOI] [PubMed] [Google Scholar]
- 2.Chakravarti A. (2001) To a future of genetic medicine. Nature, 409, 822–823. [DOI] [PubMed] [Google Scholar]
- 3.Fahrer A.M., Bazan,J.F., Papathanasiou,P., Nelms,K.A. and Goodnow,C.C. (2001) A genomic view of immunology. Nature, 409, 836–838. [DOI] [PubMed] [Google Scholar]
- 4.Jimenez-Sanchez G., Childs,B. and Valle,D. (2001) Human disease genes. Nature, 409, 853–855. [DOI] [PubMed] [Google Scholar]
- 5.Peltonen L. and McKusick,V.A. (2001) Genomics and medicine. Dissecting human disease in the postgenomic era. Science, 291, 1224–1229. [DOI] [PubMed] [Google Scholar]
- 6.Väliaho J., Riikonen,P. and Vihinen,M. (2000) Novel immunodeficiency data servers. Immunol. Rev., 178, 177–185. [DOI] [PubMed] [Google Scholar]
- 7.Cavazzana-Calvo M., Hacein-Bey,S., de Saint Basile,G., Gross,F., Yvon,E., Nusbaum,P., Selz,F., Hue,C., Certain,S., Casanova,J.L. et al. (2000) Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science, 288, 669–672. [DOI] [PubMed] [Google Scholar]
- 8.Smith C.I.E., Ochs,H.D. and Puck,J.M. (1999) Genetically determined immunodeficiency diseases: a perspective. In Ochs,H.D., Smith,C.I.E. and Puck,M. (eds), Primary Immunodeficiency Diseases. A Molecular and Genetic Approach. Oxford University Press, New York, Oxford, pp. 3–11.
- 9.Conley M.E., Notarangelo,L.D. and Etzioni,A. (1999) Diagnostic criteria for primary immunodeficiencies. Representing PAGID (Pan-American Group for Immunodeficiency) and ESID (European Society for Immunodeficiencies). Clin. Immunol., 93, 190–197. [DOI] [PubMed] [Google Scholar]