Abstract
The Gene Expression Omnibus (GEO) project was initiated in response to the growing demand for a public repository for high-throughput gene expression data. GEO provides a flexible and open design that facilitates submission, storage and retrieval of heterogeneous data sets from high-throughput gene expression and genomic hybridization experiments. GEO is not intended to replace in house gene expression databases that benefit from coherent data sets, and which are constructed to facilitate a particular analytic method, but rather complement these by acting as a tertiary, central data distribution hub. The three central data entities of GEO are platforms, samples and series, and were designed with gene expression and genomic hybridization experiments in mind. A platform is, essentially, a list of probes that define what set of molecules may be detected. A sample describes the set of molecules that are being probed and references a single platform used to generate its molecular abundance data. A series organizes samples into the meaningful data sets which make up an experiment. The GEO repository is publicly accessible through the World Wide Web at http://www.ncbi.nlm.nih.gov/geo.
BACKGROUND
Molecular biological experiments utilizing high-throughput hybridization array- and sequencing-based techniques have become extremely popular in recent years (1–3). These techniques have been used to measure the molecular abundance of mRNA and genomic DNA either in absolute or relative terms. Mainly contributing to this popularity is the highly parallel nature of these techniques and the concomitant conservation of time and resources brought about by the large number of simultaneous (or near-simultaneous) molecular sampling events performed under very similar conditions.
For a number of years there has been a growing desire for these high-throughput data sets to be made publicly available once research findings have been published in the scientific literature—similar to journal and public funding requirements for the public release of biological sequence data. There have also been calls for the establishment of a public repository for (at least the gene expression microarray subset of) these data sets (4–6), and journals and public funding agencies have begun to make public availability of high-throughput data a condition of publication (7) or funding (e.g. NINDS request for proposals BAA-RFP-NIH-NINDS-01–03, p. 76 at http://www.ninds.nih.gov/funding/2rfp_01_03.pdf), respectively. Recognizing the desire that this data should be made widely available, several laboratories and institutions have constructed primary and secondary Internet resources to distribute these high-throughput data sets (Table 1).
Table 1. A variety of public, high-throughput gene expression and genomic hybridization data resources.
| Resource name | Institution(s) | URL | |
|---|---|---|---|
| Breast Cancer Cell Line Resource | 1º | National Human Genome Research Institute, NIH | http://www.nhgri.nih.gov/DIR/CGB/CR2000 |
| CGH Database | 1º | Institute of Pathology, University Hospital Charité | http://amba.charite.de/~ksch/cghdatabase |
| Chip DB | 1º | Whitehead Institute for Biomedical Research, MIT | http://young39.wi.mit.edu/chipdb_public |
| Drug & Alcohol Abuse Microarray Data Consortium | 1º | Wake Forest University, Emory University, and Oregon Health and Science University | http://www.wfubmc.edu/microarray |
| ExpressDB | 1º | Harvard–Lipper Center for Computational Genetics | http://arep.med.harvard.edu/ExpressDB |
| Global Gene Expression Group | 1º | Science Park-Research Division, University of Texas M.D. Anderson Cancer Center | http://sciencepark.mdanderson.org/ggeg |
| MAExplorer | 1º | National Cancer Institute, NIH | http://www-lecb.ncifcrf.gov/MAExplorer |
| Microarray center | 1º | Children’s National Medical Center | http://microarray.cnmcresearch.org/ |
| Microarray project | 1º | National Human Genome Research Institute, NIH | http://www.nhgri.nih.gov/DIR/Microarray |
| Rochester Muscle Database | 1º | School of Medicine and Dentistry, University of Rochester Medical Center | http://www.urmc.rochester.edu/smd/crc/swindex.html |
| SADE | 1º | Departement de Biologie Cellulaire et Moleculaire, CEA | http://www-dsv.cea.fr/thema/get/sade.html |
| SAGENET | 1º | Johns Hopkins University School of Medicine | http://www.sagenet.org |
| Yeast Microarray Global Viewer | 1º | Laboratoire de genetique moleculaire, Ecole Normale Superieure | http://transcriptome.ens.fr/ymgv |
| RNA Abundance Database | 2º | Computational Biology and Informatics Laboratory, University of Pennsylvania | http://www.cbil.upenn.edu/RAD2 |
| SAGEmap | 2º | National Cancer Institute and National Center for Biotechnology Information, NIH | http://www.ncbi.nlm.nih.gov/sage |
| Stanford Microarray Database | 2º | Dept. of Genetics, Stanford University School of Medicine | http://www.dnachip.org |
| Gene Expression Omnibus | 3º | National Center for Biotechnology Information, NIH | http://www.ncbi.nlm.nih.gov/geo |
This list is provided to show the wide variety of public, high-throughput gene expression and genomic hybridization data currently available in a wide variety of formats. It is in no way meant to be comprehensive. Primary resources (1º) publish in-house data, and secondary resources (2º) publish both in-house and collaborator data, while tertiary resources (3º) accept data to be published from third, unrelated parties. To our knowledge, GEO is the only tertiary resource of this kind in operation.
Over the last several years, there has been an international effort to catalog the minimal set of information which is necessary in order for microarray experiments to be properly interpreted and to be comparable with one another (6). The codification and publication of this set of guidelines will be invaluable as a guide for high-throughput gene expression and genomic hybridization data producers and data repositories. We feel, however, that over-zealous application of these guidelines in setting standards and requirements must be avoided because it will stifle a rapidly developing and technically challenging field.
Therefore, our primary goal in creating the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) was to attempt to cover the broadest spectrum of high-throughput experimental methods possible and remain flexible and responsive to future trends, rather than setting rigid requirements and standards for entry. In taking this approach, however, we recognize that there are obvious, inherent limitations to functionality and analysis that can be provided on such heterogeneous data sets. Hence, GEO is not intended to replace or match primary and secondary resources that operate on homogenous data sets, but instead to serve as a complementary tertiary resource for the storage and retrieval of public high throughput gene expression and genomic hybridization data.
REPOSITORY DESIGN
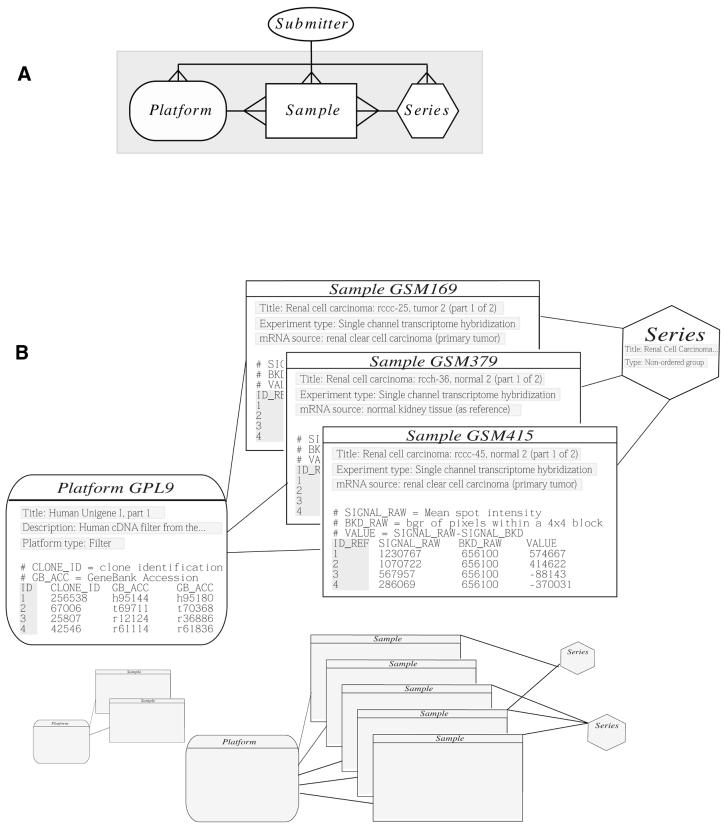
GEO segregates data into three principle components, platform, sample and series (Table 2), each of which is accessioned (i.e. given a unique and constant identifier) in a relational database (Fig. 1). To achieve an open and flexible design that allows storage and retrieval of very diverse data types, the data are not fully granulated within the database. Instead, a tab-delimited ASCII table is stored for each platform and each sample. The table consists of multiple columns with accompanying column header names. The data within this table are currently partially extracted for indexing, but may be further extracted for more extensive search and retrieval. In addition, any number of supplementary columns may be provided by the submitter for the inclusion of additional, submitter-defined information.
Table 2. Entity types and subtypes in the GEO database.
| Accession prefix | Entity types | Subtypes |
|---|---|---|
| GPL | Platform | Microarray |
| HDA | ||
| Filter | ||
| SAGEa | ||
| GSM | Sample | Dual channel, transcriptome hybridization |
| Single channel, transcriptome hybridization | ||
| Dual channel, comparative genomic hybridization | ||
| SAGE | ||
| GSE | Series | Time course |
| Dose–response | ||
| Ordered, not otherwise specified | ||
| Non-ordered |
aSAGE platforms are automatically generated, as needed, based on the organism and anchoring enzyme used to generate the SAGE library.
Figure 1.
GEO schema and example. (A) The entity-relationship diagram for the GEO database. (B) An actual example of three samples referencing one platform and contained in a single series.
An instance of a platform is, essentially, a list of probes that define what set of molecules may be detected in any experiment utilizing that platform. For example, the platform data table may contain GEO-defined columns identifying the position and biological reagent contents of each probe (spot) such as a GenBank accession number, open reading frame (ORF) name and clone identifier, as well as submitter-defined columns. Platform accession numbers have a ‘GPL’ prefix.
An instance of a sample describes the derivation of the set of molecules that are being probed and utilize platforms to generate molecular abundance data. Each sample has one, and only one, parent platform which must be previously defined. For example, a sample data table may contain columns indicating the final, relevant abundance value of the corresponding spot defined in its platform, as well as any other GEO-defined (e.g. raw signal, background signal) and submitter-defined columns. Sample accession numbers have a ‘GSM’ prefix.
An instance of a series organizes samples into the meaningful data sets which make up an experiment, and are bound together by a common attribute. Series accession numbers have a ‘GSE’ prefix.
SUBMISSIONS
Two modes of communication are available for new and update submissions, interactive or direct deposit. The interactive web form interface route is straightforward and most suited for occasional submissions of a relatively small number of samples. Bulk submissions of large data sets may be rapidly incorporated into GEO via direct deposit of files in the simple omnibus format (SOFT). SOFT is a line based, ASCII text format which allows for the representation of multiple GEO platforms, samples and series in one file. In SOFT, metadata appear as label-value pairs and are associated with the tab-delimited text tables of platforms and samples. SOFT has been designed for easy manipulation by readily available line-scanning software and may be quite readily produced from, and imported into, spreadsheet, database and analysis software. More information about SOFT and the submission process is available from the GEO web site.
Submissions may be held privately for a maximum of 6 months; this policy allows data release concordant with manuscript publication. Such submissions are given a final accession number, which may be quoted in the publication. At this point, the submissions are not curated, but are human scanned to assure that the minimal basic requirements are met. It is entirely up to the submitter to make the data useful to others by using the standard column headers in the data table, and providing adequate supplementary information.
SEARCH AND RETRIEVAL
At the time of writing, it is possible to retrieve complete platforms, samples and series submissions by accession number only. Extensive indexing and linking on the data in GEO has been performed and is queriable through a new Entrez database, named Entrez ProbeSet. The web interface to this database utilizes the same indexing and linking engine familiar through other popular NCBI resources such as PubMed and GenBank. As with any other Entrez database, a simple Boolean phrase may be entered and restricted to any number of supported attribute fields. Matches are linked to the full GEO entry as well as to other Entrez databases—currently Nucleotide, Taxonomy and PubMed—as well as related Entrez ProbeSet entries. Entrez ProbeSet is accessible through the Entrez web site (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=geo) as one of the drop-down menus used to select the Entrez database to be queried.
FUTURE DEVELOPMENTS
The GEO resource is under constant development aimed at improving its indexing, linking, searching and display capabilities in order to allow more vigorous data mining. As an extension of the GEO repository, we are currently developing a fully granulated abundance measurement database, which will allow queries and retrievals of individual abundance measurements. However, under the limitations brought about by the complexity and rapid development of current high-throughput gene expression and genomic hybridization experiments, abundance measurements may be comparable only within small groups of similarly derived data sets. We plan to exploit these comparable data subsets in order to allow as much freedom as possible to query abundance measurements, as well as provide useful synoptic views of these data.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to gratefully acknowledge the work of Vladimir Soussov, as well as the entire NCBI Entrez team, especially Grisha Starchenko, Vladimir Sirotinin, Alexey Iskhakov, and Anton Golikov. We would like to thank Jim Ostell for guidance and review of this paper, Lou Staudt for discussions during our initial planning for GEO, and the extreme patience shown by Brian Oliver, Wolfgang Huber and Gavin Sherlock when making data submissions.
REFERENCES
- 1.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 2.Lipshutz R.J., Morris,D., Chee,M., Hubbell,E., Kozal,M.J., Shah,N., Shen,N., Yang,R. and Fodor,S.P. (1995) Using oligonucleotide probe arrays to access genetic diversity. Biotechniques, 19, 442–447. [PubMed] [Google Scholar]
- 3.Velculescu V.E., Zhang,L., Vogelstein,B. and Kinzler,K.W. (1995) Serial analysis of gene expression. Science, 270, 484–487. [DOI] [PubMed] [Google Scholar]
- 4.Bassett D.E., Eisen,M.B. and Boguski,M.S. (1999) Gene expression informatics—it’s all in your mind. Nature Genet., 21 (suppl.), 51–55. [DOI] [PubMed] [Google Scholar]
- 5.Brazma A., Robinson,A., Cameron,G. and Ashburner,M. (2000) One-stop shop for microarray data. Nature, 403, 699–700. [DOI] [PubMed] [Google Scholar]
- 6.Kellam P. (2001) Microarray gene expression database: progress towards an international repository of gene expression data. Genome Biol., 2, reports4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodman L. (2001) Unlimited access—limitless success. Genome Res., 11, 637–638. [DOI] [PubMed] [Google Scholar]