Abstract
Ribosomal 5S RNA (5S rRNA) is an integral component of the large ribosomal subunit in all known organisms with the exception only of mitochondrial ribosomes of fungi and animals. It is thought to enhance protein synthesis by stabilization of a ribosome structure. This paper presents the updated database of 5S rRNA and their genes (5S rDNA). Its short characteristics are presented in the Introduction. The database contains 2280 primary structures of 5S rRNA and 5S rRNA genes. These include 536 eubacterial, 61 archaebacterial, 1611 eukaryotic and 72 organelle sequences. The database is available on line through the World Wide Web at http://biobases.ibch.poznan.pl/5SData/.
INTRODUCTION
In all organisms, messenger-directed protein synthesis is catalyzed by ribosomes. All eukaryotic and prokaryotic ribosomes consist of ribosomal ribonucleic acids (rRNAs) and ribosomal proteins. At the beginning, rRNAs were believed to play only a structural role as a scaffold for ribosomal proteins, but recently they were shown to also be the key catalytic components of peptidyl transferase (1,2).
5S rRNA has a length of ∼120 nt and a molecular weight ∼40 kDa. It is found in virtually all ribosomes with the exception of mitochondria of some fungi, higher animals and most protists (3). In prokaryotic ribosomes it was shown to bind several ribosomal proteins: L5, L18 and L25 but in eukaryotes, 5S rRNA binds only ribosomal protein L5 (4).
For many years studies on 5S rRNA have been focused on a sequence analysis and its application to the problems of molecular evolution. The size and ubiquity of 5S rRNA that enabled RNA sequencing using direct methods, made it an ideal candidate for a molecular phylogenetic marker. This led to fast accumulation of the sequence data from a variety of organisms belonging to diverse taxonomic groups. Despite many years of research, very little has been known about details of its structure and function.
Recently, the crystal structure of the large subunit of the halophile archaea Haloarcula marismortui at 2.4 Å resolution has been solved (1). It gave us an insight into the tertiary structure of rRNAs, including 5S rRNA, and their interactions with other components of the ribosome.
SECONDARY AND TERTIARY STRUCTURE OF 5S rRNA
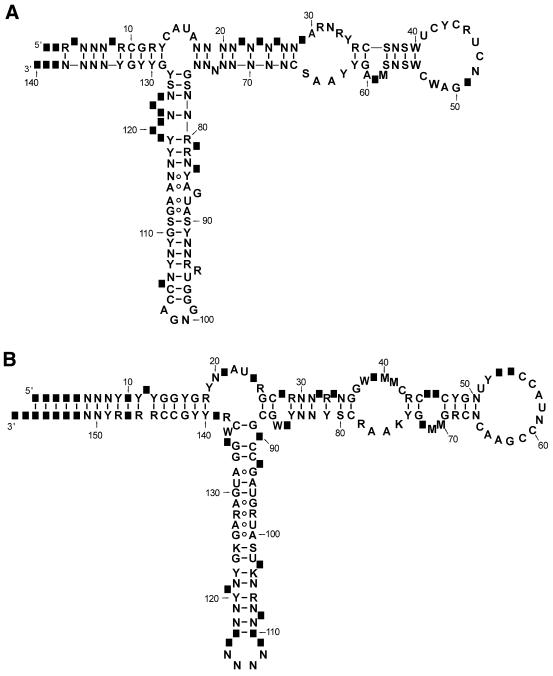
A secondary structure of 5S rRNAs is very similar in prokaryotes and eukaryotes (Fig. 1). A model of its structure model resulting from comparative sequence analysis and various structural studies has been widely accepted. It consists of five helices (I–V), two hairpin loops (C and D), two internal loops (B and E) and a hinge region (loop A) forming the three-helix junction. All 5S rRNA sequences can be folded into that structure. Because of single nucleotide insertions and deletions, in some organisms 5S rRNA secondary structure deviates from the canonical model. This can manifest itself as the presence of additional 1–3-nt bulges in double-stranded regions or the variation in the lengths of loops or stems. The sequence and structure analyses show that eukaryotic and archaeal type structures are very closely related. This is particularly the case for the structure of the internal loop E (domain γ), which is distinct from that found in eubacterial 5S rRNAs (5,6).
Figure 1.
The general secondary structures of 5S ribosomal RNA of Eukaryota (A) and Eubacteria (B). The numbering of the nucleotides corresponds to that in the multiple sequence alignments available from the database web pages. The positions of rare insertions are shown as black squares. The nucleotide symbols used in diagrams are according to IUBMB nomenclature. N: A, C, G or U; Y: C or U; R: A or G; W: A or U; S: G or C; M: A or C; K: G or U.
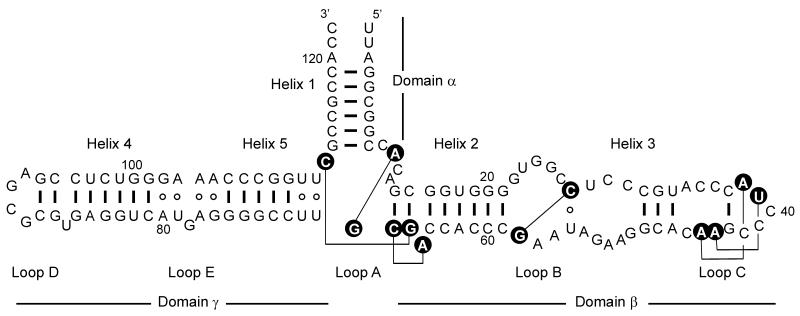
The crystal structure of the large ribosomal subunit of H.marismortui (1), allowed comparison of the structure present in the ribosome (Fig. 2) with the model secondary structure. The 5S rRNA of H.marismortui is 122 nt long and its structure observed in the crystal of the large ribosomal subunit consists of three stems projecting out from loop A. The domain β containing helices II and III of the 5S rRNA molecule is almost coaxial with the domain γ consisting of helices IV and V. Helix I (domain α) is folded in the central part of the molecule. The comparison with the secondary structure model based on comparative sequence analysis shows that some of the earlier predicted base pairs do not exist in a structure of ribosome-bound 5S rRNA. On the other hand, a few base pairs found in the crystal structure of 5S rRNA have not been predicted by a comparative sequence analysis. The ribosome crystal structure also confirmed the array of non-canonical base pairs observed in earlier crystallographic and NMR studies within the loop E of 5S rRNA (7).
Figure 2.
The secondary structure and tertiary interactions of H.marismortui 5S rRNA as observed in the crystal structure of large ribosomal subunit. Watson–Crick and wobble base pairs are shown as thick lines, non-canonical base pairs are shown as open circles. Nucleotides involved in the tertiary interactions (thin lines) are shown as black circles.
Solving of the crystal structure of the large ribosomal subunit of H.marismortui gave us for the first time a three-dimensional (3D) picture of 5S rRNA at high resolution. Earlier efforts of crystallization of full-length 5S rRNAs alone or in complexes with proteins did not give satisfactory results (8). However, there were several successful attempts at solving the 3D structure of the single domains of the 5S rRNA (7–10) and the whole ribosome of Thermus thermophilus (11).
The 3D structure of the large ribosomal subunit also gave us a detailed insight into interactions of 5S rRNA with ribosomal proteins and 23S rRNA. The structure shows a number of interactions between 5S rRNA and ribosomal proteins L5, L10e, L18, L21e and L30. There is only one direct interaction between 5S rRNA domain γ and helix 38 of 23S ribosomal RNA.
CONTENTS OF THE DATABASE
The database is a collection of nucleotide sequences of 5S rRNAs and 5S rRNA genes. On the web pages we also included additional information about secondary structure and a review of current state of research on functions and interactions of 5S rRNA with proteins. Currently the database contains over 2000 sequences from Archaea, Eubacteria, Eukaryota and organelles. The numbers of entries in particular groups is shown in Table 1. The entries in the database also include partial sequences that have been determined in the studies on 5S rRNA intergenic regions and deposited in EMBL/GenBank under separate accession numbers. This is the case for most of the eukaryotic genes where the primers used to amplify intergenic regions cover the interior of the 5S rRNA-coding sequences. Our previous analyses of plant genes have shown that in many cases the putative 5S rRNA gene sequences determined by these means cannot be folded into general secondary structure model and may therefore represent pseudogenes (12). Often, their sequences also do not correspond to 5S rRNA sequences determined on the RNA level using enzymatic or chemical methods. These observations prompted us to substract the eukaryotic genes and they are not included in the 5S rRNA alignments.
Table 1. Distribution of the 5S rRNA database entries within major taxonomic groups.
| Taxonomic group | Number of entries |
|---|---|
| Eubacteria | 536 |
| Archaea | 61 |
| Organelles | 72 |
| Mitochondria | 9 |
| Chloroplasts | 63 |
| Eukaryota | 1611 |
| Protista | 57 |
| Fungi | 243 |
| Animals | 504 |
| Plants | 807 |
| Total | 2280 |
The individual sequence files can be retrieved using the phylogenetic browser, where the organisms are grouped according to their taxonomic position. All entries use the format of the EMBL Nucleotide Sequence Data Bank and in addition to the sequence data contain literature information and the original accession number. Some of the sequences presented in the database that were not submitted to the EMBL or GenBank databases have been taken from the original papers.
In a few cases 5S rRNAs from Archaea and some Eukaryota have been found to contain modified nucleotides. The database includes information on the nature and localization of these modifications.
The 5S rRNAs can also be downloaded as multiple sequence alignments for Archaea, Eubacteria and Eukaryota. The alignment files are available in FASTA and Tab-delimited formats. The alignments were created using CLUSTALW (13) and manually corrected to reflect general secondary structure models for respective groups. There are also tables of modified bases occurring in 5S rRNA.
The database can be accessed online at http://biobases.ibch.poznan.pl/5SData/.
Acknowledgments
ACKNOWLEDGEMENTS
This work has been supported by the Polish State Committee for Scientific Research and Deutsche Forschungsgemeinschaft (Gottfried Wilhelm Leibnitz Prize to V.A.E., the Sonderforschungsbereich 344-C8, the Deutsche Agentur fur Raumfahrtangelegenheiten GmbH, the Fonds der Chemischen Industrie e.V.
REFERENCES
- 1.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 2.Nissen P., Hansen,J., Ban,N., Moore,P.B. and Steitz,T.A. (2000) The structural basis of ribosome activity in peptide bond synthesis. Science, 289, 920–930. [DOI] [PubMed] [Google Scholar]
- 3.Gray M.W., Burger,G. and Lang,B.F. (1999) Mitochondrial evolution. Science, 283, 1476–1481. [DOI] [PubMed] [Google Scholar]
- 4.Moore P.B. (2001) The ribosome at atomic resolution. Biochemistry, 20, 3243–3250. [DOI] [PubMed] [Google Scholar]
- 5.Leontis N.B. and Westhof,E. (1998) The 5S rRNA loop E: chemical probing and phylogenetic data versus crystal structure. RNA, 4, 1134–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leontis N.B. and Westhof,E. (1999) Recurrent RNA motifs. In Barciszewski,J. and Clark,B.F.C. (eds), RNA Biochemistry and Biotechnology. Kluver Academic Publishers, 45–61.
- 7.Wimberly B., Varani,G. and Tinoco,I.,Jr (1993) The conformation of loop E of eukaryotic 5S ribosomal RNA. Biochemistry, 32, 1078–1087. [DOI] [PubMed] [Google Scholar]
- 8.Lorenz S., Perbandt,M., Lippmann,C., Moore,K., DeLucas,L.J., Betzel,C. and Erdmann,V.A. (2000) Crystallization of engineered Thermus flavus 5S rRNA under earth and microgravity conditions. Acta Crystallogr. D. Biol. Crystallogr., 56, 498–500. [DOI] [PubMed] [Google Scholar]
- 9.Xiong Y. and Sundaralingham,M. (2001) Two crystal forms of helix II of Xenopus laevis 5S rRNA with cytosine bulge. RNA, 6, 1316–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu M. and Steitz,T.A. (2000) Structure of Escherichia coli ribosomal protein L25 complexed with a 5S rRNA fragment at 1.8 Å resolution. Proc. Natl Acad. Sci. USA, 97, 2023–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yusupov M.M., Yusupova,G.Z., Baucom,A., Lieberman,K., Earnest,T.N., Cate,J.H.D. and Noller,H.F. (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science, 292, 883–896. [DOI] [PubMed] [Google Scholar]
- 12.Barciszewska M., Szymaáski,M., Specht,T., Erdmann,V.A. and Barciszewski,J. (1994) Compilation of plant 5S ribosomal RNA sequences on RNA and DNA levels. Plant Sci., 100, 117–128. [Google Scholar]
- 13.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]