The syntheses and crystal structures of two thiazinone compounds, in an enantiopure form, are reported. The thiazine rings in the two structures differ in their puckering, as a half-chair in the first and a boat pucker in the second.
Keywords: crystal structure, thiazinone, sulfone, enantiopure
Abstract
The syntheses and crystal structures of two thiazinone compounds, namely, rac-2,3-diphenyl-2,3,5,6-tetrahydro-4H-1,3-thiazine-1,1,4-trione, C16H15NO3S, in its racemic form, and N-[(2S,5R)-1,1,4-trioxo-2,3-diphenyl-1,3-thiazinan-5-yl]acetamide, C18H18N2O4S, in an enantiopure form, are reported. The thiazine rings in the two structures differ in their puckering, as a half-chair in the first and a boat pucker in the second. The extended structures for both compounds have only C—H⋯O-type interactions between symmetry-related molecules, and exhibit no π–π stacking interactions in spite of each having two phenyl rings.
1. Chemical context
The 2,3-dihydro-4H-1,3-thiazin-4-ones are a group of six-membered heterocycles with a wide range of biological activity (Ryabukhin et al., 1996 ▸; Silverberg & Moyer, 2019 ▸). Surrey’s research (Surrey et al., 1958 ▸; Surrey, 1963a
▸,b
▸) resulted in the discovery of two drugs, the antianxiety and muscle relaxant chlormezanone, C11H12ClNO3S, [2-(4-chlorophenyl)-3-methyl-2,3,5,6-tetrahydro-4H-1,3-thiazin-4-one 1,1-dioxide] (O’Neil, 2006 ▸; Tanaka & Horayama, 2005 ▸) and the muscle relaxant dichlormezanone, C11H11Cl2NO3S, [2-(3,4-dichlorophenyl)-3-methyl-2,3,5,6-tetrahydro-4H-1,3-thiazin-4-one 1,1-dioxide] (Elks & Ganellin, 1990 ▸). These sulfones showed greater activity than the sulfides from which they were synthesized (Surrey et al., 1958 ▸).
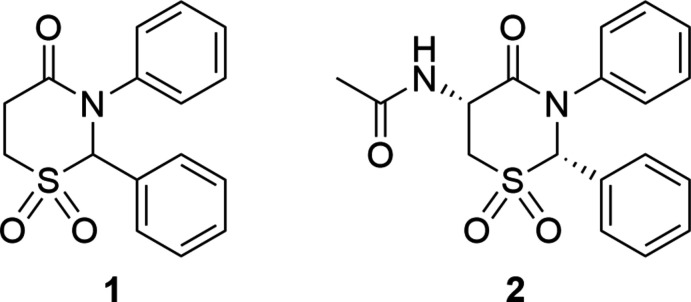
We have previously reported the preparation of the sulfones rac-2,3-dihydro-2,3-diphenyl-4H-1,3-thiazin-4-one 1,1-dioxide and N-[(2S,5R)-1,1-dioxido-4-oxo-2,3-diphenyl-1,3-thiazinan-5-yl]acetamide (Silverberg, 2020 ▸). We have also reported X-ray crystal structures of the corresponding sulfides and sulfoxides (Yennawar & Silverberg, 2014 ▸, 2015 ▸; Yennawar et al., 2015 ▸, 2016 ▸, 2017 ▸). The crystal structure of chlormezanone has been reported (Tanaka & Horayama, 2005 ▸). Herein we report the crystal structures of rac-2,3-diphenyl-2,3,5,6-tetrahydro-4H-1,3-thiazine-1,1,4-trione, 1, and N-[(2S,5R)-1,1,4-trioxo-2,3-diphenyl-1,3-thiazinan-5-yl]acetamide, 2.
2. Structural commentary
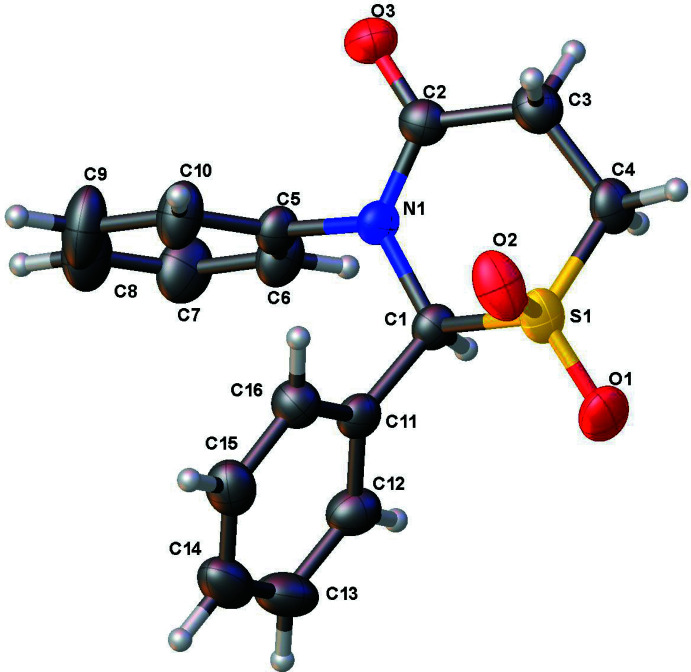
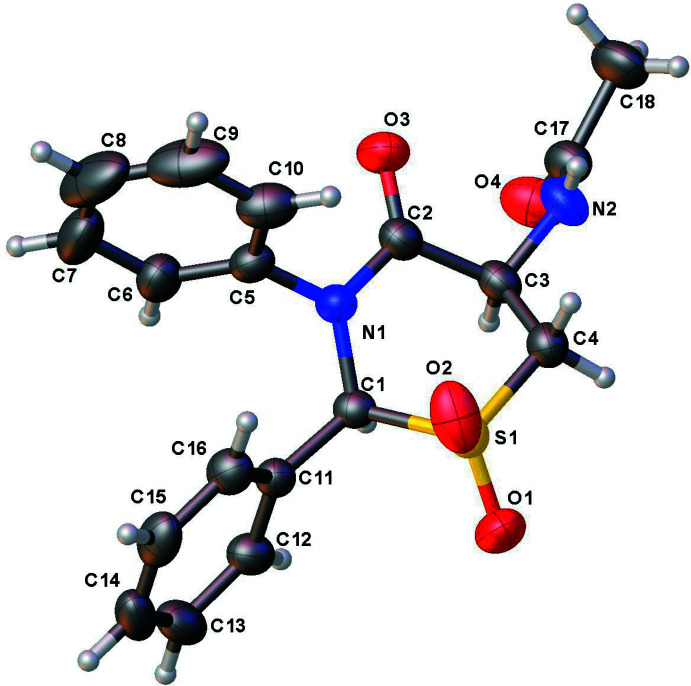
Compound 1 has one chiral center at C1 with an S configuration in the arbitrarily chosen asymmetric unit but crystal symmetry generates a racemic mixture (space group P21/c). Compound 2 has two chiral centers, at C1 and C3 (S and R respectively), synthesized as such, and crystallizes in space group P212121. In 1, the dihedral angles between the thiazine ring (all atoms) and the pendant C5–C10 and C11–C16 phenyl groups are 84.02 (14) and 79.56 (12)°, respectively; the dihedral angle between the pendant rings is 61.26 (15)°. The equivalent angles in 2 are 81.25 (15), 82.58 (13) and 50.40 (15)°, respectively.
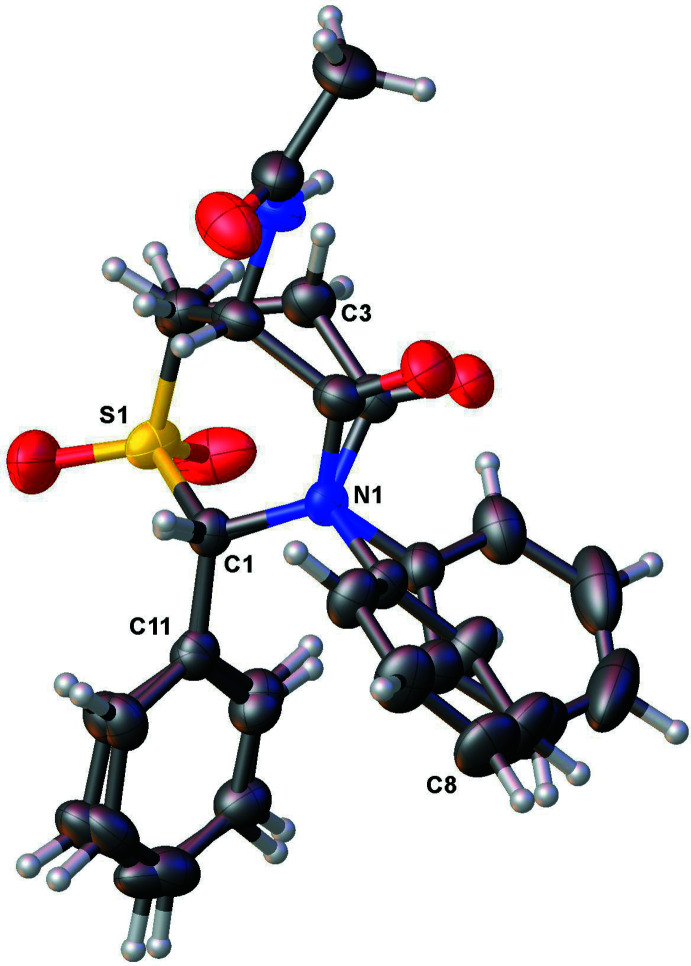
The structure of 1 (Fig. 1 ▸) has a half-chair puckering of the thiazine ring with puckering amplitude Q = 0.605 (2) Å, θ = 47.2 (2)°, φ = 346.7 (3)°, while in 2 (Fig. 2 ▸) the ring has a boat pucker [Q = 0.770 (2) Å, θ = 85.31 (15)°, φ = 61.89 (17)°]. This change in the puckering of the central ring system of the two molecules leads to differing orientations of one of the phenyl rings, which is clear from the overlay diagram (Fig. 3 ▸).
Figure 1.
The asymmetric unit of 1 with displacement ellipsoids drawn at 50% probability level.
Figure 2.
The asymmetric unit of 2 with displacement ellipsoids drawn at 50% probability level.
Figure 3.
Overlay plot of 1 and 2 where the three atoms S1, N1, and C11 are matched. Atoms C3 and C8 of compound 1 are labeled.
3. Supramolecular features
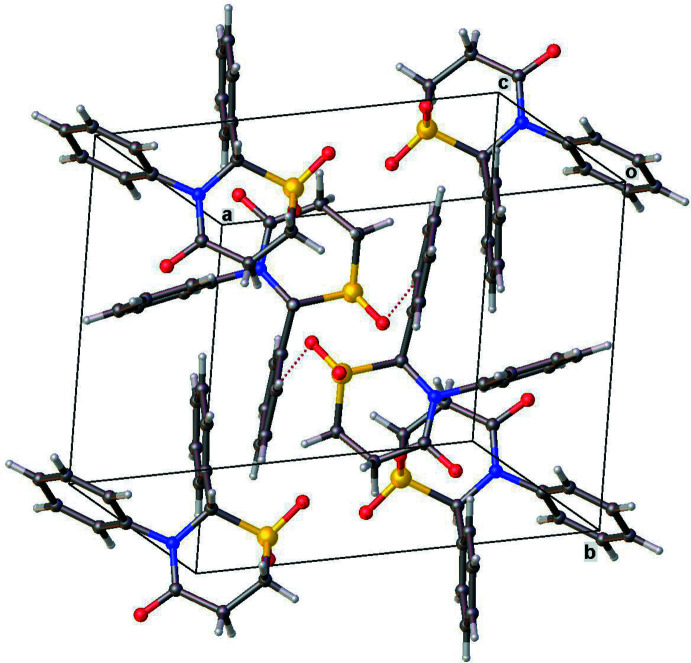
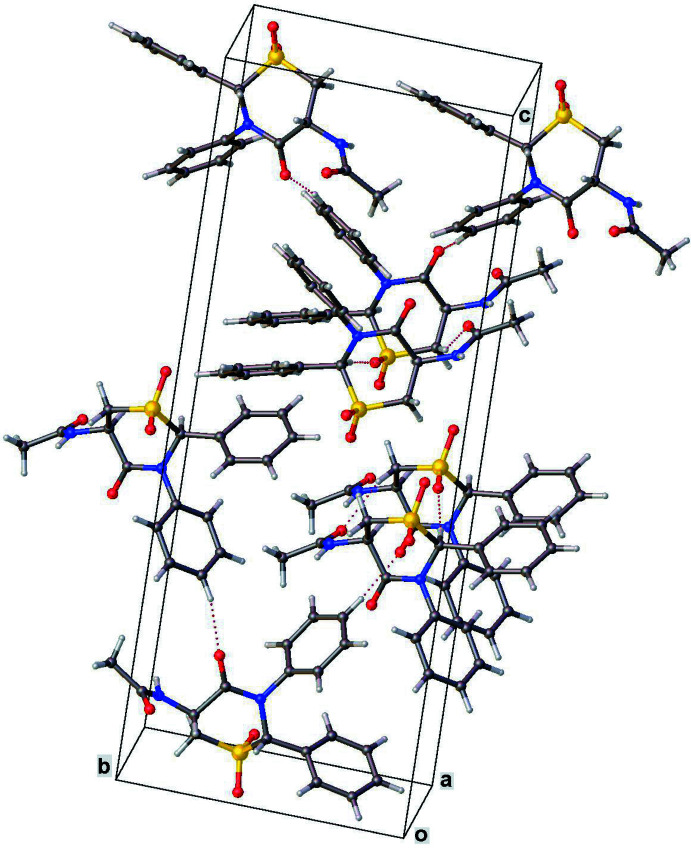
In both structures, only C—H⋯O-type hydrogen-bond interactions between symmetry-related molecules are observed (Tables 1 ▸ and 2 ▸). In 1, a single hydrogen bond [C12—H12⋯O1 = 3.454 (4) Å, 157°] and its symmetry-equivalent form a pair of parallel interactions (Fig. 4 ▸). In 2 (Fig. 5 ▸), the carbon atoms C1 and C4, both of the thiazine ring, as well as C8 of one of the phenyl rings each donate an H atom for three distinct interactions involving three of the four oxygen atoms in the molecule. Although both compounds each have two phenyl rings, neither of the lattices exhibit any π–π stacking interactions.
Table 1. Hydrogen-bond geometry (Å, °) for 1 .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C12—H12⋯O1i | 0.95 | 2.56 | 3.454 (4) | 157 |
Symmetry code: (i)
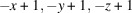 .
.
Table 2. Hydrogen-bond geometry (Å, °) for 2 .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C1—H1⋯O2i | 0.98 | 2.39 | 3.365 (3) | 173 |
| C4—H4A⋯O4ii | 0.97 | 2.29 | 3.185 (4) | 153 |
| C8—H8⋯O3iii | 0.93 | 2.51 | 3.378 (5) | 155 |
Symmetry codes: (i)
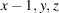 ; (ii)
; (ii)
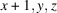 ; (iii)
; (iii)
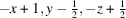 .
.
Figure 4.
Crystal packing diagram for 1 showing intermolecular pairs of C—H⋯O hydrogen bonds.
Figure 5.
Crystal packing diagram for 2 showing intermolecular C—H⋯O hydrogen bonds.
4. Database survey
Searches undertaken using the American Chemical Society’s Chemical Abstract Service (CAS) Scifinder platform did not find crystal structures of any 1,3-thiazin-4-one sulfones other than chlormezanone (CSD refcode KAPNAR; Tanaka & Horayama, 2005 ▸).
5. Synthesis and crystallization
General oxidation procedure (Surrey et al., 1958 ▸; Silverberg, 2020 ▸; Cannon et al. 2015 ▸): the heterocycle (0.267 mmol) was dissolved in glacial acetic acid (1.2 ml). An aqueous solution of KMnO4 (0.535 mmol in 1.45 ml water) was added dropwise at room temperature with vigorous stirring. The reaction was followed by TLC. Solid sodium bisulfite (NaHSO3/Na2S2O5) was added until the mixture remained colorless and then 1.45 ml of water were added and stirred for 10 min. The mixture was extracted with CH2Cl2 (3 × 5 ml). The organics were combined and washed once with sat. NaCl. The solution was dried over Na2SO4 and filtered. The product was purified by chromatography in a silica gel micro-column.
rac-2,3-diphenyl-2,3,5,6-tetrahydro-4H-1,3-thiazine-1,1,4-trione, 1: Eluted with mixtures of ethyl acetate and hexanes. White solid (0.053 g, 70%). m.p.: 418–421 K. Crystals for X-ray diffraction studies were grown by slow evaporation from toluene solution.
N-[(2S,5R)-1,1,4-trioxo-2,3-diphenyl-1,3-thiazinan-5-yl]acetamide, 2: Eluted with a mixture of 10% acetone and 90% ethyl acetate. White solid (0.076 g, 80%). m.p.: 443–467 K (decomposition). Crystals were grown by slow evaporation from ethanol solution.
6. Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. The hydrogen atoms were placed in their geometrically calculated positions and their coordinates refined using the riding model with parent-atom—H lengths of 0.93 Å (CH), 0.98 Å (chiral-CH), 0.96 Å (CH3), 0.97 Å (CH2). Isotropic displacement parameters for these atoms were set to 1.2 (CH) or 1.5 (CH3) times U eq of the parent atom.
Table 3. Experimental details.
| 1 | 2 | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C16H15NO3S | C18H18N2O4S |
| M r | 301.35 | 358.40 |
| Crystal system, space group | Monoclinic, P21/c | Orthorhombic, P212121 |
| Temperature (K) | 173 | 298 |
| a, b, c (Å) | 14.4485 (6), 10.2031 (5), 10.4950 (4) | 5.5230 (4), 10.6857 (9), 28.430 (2) |
| α, β, γ (°) | 90, 107.179 (4), 90 | 90, 90, 90 |
| V (Å3) | 1478.13 (11) | 1677.8 (2) |
| Z | 4 | 4 |
| Radiation type | Cu Kα | Mo Kα |
| μ (mm−1) | 2.03 | 0.22 |
| Crystal size (mm) | 0.2 × 0.18 × 0.09 | 0.22 × 0.06 × 0.06 |
| Data collection | ||
| Diffractometer | Rigaku Oxford Diffraction Synergy Custom system, HyPix-Arc 150 | Bruker SMART CCD area detector |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku OD, 2022 ▸) | Multi-scan (SADABS; Krause et al., 2015 ▸) |
| T min, T max | 0.668, 1.000 | 0.656, 0.900 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 7437, 2856, 2139 | 13301, 4037, 3460 |
| R int | 0.056 | 0.035 |
| (sin θ/λ)max (Å−1) | 0.628 | 0.667 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.052, 0.150, 1.10 | 0.042, 0.103, 1.04 |
| No. of reflections | 2856 | 4037 |
| No. of parameters | 191 | 231 |
| H-atom treatment | H-atom parameters constrained | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.32, −0.34 | 0.24, −0.14 |
| Absolute structure | – | Flack x determined using 1213 quotients [(I +)−(I −)]/[(I +)+(I −)] (Parsons et al., 2013 ▸) |
| Absolute structure parameter | – | 0.07 (4) |
Supplementary Material
Crystal structure: contains datablock(s) 1, 2. DOI: 10.1107/S2056989023000695/hb8050sup1.cif
Structure factors: contains datablock(s) 1. DOI: 10.1107/S2056989023000695/hb80501sup2.hkl
Supporting information file. DOI: 10.1107/S2056989023000695/hb80501sup4.mol
Structure factors: contains datablock(s) 2. DOI: 10.1107/S2056989023000695/hb80502sup3.hkl
Supporting information file. DOI: 10.1107/S2056989023000695/hb80502sup5.mol
Supporting information file. DOI: 10.1107/S2056989023000695/hb80501sup6.cml
Supporting information file. DOI: 10.1107/S2056989023000695/hb80502sup7.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
rac-2,3-Diphenyl-2,3,5,6-tetrahydro-4H-1,3-thiazine-1,1,4-trione (1) . Crystal data
| C16H15NO3S | F(000) = 632 |
| Mr = 301.35 | Dx = 1.354 Mg m−3 |
| Monoclinic, P21/c | Cu Kα radiation, λ = 1.54184 Å |
| a = 14.4485 (6) Å | Cell parameters from 3416 reflections |
| b = 10.2031 (5) Å | θ = 3.2–73.3° |
| c = 10.4950 (4) Å | µ = 2.03 mm−1 |
| β = 107.179 (4)° | T = 173 K |
| V = 1478.13 (11) Å3 | Block, clear colourless |
| Z = 4 | 0.2 × 0.18 × 0.09 mm |
rac-2,3-Diphenyl-2,3,5,6-tetrahydro-4H-1,3-thiazine-1,1,4-trione (1) . Data collection
| Rigaku Oxford Diffraction Synergy Custom system, HyPix-Arc 150 diffractometer | 2856 independent reflections |
| Radiation source: Rotating-anode X-ray tube, Rigaku (Cu) X-ray Source | 2139 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.056 |
| Detector resolution: 10.0000 pixels mm-1 | θmax = 75.6°, θmin = 3.2° |
| ω scans | h = −15→17 |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku OD, 2022) | k = −11→12 |
| Tmin = 0.668, Tmax = 1.000 | l = −12→13 |
| 7437 measured reflections |
rac-2,3-Diphenyl-2,3,5,6-tetrahydro-4H-1,3-thiazine-1,1,4-trione (1) . Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.052 | w = 1/[σ2(Fo2) + (0.0712P)2 + 0.268P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.150 | (Δ/σ)max < 0.001 |
| S = 1.10 | Δρmax = 0.32 e Å−3 |
| 2856 reflections | Δρmin = −0.34 e Å−3 |
| 191 parameters | Extinction correction: SHELXL2018/3 (Sheldrick 2015b), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 0 restraints | Extinction coefficient: 0.0023 (5) |
| Primary atom site location: dual |
rac-2,3-Diphenyl-2,3,5,6-tetrahydro-4H-1,3-thiazine-1,1,4-trione (1) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
rac-2,3-Diphenyl-2,3,5,6-tetrahydro-4H-1,3-thiazine-1,1,4-trione (1) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.45082 (4) | 0.66477 (7) | 0.66401 (5) | 0.0380 (2) | |
| O1 | 0.52700 (12) | 0.5704 (2) | 0.67907 (19) | 0.0514 (6) | |
| O2 | 0.41925 (14) | 0.6961 (2) | 0.77812 (16) | 0.0519 (6) | |
| O3 | 0.22170 (13) | 0.91599 (19) | 0.44575 (18) | 0.0455 (5) | |
| N1 | 0.27444 (13) | 0.7076 (2) | 0.48536 (18) | 0.0306 (5) | |
| C1 | 0.35000 (16) | 0.6063 (3) | 0.5290 (2) | 0.0310 (5) | |
| H1 | 0.374440 | 0.583984 | 0.451921 | 0.037* | |
| C4 | 0.47674 (18) | 0.8100 (3) | 0.5937 (2) | 0.0396 (6) | |
| H4A | 0.532525 | 0.854461 | 0.656910 | 0.048* | |
| H4B | 0.493981 | 0.790381 | 0.511127 | 0.048* | |
| C3 | 0.38813 (19) | 0.8988 (3) | 0.5617 (3) | 0.0425 (6) | |
| H3A | 0.399282 | 0.971486 | 0.505438 | 0.051* | |
| H3B | 0.384838 | 0.938047 | 0.646531 | 0.051* | |
| C2 | 0.28912 (18) | 0.8402 (3) | 0.4920 (2) | 0.0348 (6) | |
| C5 | 0.18322 (17) | 0.6594 (3) | 0.3978 (2) | 0.0333 (6) | |
| C6 | 0.17961 (19) | 0.6182 (3) | 0.2711 (2) | 0.0440 (7) | |
| H6 | 0.235336 | 0.625452 | 0.241121 | 0.053* | |
| C7 | 0.0947 (2) | 0.5664 (4) | 0.1879 (3) | 0.0559 (9) | |
| H7 | 0.091908 | 0.537900 | 0.100655 | 0.067* | |
| C8 | 0.0141 (2) | 0.5563 (4) | 0.2324 (3) | 0.0633 (10) | |
| H8 | −0.044126 | 0.519958 | 0.175939 | 0.076* | |
| C9 | 0.0179 (2) | 0.5990 (4) | 0.3588 (3) | 0.0682 (11) | |
| H9 | −0.038042 | 0.592964 | 0.388363 | 0.082* | |
| C10 | 0.10296 (19) | 0.6506 (3) | 0.4427 (3) | 0.0496 (8) | |
| H10 | 0.105807 | 0.679455 | 0.529797 | 0.060* | |
| C11 | 0.31292 (16) | 0.4818 (3) | 0.5756 (2) | 0.0334 (6) | |
| C12 | 0.3255 (2) | 0.3631 (3) | 0.5188 (3) | 0.0437 (7) | |
| H12 | 0.356067 | 0.360439 | 0.450153 | 0.052* | |
| C13 | 0.2933 (2) | 0.2481 (3) | 0.5624 (3) | 0.0550 (8) | |
| H13 | 0.301505 | 0.166705 | 0.523015 | 0.066* | |
| C14 | 0.2495 (2) | 0.2515 (3) | 0.6624 (3) | 0.0541 (8) | |
| H14 | 0.227909 | 0.172599 | 0.692365 | 0.065* | |
| C15 | 0.2371 (2) | 0.3693 (3) | 0.7189 (3) | 0.0468 (7) | |
| H15 | 0.207209 | 0.371115 | 0.788272 | 0.056* | |
| C16 | 0.26749 (17) | 0.4849 (3) | 0.6758 (2) | 0.0391 (6) | |
| H16 | 0.257539 | 0.566039 | 0.714104 | 0.047* |
rac-2,3-Diphenyl-2,3,5,6-tetrahydro-4H-1,3-thiazine-1,1,4-trione (1) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0329 (4) | 0.0492 (5) | 0.0252 (3) | −0.0062 (3) | −0.0018 (2) | 0.0054 (3) |
| O1 | 0.0332 (10) | 0.0563 (14) | 0.0540 (11) | 0.0048 (9) | −0.0038 (8) | 0.0172 (10) |
| O2 | 0.0568 (12) | 0.0699 (15) | 0.0249 (9) | −0.0242 (11) | 0.0060 (8) | −0.0045 (9) |
| O3 | 0.0431 (11) | 0.0385 (11) | 0.0531 (11) | 0.0069 (9) | 0.0117 (8) | 0.0068 (9) |
| N1 | 0.0264 (10) | 0.0339 (12) | 0.0271 (9) | 0.0014 (9) | 0.0010 (7) | 0.0010 (9) |
| C1 | 0.0288 (12) | 0.0369 (14) | 0.0242 (11) | 0.0019 (11) | 0.0029 (8) | 0.0001 (10) |
| C4 | 0.0332 (13) | 0.0488 (18) | 0.0306 (12) | −0.0078 (12) | −0.0002 (9) | −0.0007 (12) |
| C3 | 0.0429 (15) | 0.0410 (16) | 0.0409 (13) | −0.0035 (13) | 0.0085 (11) | −0.0046 (12) |
| C2 | 0.0387 (13) | 0.0361 (15) | 0.0287 (12) | 0.0004 (12) | 0.0088 (9) | 0.0008 (11) |
| C5 | 0.0266 (12) | 0.0412 (16) | 0.0273 (11) | 0.0008 (11) | 0.0006 (9) | 0.0025 (11) |
| C6 | 0.0344 (13) | 0.061 (2) | 0.0318 (13) | 0.0004 (13) | 0.0024 (10) | −0.0038 (13) |
| C7 | 0.0457 (16) | 0.075 (2) | 0.0375 (14) | −0.0016 (16) | −0.0020 (11) | −0.0111 (15) |
| C8 | 0.0393 (16) | 0.083 (3) | 0.0542 (18) | −0.0118 (17) | −0.0070 (13) | −0.0056 (18) |
| C9 | 0.0319 (15) | 0.118 (3) | 0.0511 (18) | −0.0105 (19) | 0.0072 (12) | 0.003 (2) |
| C10 | 0.0347 (14) | 0.079 (2) | 0.0340 (13) | −0.0036 (15) | 0.0083 (10) | −0.0019 (14) |
| C11 | 0.0284 (12) | 0.0365 (15) | 0.0299 (11) | 0.0023 (11) | 0.0005 (9) | 0.0008 (11) |
| C12 | 0.0441 (15) | 0.0421 (17) | 0.0402 (14) | 0.0065 (13) | 0.0051 (11) | −0.0036 (13) |
| C13 | 0.063 (2) | 0.0324 (17) | 0.0579 (18) | 0.0028 (15) | 0.0007 (15) | 0.0000 (14) |
| C14 | 0.0525 (17) | 0.0420 (19) | 0.0573 (18) | −0.0064 (15) | −0.0001 (14) | 0.0102 (15) |
| C15 | 0.0428 (15) | 0.0516 (19) | 0.0431 (15) | −0.0038 (14) | 0.0083 (11) | 0.0098 (14) |
| C16 | 0.0398 (14) | 0.0400 (16) | 0.0360 (13) | −0.0006 (12) | 0.0089 (10) | 0.0013 (12) |
rac-2,3-Diphenyl-2,3,5,6-tetrahydro-4H-1,3-thiazine-1,1,4-trione (1) . Geometric parameters (Å, º)
| S1—O1 | 1.435 (2) | C6—C7 | 1.383 (4) |
| S1—O2 | 1.438 (2) | C7—H7 | 0.9500 |
| S1—C1 | 1.807 (2) | C7—C8 | 1.381 (4) |
| S1—C4 | 1.744 (3) | C8—H8 | 0.9500 |
| O3—C2 | 1.226 (3) | C8—C9 | 1.382 (5) |
| N1—C1 | 1.475 (3) | C9—H9 | 0.9500 |
| N1—C2 | 1.368 (3) | C9—C10 | 1.387 (4) |
| N1—C5 | 1.451 (3) | C10—H10 | 0.9500 |
| C1—H1 | 1.0000 | C11—C12 | 1.385 (4) |
| C1—C11 | 1.514 (4) | C11—C16 | 1.394 (3) |
| C4—H4A | 0.9900 | C12—H12 | 0.9500 |
| C4—H4B | 0.9900 | C12—C13 | 1.389 (4) |
| C4—C3 | 1.523 (4) | C13—H13 | 0.9500 |
| C3—H3A | 0.9900 | C13—C14 | 1.377 (5) |
| C3—H3B | 0.9900 | C14—H14 | 0.9500 |
| C3—C2 | 1.524 (4) | C14—C15 | 1.375 (4) |
| C5—C6 | 1.381 (3) | C15—H15 | 0.9500 |
| C5—C10 | 1.377 (4) | C15—C16 | 1.381 (4) |
| C6—H6 | 0.9500 | C16—H16 | 0.9500 |
| O1—S1—O2 | 118.62 (12) | C5—C6—H6 | 120.0 |
| O1—S1—C1 | 106.20 (12) | C5—C6—C7 | 119.9 (3) |
| O1—S1—C4 | 111.28 (13) | C7—C6—H6 | 120.0 |
| O2—S1—C1 | 110.23 (11) | C6—C7—H7 | 120.2 |
| O2—S1—C4 | 108.92 (14) | C8—C7—C6 | 119.6 (3) |
| C4—S1—C1 | 99.96 (11) | C8—C7—H7 | 120.2 |
| C2—N1—C1 | 126.0 (2) | C7—C8—H8 | 119.9 |
| C2—N1—C5 | 117.7 (2) | C7—C8—C9 | 120.2 (3) |
| C5—N1—C1 | 114.2 (2) | C9—C8—H8 | 119.9 |
| S1—C1—H1 | 108.3 | C8—C9—H9 | 119.8 |
| N1—C1—S1 | 111.31 (17) | C8—C9—C10 | 120.4 (3) |
| N1—C1—H1 | 108.3 | C10—C9—H9 | 119.8 |
| N1—C1—C11 | 112.84 (19) | C5—C10—C9 | 119.1 (3) |
| C11—C1—S1 | 107.70 (15) | C5—C10—H10 | 120.5 |
| C11—C1—H1 | 108.3 | C9—C10—H10 | 120.5 |
| S1—C4—H4A | 109.9 | C12—C11—C1 | 119.3 (2) |
| S1—C4—H4B | 109.9 | C12—C11—C16 | 119.7 (3) |
| H4A—C4—H4B | 108.3 | C16—C11—C1 | 121.0 (2) |
| C3—C4—S1 | 109.11 (19) | C11—C12—H12 | 120.1 |
| C3—C4—H4A | 109.9 | C11—C12—C13 | 119.8 (3) |
| C3—C4—H4B | 109.9 | C13—C12—H12 | 120.1 |
| C4—C3—H3A | 107.6 | C12—C13—H13 | 119.9 |
| C4—C3—H3B | 107.6 | C14—C13—C12 | 120.2 (3) |
| C4—C3—C2 | 118.7 (2) | C14—C13—H13 | 119.9 |
| H3A—C3—H3B | 107.1 | C13—C14—H14 | 120.0 |
| C2—C3—H3A | 107.6 | C15—C14—C13 | 119.9 (3) |
| C2—C3—H3B | 107.6 | C15—C14—H14 | 120.0 |
| O3—C2—N1 | 120.7 (2) | C14—C15—H15 | 119.7 |
| O3—C2—C3 | 117.7 (2) | C14—C15—C16 | 120.7 (3) |
| N1—C2—C3 | 121.5 (2) | C16—C15—H15 | 119.7 |
| C6—C5—N1 | 118.8 (2) | C11—C16—H16 | 120.2 |
| C10—C5—N1 | 120.3 (2) | C15—C16—C11 | 119.6 (3) |
| C10—C5—C6 | 120.9 (2) | C15—C16—H16 | 120.2 |
| S1—C1—C11—C12 | −111.1 (2) | C4—C3—C2—O3 | −167.2 (2) |
| S1—C1—C11—C16 | 68.1 (2) | C4—C3—C2—N1 | 15.3 (4) |
| S1—C4—C3—C2 | −45.9 (3) | C2—N1—C1—S1 | 29.3 (3) |
| O1—S1—C1—N1 | −168.32 (16) | C2—N1—C1—C11 | 150.5 (2) |
| O1—S1—C1—C11 | 67.50 (19) | C2—N1—C5—C6 | 96.5 (3) |
| O1—S1—C4—C3 | 172.01 (17) | C2—N1—C5—C10 | −86.0 (3) |
| O2—S1—C1—N1 | 62.00 (19) | C5—N1—C1—S1 | −167.57 (16) |
| O2—S1—C1—C11 | −62.2 (2) | C5—N1—C1—C11 | −46.3 (3) |
| O2—S1—C4—C3 | −55.4 (2) | C5—N1—C2—O3 | 13.2 (3) |
| N1—C1—C11—C12 | 125.6 (2) | C5—N1—C2—C3 | −169.4 (2) |
| N1—C1—C11—C16 | −55.2 (3) | C5—C6—C7—C8 | 0.0 (5) |
| N1—C5—C6—C7 | 177.1 (3) | C6—C5—C10—C9 | 0.3 (5) |
| N1—C5—C10—C9 | −177.2 (3) | C6—C7—C8—C9 | 0.7 (6) |
| C1—S1—C4—C3 | 60.16 (19) | C7—C8—C9—C10 | −0.9 (6) |
| C1—N1—C2—O3 | 175.8 (2) | C8—C9—C10—C5 | 0.4 (6) |
| C1—N1—C2—C3 | −6.8 (3) | C10—C5—C6—C7 | −0.5 (4) |
| C1—N1—C5—C6 | −68.1 (3) | C11—C12—C13—C14 | −0.4 (4) |
| C1—N1—C5—C10 | 109.4 (3) | C12—C11—C16—C15 | 1.1 (4) |
| C1—C11—C12—C13 | 178.9 (2) | C12—C13—C14—C15 | 0.4 (4) |
| C1—C11—C16—C15 | −178.1 (2) | C13—C14—C15—C16 | 0.4 (4) |
| C4—S1—C1—N1 | −52.56 (18) | C14—C15—C16—C11 | −1.2 (4) |
| C4—S1—C1—C11 | −176.73 (18) | C16—C11—C12—C13 | −0.3 (4) |
rac-2,3-Diphenyl-2,3,5,6-tetrahydro-4H-1,3-thiazine-1,1,4-trione (1) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C12—H12···O1i | 0.95 | 2.56 | 3.454 (4) | 157 |
Symmetry code: (i) −x+1, −y+1, −z+1.
N-[(2S,5R)-1,1,4-Trioxo-2,3-diphenyl-1,3-thiazinan-5-yl]acetamide (2) . Crystal data
| C18H18N2O4S | Dx = 1.419 Mg m−3 |
| Mr = 358.40 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, P212121 | Cell parameters from 4224 reflections |
| a = 5.5230 (4) Å | θ = 2.4–25.0° |
| b = 10.6857 (9) Å | µ = 0.22 mm−1 |
| c = 28.430 (2) Å | T = 298 K |
| V = 1677.8 (2) Å3 | Rod, colorless |
| Z = 4 | 0.22 × 0.06 × 0.06 mm |
| F(000) = 752 |
N-[(2S,5R)-1,1,4-Trioxo-2,3-diphenyl-1,3-thiazinan-5-yl]acetamide (2) . Data collection
| Bruker SMART CCD area detector diffractometer | 3460 reflections with I > 2σ(I) |
| phi and ω scans | Rint = 0.035 |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | θmax = 28.3°, θmin = 1.4° |
| Tmin = 0.656, Tmax = 0.900 | h = −7→5 |
| 13301 measured reflections | k = −12→14 |
| 4037 independent reflections | l = −36→37 |
N-[(2S,5R)-1,1,4-Trioxo-2,3-diphenyl-1,3-thiazinan-5-yl]acetamide (2) . Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.042 | w = 1/[σ2(Fo2) + (0.0564P)2 + 0.035P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.103 | (Δ/σ)max < 0.001 |
| S = 1.04 | Δρmax = 0.24 e Å−3 |
| 4037 reflections | Δρmin = −0.14 e Å−3 |
| 231 parameters | Absolute structure: Flack x determined using 1213 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons et al., 2013) |
| 0 restraints | Absolute structure parameter: 0.07 (4) |
| Primary atom site location: structure-invariant direct methods |
N-[(2S,5R)-1,1,4-Trioxo-2,3-diphenyl-1,3-thiazinan-5-yl]acetamide (2) . Special details
| Experimental. The data collection nominally covered a full sphere of reciprocal space by a combination of 4 sets of ω scans each set at different φ and/or 2θ angles and each scan (10 s exposure) covering -0.300° degrees in ω. The crystal to detector distance was 5.82 cm. |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
N-[(2S,5R)-1,1,4-Trioxo-2,3-diphenyl-1,3-thiazinan-5-yl]acetamide (2) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.29097 (13) | 0.61561 (6) | 0.04750 (2) | 0.04039 (18) | |
| O1 | 0.2013 (5) | 0.5960 (2) | 0.00084 (6) | 0.0665 (7) | |
| O2 | 0.5430 (4) | 0.5943 (2) | 0.05600 (8) | 0.0655 (6) | |
| O3 | 0.0948 (4) | 0.72050 (18) | 0.18769 (6) | 0.0575 (6) | |
| O4 | −0.3983 (4) | 0.8940 (2) | 0.13514 (8) | 0.0659 (6) | |
| N1 | 0.1594 (4) | 0.56459 (18) | 0.13538 (6) | 0.0394 (5) | |
| N2 | 0.0062 (5) | 0.8977 (2) | 0.12438 (9) | 0.0485 (6) | |
| C1 | 0.1124 (4) | 0.5198 (2) | 0.08754 (7) | 0.0333 (5) | |
| H1 | −0.058432 | 0.536713 | 0.080716 | 0.040* | |
| C2 | 0.0930 (5) | 0.6841 (2) | 0.14753 (8) | 0.0409 (6) | |
| C3 | 0.0251 (5) | 0.7713 (2) | 0.10656 (8) | 0.0398 (6) | |
| H3 | −0.132274 | 0.745690 | 0.093867 | 0.048* | |
| C4 | 0.2134 (6) | 0.7692 (2) | 0.06686 (8) | 0.0424 (6) | |
| H4A | 0.359008 | 0.811013 | 0.077660 | 0.051* | |
| H4B | 0.150687 | 0.816206 | 0.040330 | 0.051* | |
| C11 | 0.1560 (4) | 0.3832 (2) | 0.07738 (7) | 0.0350 (5) | |
| C16 | 0.3688 (5) | 0.3203 (3) | 0.08898 (9) | 0.0448 (6) | |
| H16 | 0.493371 | 0.362002 | 0.104412 | 0.054* | |
| C15 | 0.3932 (6) | 0.1953 (3) | 0.07738 (10) | 0.0559 (8) | |
| H15 | 0.533040 | 0.152566 | 0.085968 | 0.067* | |
| C14 | 0.2132 (7) | 0.1331 (3) | 0.05326 (10) | 0.0596 (9) | |
| H14 | 0.232717 | 0.049213 | 0.045356 | 0.072* | |
| C13 | 0.0065 (7) | 0.1949 (3) | 0.04104 (10) | 0.0565 (8) | |
| H13 | −0.114479 | 0.153289 | 0.024516 | 0.068* | |
| C12 | −0.0235 (5) | 0.3194 (2) | 0.05317 (8) | 0.0434 (6) | |
| H12 | −0.165681 | 0.360621 | 0.044990 | 0.052* | |
| C5 | 0.2702 (5) | 0.4889 (2) | 0.17177 (7) | 0.0393 (6) | |
| C6 | 0.1536 (6) | 0.3843 (3) | 0.18873 (9) | 0.0535 (8) | |
| H6 | 0.004073 | 0.360301 | 0.176670 | 0.064* | |
| C7 | 0.2639 (10) | 0.3152 (3) | 0.22429 (10) | 0.0815 (13) | |
| H7 | 0.188747 | 0.243530 | 0.235747 | 0.098* | |
| C8 | 0.4806 (12) | 0.3516 (5) | 0.24243 (12) | 0.0976 (17) | |
| H8 | 0.553113 | 0.304469 | 0.266063 | 0.117* | |
| C9 | 0.5929 (8) | 0.4575 (5) | 0.22597 (12) | 0.0839 (13) | |
| H9 | 0.738587 | 0.483351 | 0.239181 | 0.101* | |
| C10 | 0.4902 (6) | 0.5261 (3) | 0.18983 (10) | 0.0560 (8) | |
| H10 | 0.568552 | 0.596319 | 0.177902 | 0.067* | |
| C17 | −0.2027 (7) | 0.9434 (3) | 0.14185 (9) | 0.0490 (7) | |
| C18 | −0.1753 (8) | 1.0631 (3) | 0.16996 (11) | 0.0678 (10) | |
| H18A | −0.330040 | 1.103268 | 0.172852 | 0.102* | |
| H18B | −0.064566 | 1.118103 | 0.154150 | 0.102* | |
| H18C | −0.113853 | 1.043699 | 0.200700 | 0.102* | |
| H2 | 0.139 (5) | 0.924 (3) | 0.1348 (9) | 0.042 (8)* |
N-[(2S,5R)-1,1,4-Trioxo-2,3-diphenyl-1,3-thiazinan-5-yl]acetamide (2) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0452 (4) | 0.0358 (3) | 0.0402 (3) | 0.0022 (3) | 0.0046 (3) | 0.0016 (2) |
| O1 | 0.1092 (19) | 0.0556 (12) | 0.0347 (9) | −0.0001 (14) | 0.0030 (11) | −0.0010 (8) |
| O2 | 0.0404 (11) | 0.0512 (13) | 0.1048 (17) | 0.0035 (10) | 0.0137 (12) | 0.0130 (11) |
| O3 | 0.0835 (16) | 0.0502 (12) | 0.0387 (9) | 0.0055 (11) | 0.0013 (10) | −0.0095 (8) |
| O4 | 0.0562 (14) | 0.0671 (15) | 0.0742 (14) | 0.0093 (13) | −0.0051 (11) | −0.0223 (12) |
| N1 | 0.0536 (15) | 0.0329 (10) | 0.0317 (9) | 0.0008 (10) | −0.0029 (9) | −0.0018 (8) |
| N2 | 0.0530 (16) | 0.0315 (12) | 0.0611 (14) | −0.0018 (12) | −0.0034 (12) | −0.0078 (10) |
| C1 | 0.0321 (12) | 0.0353 (12) | 0.0324 (10) | 0.0017 (10) | −0.0035 (9) | −0.0027 (9) |
| C2 | 0.0461 (16) | 0.0349 (13) | 0.0415 (12) | −0.0033 (12) | 0.0008 (12) | −0.0033 (10) |
| C3 | 0.0431 (15) | 0.0303 (12) | 0.0460 (13) | −0.0002 (11) | −0.0029 (11) | −0.0054 (10) |
| C4 | 0.0506 (15) | 0.0347 (12) | 0.0419 (12) | −0.0013 (13) | −0.0016 (12) | 0.0019 (9) |
| C11 | 0.0406 (14) | 0.0327 (11) | 0.0317 (10) | −0.0004 (11) | 0.0015 (9) | −0.0009 (9) |
| C16 | 0.0480 (16) | 0.0434 (14) | 0.0431 (13) | 0.0080 (13) | −0.0010 (11) | 0.0002 (11) |
| C15 | 0.072 (2) | 0.0458 (16) | 0.0498 (15) | 0.0223 (16) | 0.0092 (15) | 0.0110 (12) |
| C14 | 0.095 (3) | 0.0319 (13) | 0.0516 (15) | −0.0005 (17) | 0.0225 (18) | 0.0014 (11) |
| C13 | 0.072 (2) | 0.0451 (16) | 0.0521 (15) | −0.0183 (17) | 0.0068 (15) | −0.0108 (12) |
| C12 | 0.0463 (15) | 0.0445 (15) | 0.0394 (12) | −0.0049 (13) | 0.0010 (12) | −0.0051 (11) |
| C5 | 0.0445 (15) | 0.0420 (13) | 0.0315 (10) | 0.0043 (12) | −0.0011 (11) | −0.0019 (9) |
| C6 | 0.071 (2) | 0.0473 (15) | 0.0422 (13) | −0.0011 (16) | 0.0077 (13) | 0.0033 (12) |
| C7 | 0.138 (4) | 0.060 (2) | 0.0462 (16) | 0.015 (3) | 0.012 (2) | 0.0154 (14) |
| C8 | 0.147 (5) | 0.100 (4) | 0.0458 (18) | 0.058 (3) | −0.020 (2) | 0.0019 (19) |
| C9 | 0.073 (3) | 0.118 (4) | 0.060 (2) | 0.039 (3) | −0.0289 (19) | −0.029 (2) |
| C10 | 0.0483 (18) | 0.071 (2) | 0.0483 (14) | 0.0058 (16) | −0.0050 (13) | −0.0119 (14) |
| C17 | 0.065 (2) | 0.0384 (14) | 0.0435 (13) | 0.0077 (15) | −0.0061 (15) | −0.0038 (10) |
| C18 | 0.095 (3) | 0.0437 (16) | 0.0649 (18) | 0.0125 (19) | −0.0055 (19) | −0.0156 (13) |
N-[(2S,5R)-1,1,4-Trioxo-2,3-diphenyl-1,3-thiazinan-5-yl]acetamide (2) . Geometric parameters (Å, º)
| S1—O1 | 1.431 (2) | C15—H15 | 0.9300 |
| S1—O2 | 1.431 (2) | C15—C14 | 1.378 (5) |
| S1—C1 | 1.821 (2) | C14—H14 | 0.9300 |
| S1—C4 | 1.783 (3) | C14—C13 | 1.364 (5) |
| O3—C2 | 1.206 (3) | C13—H13 | 0.9300 |
| O4—C17 | 1.218 (4) | C13—C12 | 1.384 (4) |
| N1—C1 | 1.465 (3) | C12—H12 | 0.9300 |
| N1—C2 | 1.373 (3) | C5—C6 | 1.377 (4) |
| N1—C5 | 1.449 (3) | C5—C10 | 1.378 (4) |
| N2—C3 | 1.446 (3) | C6—H6 | 0.9300 |
| N2—C17 | 1.348 (4) | C6—C7 | 1.393 (5) |
| N2—H2 | 0.84 (3) | C7—H7 | 0.9300 |
| C1—H1 | 0.9800 | C7—C8 | 1.360 (7) |
| C1—C11 | 1.507 (3) | C8—H8 | 0.9300 |
| C2—C3 | 1.538 (3) | C8—C9 | 1.373 (6) |
| C3—H3 | 0.9800 | C9—H9 | 0.9300 |
| C3—C4 | 1.535 (4) | C9—C10 | 1.384 (5) |
| C4—H4A | 0.9700 | C10—H10 | 0.9300 |
| C4—H4B | 0.9700 | C17—C18 | 1.515 (4) |
| C11—C16 | 1.394 (4) | C18—H18A | 0.9600 |
| C11—C12 | 1.386 (3) | C18—H18B | 0.9600 |
| C16—H16 | 0.9300 | C18—H18C | 0.9600 |
| C16—C15 | 1.382 (4) | ||
| O1—S1—C1 | 108.03 (13) | C16—C15—H15 | 119.5 |
| O1—S1—C4 | 109.74 (13) | C14—C15—C16 | 121.0 (3) |
| O2—S1—O1 | 118.01 (15) | C14—C15—H15 | 119.5 |
| O2—S1—C1 | 109.38 (12) | C15—C14—H14 | 120.1 |
| O2—S1—C4 | 109.15 (14) | C13—C14—C15 | 119.8 (3) |
| C4—S1—C1 | 101.20 (11) | C13—C14—H14 | 120.1 |
| C2—N1—C1 | 119.34 (19) | C14—C13—H13 | 119.9 |
| C2—N1—C5 | 116.91 (18) | C14—C13—C12 | 120.2 (3) |
| C5—N1—C1 | 123.75 (19) | C12—C13—H13 | 119.9 |
| C3—N2—H2 | 112 (2) | C11—C12—H12 | 119.6 |
| C17—N2—C3 | 122.0 (3) | C13—C12—C11 | 120.7 (3) |
| C17—N2—H2 | 120 (2) | C13—C12—H12 | 119.6 |
| S1—C1—H1 | 107.1 | C6—C5—N1 | 120.4 (2) |
| N1—C1—S1 | 107.50 (16) | C10—C5—N1 | 118.5 (3) |
| N1—C1—H1 | 107.1 | C10—C5—C6 | 121.1 (3) |
| N1—C1—C11 | 117.76 (19) | C5—C6—H6 | 120.7 |
| C11—C1—S1 | 109.76 (16) | C5—C6—C7 | 118.7 (3) |
| C11—C1—H1 | 107.1 | C7—C6—H6 | 120.7 |
| O3—C2—N1 | 122.4 (2) | C6—C7—H7 | 119.7 |
| O3—C2—C3 | 121.6 (2) | C8—C7—C6 | 120.6 (4) |
| N1—C2—C3 | 115.99 (19) | C8—C7—H7 | 119.7 |
| N2—C3—C2 | 108.5 (2) | C7—C8—H8 | 119.9 |
| N2—C3—H3 | 109.0 | C7—C8—C9 | 120.2 (4) |
| N2—C3—C4 | 108.7 (2) | C9—C8—H8 | 119.9 |
| C2—C3—H3 | 109.0 | C8—C9—H9 | 119.8 |
| C4—C3—C2 | 112.5 (2) | C8—C9—C10 | 120.3 (4) |
| C4—C3—H3 | 109.0 | C10—C9—H9 | 119.8 |
| S1—C4—H4A | 108.8 | C5—C10—C9 | 119.0 (4) |
| S1—C4—H4B | 108.8 | C5—C10—H10 | 120.5 |
| C3—C4—S1 | 113.79 (17) | C9—C10—H10 | 120.5 |
| C3—C4—H4A | 108.8 | O4—C17—N2 | 123.0 (2) |
| C3—C4—H4B | 108.8 | O4—C17—C18 | 122.5 (3) |
| H4A—C4—H4B | 107.7 | N2—C17—C18 | 114.5 (3) |
| C16—C11—C1 | 123.8 (2) | C17—C18—H18A | 109.5 |
| C12—C11—C1 | 117.2 (2) | C17—C18—H18B | 109.5 |
| C12—C11—C16 | 118.9 (2) | C17—C18—H18C | 109.5 |
| C11—C16—H16 | 120.3 | H18A—C18—H18B | 109.5 |
| C15—C16—C11 | 119.5 (3) | H18A—C18—H18C | 109.5 |
| C15—C16—H16 | 120.3 | H18B—C18—H18C | 109.5 |
| S1—C1—C11—C16 | 73.2 (2) | C2—N1—C5—C6 | 114.0 (3) |
| S1—C1—C11—C12 | −103.8 (2) | C2—N1—C5—C10 | −64.0 (3) |
| O1—S1—C1—N1 | −165.13 (17) | C2—C3—C4—S1 | 50.6 (3) |
| O1—S1—C1—C11 | 65.65 (19) | C3—N2—C17—O4 | −15.9 (4) |
| O1—S1—C4—C3 | 111.3 (2) | C3—N2—C17—C18 | 165.1 (2) |
| O2—S1—C1—N1 | 65.22 (19) | C4—S1—C1—N1 | −49.88 (19) |
| O2—S1—C1—C11 | −64.00 (19) | C4—S1—C1—C11 | −179.10 (17) |
| O2—S1—C4—C3 | −117.9 (2) | C11—C16—C15—C14 | 2.0 (4) |
| O3—C2—C3—N2 | 8.7 (4) | C16—C11—C12—C13 | 0.6 (4) |
| O3—C2—C3—C4 | 129.0 (3) | C16—C15—C14—C13 | −0.7 (4) |
| N1—C1—C11—C16 | −50.2 (3) | C15—C14—C13—C12 | −0.6 (4) |
| N1—C1—C11—C12 | 132.9 (2) | C14—C13—C12—C11 | 0.7 (4) |
| N1—C2—C3—N2 | −169.1 (2) | C12—C11—C16—C15 | −1.9 (4) |
| N1—C2—C3—C4 | −48.8 (3) | C5—N1—C1—S1 | −117.4 (2) |
| N1—C5—C6—C7 | −178.9 (3) | C5—N1—C1—C11 | 7.2 (3) |
| N1—C5—C10—C9 | 177.3 (3) | C5—N1—C2—O3 | −9.9 (4) |
| N2—C3—C4—S1 | 170.82 (19) | C5—N1—C2—C3 | 167.9 (2) |
| C1—S1—C4—C3 | −2.7 (2) | C5—C6—C7—C8 | 1.1 (5) |
| C1—N1—C2—O3 | 169.2 (3) | C6—C5—C10—C9 | −0.8 (4) |
| C1—N1—C2—C3 | −13.1 (4) | C6—C7—C8—C9 | 0.3 (6) |
| C1—N1—C5—C6 | −64.9 (3) | C7—C8—C9—C10 | −2.0 (6) |
| C1—N1—C5—C10 | 117.0 (3) | C8—C9—C10—C5 | 2.2 (5) |
| C1—C11—C16—C15 | −178.8 (2) | C10—C5—C6—C7 | −0.9 (4) |
| C1—C11—C12—C13 | 177.7 (2) | C17—N2—C3—C2 | −88.7 (3) |
| C2—N1—C1—S1 | 63.7 (3) | C17—N2—C3—C4 | 148.7 (3) |
| C2—N1—C1—C11 | −171.8 (2) |
N-[(2S,5R)-1,1,4-Trioxo-2,3-diphenyl-1,3-thiazinan-5-yl]acetamide (2) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C1—H1···O2i | 0.98 | 2.39 | 3.365 (3) | 173 |
| C4—H4A···O4ii | 0.97 | 2.29 | 3.185 (4) | 153 |
| C8—H8···O3iii | 0.93 | 2.51 | 3.378 (5) | 155 |
Symmetry codes: (i) x−1, y, z; (ii) x+1, y, z; (iii) −x+1, y−1/2, −z+1/2.
Funding Statement
Research reported here was conducted on instrumentation funded by NSF (for Bruker AXS system) CHEM-0131112, and SIG S10 grants of the National Institutes of Health (for the Rigaku rotating anode system) under award numbers 1S10OD028589–01 and 1S10RR023439–01 to Dr Neela Yennawar.
References
- Bruker (2016). SMART and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Cannon, K., Gandla, D., Lauro, S., Silverberg, L., Tierney, J. & Lagalante, A. (2015). Intl. J. Chem. 7, 73–84.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Elks, J. & Ganellin, C. R. (1990). Editors. Dictionary of Drugs, p. 382. Cambridge: Chapman and Hall.
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- O’Neil, M. J. (2006). Editor. The Merck Index, 14th ed., p. 349. Whitehouse Station, NJ: Merck & Co. Inc.
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Rigaku OD (2022). CrysAlis PRO. Rigaku Oxford Diffraction, Yarnton, England.
- Ryabukhin, Y. I., Korzhavina, O. B. & Suzdalev, K. F. (1996). Adv. Heterocycl. Chem. 66, 131–190.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Silverberg, L. J. (2020). World patent WO2020231873 A1.
- Silverberg, L. J. & Moyer, Q. J. (2019). Arkivoc, (i), 139-227.
- Surrey, A. R. (1963a). US Patent 3082209.
- Surrey, A. R. (1963b). US Patent 3093639.
- Surrey, A. R., Webb, W. G. & Gesler, R. M. (1958). J. Am. Chem. Soc. 80, 3469–3471.
- Tanaka, R. & Horayama, N. (2005). Anal. Sci. X, 21, X57–X58.
- Yennawar, H. P., Noble, D. J. & Silverberg, L. J. (2017). Acta Cryst. E73, 1417–1420. [DOI] [PMC free article] [PubMed]
- Yennawar, H. P. & Silverberg, L. J. (2014). Acta Cryst. E70, o133. [DOI] [PMC free article] [PubMed]
- Yennawar, H. P. & Silverberg, L. J. (2015). Acta Cryst. E71, e5. [DOI] [PMC free article] [PubMed]
- Yennawar, H. P., Singh, H. & Silverberg, L. J. (2015). Acta Cryst. E71, 62–64. [DOI] [PMC free article] [PubMed]
- Yennawar, H. P., Yang, Z. & Silverberg, L. J. (2016). Acta Cryst. E72, 1541–1543. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) 1, 2. DOI: 10.1107/S2056989023000695/hb8050sup1.cif
Structure factors: contains datablock(s) 1. DOI: 10.1107/S2056989023000695/hb80501sup2.hkl
Supporting information file. DOI: 10.1107/S2056989023000695/hb80501sup4.mol
Structure factors: contains datablock(s) 2. DOI: 10.1107/S2056989023000695/hb80502sup3.hkl
Supporting information file. DOI: 10.1107/S2056989023000695/hb80502sup5.mol
Supporting information file. DOI: 10.1107/S2056989023000695/hb80501sup6.cml
Supporting information file. DOI: 10.1107/S2056989023000695/hb80502sup7.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report