Abstract
The apolipoprotein E (APOE) ε4 allele is strongly linked with cerebral β-amyloidosis, but its relationship with tauopathy is less established. We investigated the relationship between APOE ε4 carrier status, regional amyloid-β (Aβ), MRI volumetrics, tau positron emission tomography (PET), APOE mRNA expression maps, and cerebrospinal fluid phosphorylated tau (CSF ptau181). 350 participants underwent imaging and 270 had ptau181. Models evaluated the main effect of APOE ε4 carrier status on regional neuroimaging values and then the interaction of ε4 status and global Aβ on regional tau PET and brain volumes as well as CSF ptau181 values. A final model examined the additional interactive influence of sex. We found, for the same level of Aβ burden, APOE ε4 carriers showed greater tau PET signal relative to non-carriers in temporal regions, but no interaction was present for MRI volumes or CSF ptau181. This potentiation of tau aggregation irrespective of sex occurred in brain regions with high APOE mRNA expression, suggesting local vulnerabilities to tauopathy. There were greater effects of APOE genotype in females, although the interactive sex effects did not strongly mirror mRNA expression.
Keywords: Alzheimer Disease, Tau PET, APOE, Amyloid PET, Apoliporotein E, CSF, Genetic
One Sentence Summary:
APOE ε4 genotype is associated with greater tau PET levels for the same level of Aβ PET in regions of high APOE mRNA expression, but not for CSF tau.
1. Introduction
The apolipoprotein E (APOE) ε4 allele is the strongest genetic risk factor for late-onset Alzheimer disease (AD) (1). APOE has three alleles: ε3 is the most common, followed by ε4 and ε2. The ε2 allele is associated with a decreased risk of AD relative to ε3 whereas the presence of the ε4 allele increases the risk of AD dementia and is associated with an earlier age of symptom onset (2, 3). Moreover, the gene acts in a dose-dependent manner, with one ε4 allele increasing the risk of AD by ~3–4 fold and two ε4 alleles increasing the risk by ~12 fold (2, 4).
Given its prominent role in AD, it is critical to understand the underlying mechanism of the APOE ε4 allele and its effects on AD pathophysiology. The apoe protein has long been linked with amyloid-β (Aβ) plaques, one of the hallmarks of AD, with early research showing that apoe binds to Aβ peptides (5, 6) and that individuals carrying the APOE ε4 allele have greater Aβ plaque pathology compared to non-carriers in postmortem studies (7–10). Later in vivo work, utilizing positron emission tomography (PET) and cerebrospinal fluid (CSF) assays, mirror these postmortem observations. APOE ε4 carriers with the ε4 allele have consistently been shown to have elevated Aβ PET levels relative to non-carriers in a dose-dependent fashion (11) as well as lower CSF Aβ1–42 levels (12–18) indicative of the presence of amyloid (19). Individuals with the APOE ε4 allele also begin accumulating pathology at an earlier age compared to non-carriers (14, 20).
Along with Aβ pathology, neurofibrillary tangles composed of hyperphosphorylated tau protein are a hallmark of AD. Previous work has shown that APOE ε4 carrier status is associated with tau pathology, but the underlying mechanisms of this relationship are still unclear. Specifically, individuals carrying the APOE ε4 allele have greater neurofibrillary tau pathology compared to non-carriers in postmortem studies when Aβ pathology is present (10), but this association disappeared when Aβ pathology was not present (21, 22). Research utilizing CSF tau measures have reported both significant effects of the APOE ε4 allele (18, 23) even when controlling for CSF Aβ1–42 levels (24), as well as weak or no effects (14, 17, 18, 20). Using animal models, it has also been observed that APOE ε4 carrier status exacerbates tau pathology and tau-mediated neurodegeneration independent of Aβ (25). This finding is consistent within several PET studies that show the APOE ε4 is associated with greater levels of tauopathy (26–33), although a minority of work has found the opposite effect (34). These prior analyses have provided important contributions to the literature but have limitations. The PET analyses do not typically examine if there is an interaction between genotype and Aβ (32), rarely includes both CSF and imaging measures of tau (32, 33), and when multiple modalities are included they may come from different cohorts (33). These caveats limit the ability to detect a APOE mediated potentiation of tauopathy as predicted by animal work.
Although genetic polymorphisms such as APOE genotype are often viewed holistically, the relative expression of genes including APOE varies in different brain regions (35, 36). The availability of gene expression data such as the Allen Human Brain Atlas (AHBA) now make it possible to relate AD pathology observed using neuroimaging to underlying topologies of genetic expression that they may reflect (35–40).
The goal of the current study is to evaluate whether APOE ε4 carrier status represents an additive or interaction effect with levels of Aβ in predicting tau PET. By comparing PET and CSF measures, we examined how APOE ε4 carrier status influences both soluble and insoluble forms of tau. Finally, we examined the relationship between APOE ε4 carrier status, regional Aβ and tau PET, and APOE mRNA expression patterns to relate APOE ε4 carrier status with the spatial distribution of APOE mRNA expression, Aβ, and tau in the human brain.
2. Results
2.1. Demographics
In the three hundred fifty individuals, age, sex, education, and racial makeup did not differ between APOE ε4 carrier and noncarrier groups. APOE ε4 carriers had a greater percentage of CDR > 0 participants, lower MMSE scores, higher CDR sum of boxes, as well as higher frequencies of tau and Aβ PET positivity compared to noncarriers (Table 1). For the individuals that had a CDR>0, 14 individuals had a clinical diagnosis of uncertain etiology and 31 had a primary AD diagnosis. The median absolute interval between the Aβ PET scan and the tau PET scan was 28 days (range 1–365), the median absolute lag with the lumbar puncture was 36 days (range 1–688), and the median absolute lag with the clinical visit was 121 days (range 0–562).
Table 1.
Demographic summary for included participants.
| Participant Demographics | APOE ε4 – (n=223) | APOE ε4 + (n=127) | p-value |
|---|---|---|---|
|
| |||
| Age, years | 70.2 (7.9) | 68.9 (8.3) | t = −1.43, p=0.15 |
| Male, n (%) | 103 (46.2) | 52 (41.0) | χ2=0.70, p=0.40 |
| Education, years | 16.3 (2.4) | 16.3 (2.3) | t = −0.07, p=0.95 |
| CDR > 0, n (%) | 18 (8.1) | 27 (21.2) | χ2=11.42, p=0.0007 |
| CDR = 0.5 | 14 (6.3) | 22 (17.3) | |
| CDR > 0.5 | 4 (1.8) | 5 (4.0) | |
| CDR Sum of Boxes | 0.25 (1.04) | 0.65 (1.52) | t = 2.96, p=0.003 |
| MMSE | 29.0 (1.7) | 28.5 (2.5) | t = − 2.30, p=0.02 |
| White, n (%) | 200 (89.7) | 110 (86.6) | χ2 =0.48, p=0.49 |
| Tau PET +, n (%) | 85 (38.1) | 64 (50.4) | χ2 =6.34, p=0.01 |
| Aβ PET +, n (%) | 45 (20.2) | 61 (48.0) | χ2 =26.56, p<0.0001 |
| Aβ Summary SUVR | 1.12 (0.50) | 1.49 (0.73) | t = 5.56, p<0.0001 |
| Tau Summary SUVR | 1.24 (0.31) | 1.38 (0.41) | t = 3.47, p=0.0006 |
Mean (standard deviation) unless otherwise noted.
APOE = apolipoprotein E; CDR = Clinical Dementia Rating; Aβ = amyloid-beta.
2.2. APOE ε4 carrier status, Aβ, tau PET, CSF ptau181, and MRI relationships
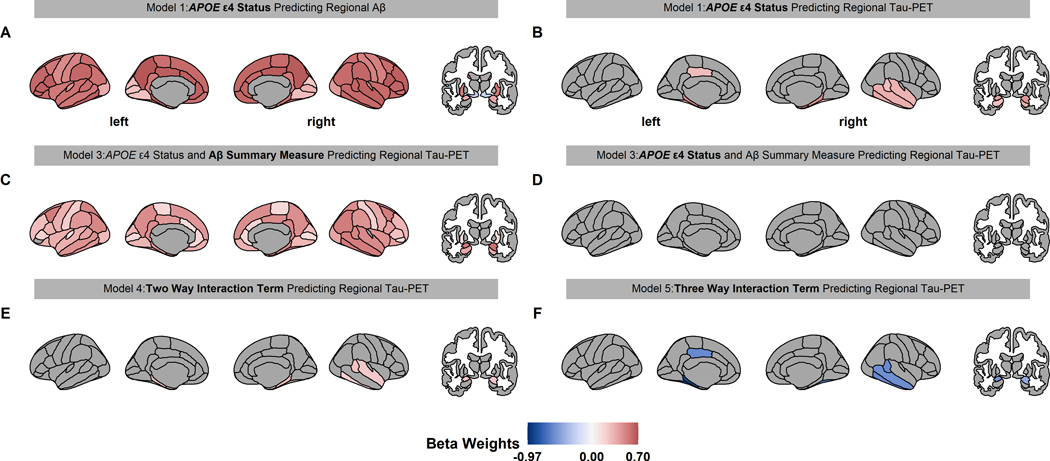
APOE ε4 carriers had higher Aβ PET throughout the cerebral cortex and in the amygdala, hippocampus, and putamen. (Fig. 1A, Table S1). APOE ε4 carriers had higher tau PET primarily in temporal, hippocampus, and amygdala regions (Fig. 1B, Table S2). When APOE ε4 carrier status and Aβ summary measure were included concurrently in models of regional tau PET, we found that the Aβ summary measure, but not APOE ε4 carrier status, significantly predicted tau PET levels (Fig. 1C, Fig. 1D, Table S3). This result suggests shared variance between these measures. Notably, there was significant interaction between APOE ε4 carrier status and the Aβ summary measure, with the ε4 carriers having elevated tau PET relative to non-carriers for the same level of Aβ pathology. This effect was predominately observed in the bilateral entorhinal, parahippocampal, and amygdala regions, as well as the right hemisphere temporal regions, (Fig. 1E, Fig. S1A, Table S4). When the three-way interaction was modeled, a number of regions demonstrated significant interaction between sex, genotype, and Aβ PET levels (Fig. 1F, Fig. 2A). The relationship was such that females had a larger interaction between genotype and continuous Aβ PET values. Full regional results from the tau PET models are in Table S2, S3, S4 and Fig S2.
Fig 1.
Significant beta weights predicting regional Aβ and tau PET for linear models 1–5. The associated linear model is displayed above each brain with the beta weight term in bold text. Red and blue color indicate a positive and negative value, respectively. Only the regions that were statistically significant in figures 1A and B (conjunction) were analyzed in E and F. Aβ PET is increased throughout the brain in APOE ε4 carrier (A). APOE ε4 carriers have higher tau PET in the temporal, amygdala, and hippocampus regions (B). When APOE ε4 carrier status and Aβ PET were both in a model predicting regional tau PET, the Aβ summary measure was associated with tau PET throughout the brain (C), but APOE ε4 carrier status was not associated with tau PET (D). Importantly, the interaction between Aβ summary measure and APOE ε4 carrier status in predicting regional tau PET were significant in the entorhinal, temporal, and amygdala regions (E) and this potentiation varies by sex (F).
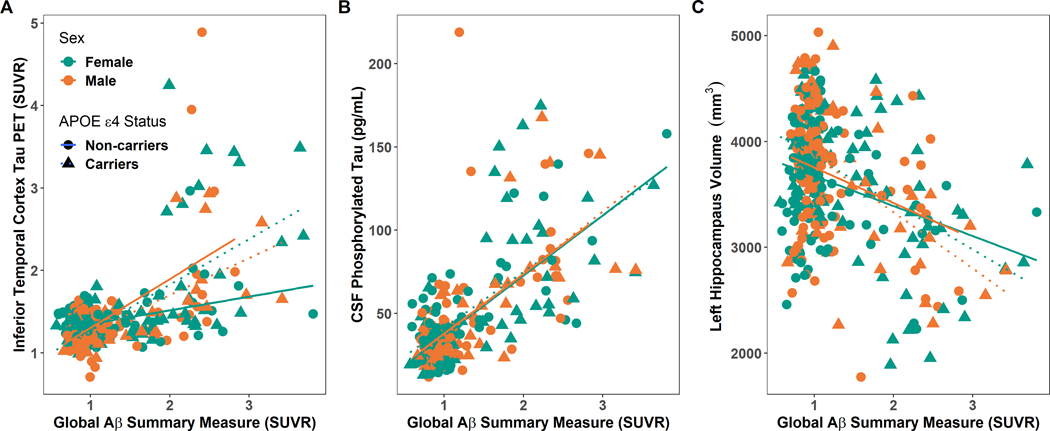
Fig 2.
Comparison between entorhinal tau PET SUVR (A), CSF ptau181 (B) and regional volume (C) relationship relative to Aβ summary measure levels, APOE ε4 carrier status, and sex. The green color represents females and brown color represents males. and blue non-carriers. Triangles and dotted lines represent the fit for APOE ε4 carriers while circles and solid lines represent the fit for APOE ε4 non-carriers.
APOE ε4 carrier status (model 1: β = 0.51, p = 8.11E-6) and Aβ summary measure (model 2: β = 0.60, p = 2.82E-28) separately predicted significant elevations in CSF ptau181. As with tau PET, in the concurrent model, Aβ (model 3: β = 0.57, p = 2.12E-24) but not APOE ε4 carrier status (model 3: β = 0.16, p = 0.11) predicted CSF ptau181 values. Unlike tau PET, there was no two-way interaction between Aβ PET levels and APOE ε4 carrier status on CSF ptau181 (model 4: β = −0.04, p = 0.65, Fig. S1B) or three-way interaction between sex, Aβ PET levels, and APOE ε4 carrier status (model 5: β = 0.02, p = 0.91, Fig. 2B).
When examining regional volumes there were no significant main effects of APOE ε4 carrier status. Higher levels of the Aβ summary measure predicted lower volumes in the left and right hippocampal volumes (Fig. S1C) as well as the right amygdala volume (Table S5). This effect of global Aβ levels in these regions remained significant even in the joint model that included additive main effects of ε4 status (Table S6). There were no significant two-way or three-way interactions (Fig. 2C, Table S7).
2.3. APOE mRNA expression and APOE ε4 carrier status spatial relationship
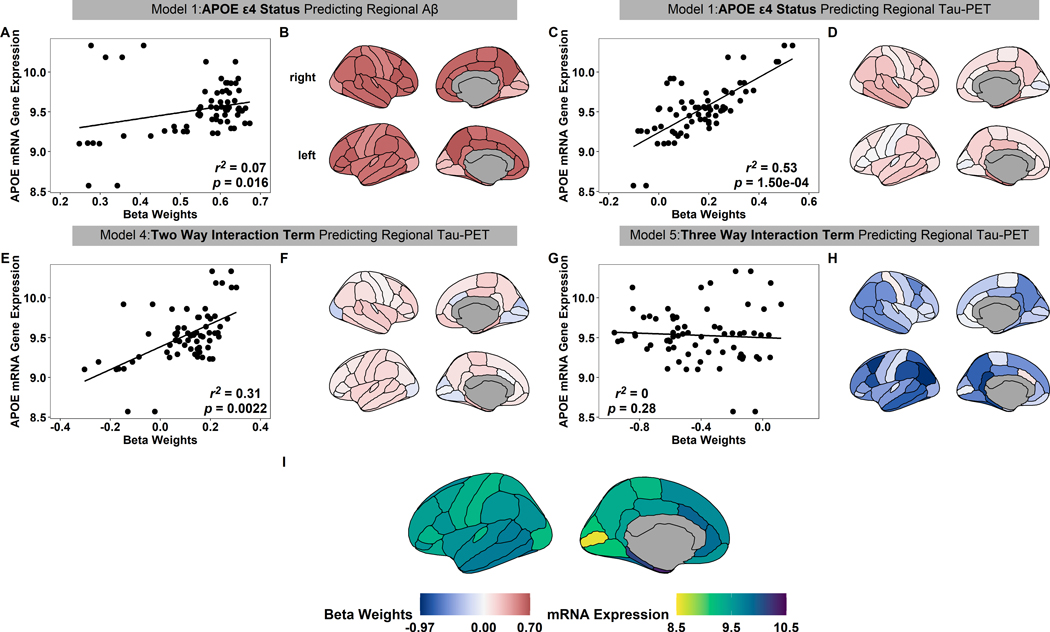
To analyze the spatial association between APOE mRNA and effects of APOE ε4 carriers status, we associated all regional beta weights estimates, non-significant and significant, to APOE mRNA gene expression (Fig. 3I) from the following terms: APOE ε4 carrier status beta weights predicting regional Aβ PET (Fig. 3B), APOE ε4 carrier status beta weights predicting regional tau PET (Fig. 3D), the beta weights for the APOE ε4 carrier status and Aβ summary measure interaction estimated using regional tau PET (Fig. 3F), and the beta weights from the three way-interaction (Fig. 3G). Interestingly, the APOE ε4 carrier status beta weight for regional tau PET (r2 = 0.53, p = 1.50e-04, Fig. 3C) and the interaction between Aβ summary measure and APOE ε4 carrier status beta weight for regional tau PET (r2 = 0.31, p = 0.0022, Fig. 3E) were both significantly associated with the spatial APOE mRNA expression pattern. The spatial pattern of APOE mRNA gene expression has a reduced, albeit still significant, association with the spatial pattern of the main effect of APOE ε4 carrier status estimated on regional Aβ -PET (r2 = 0.07, p = 0.016, Fig. 3A). There was no significant association between the spatial pattern of the three-way interaction between sex, genotype, and Aβ-PET (r2 = 0, p = 0.28, Fig. 3G). Although there were minimal significant main effects of either Aβ or genotype on regional volumes, the relatively pattern of these non-significant effects was strongly related to the spatial pattern of APOE mRNA expression (Fig. S4). This suggests a structured, albeit weak, effect on volumes.
Fig 3.
The spatial relationship between APOE ε4 carrier status and APOE mRNA gene expression. The regional beta weights estimates from the APOE ε4 carrier status beta weights estimated on regional Aβ PET (B), APOE ε4 carrier status beta weights estimated on regional tau PET (D), the APOE ε4 carrier status and Aβ summary measure interaction estimated on regional tau PET (F), and the weights from the three-way interaction between sex, Aβ, and regional tau (H). The relationship between each beta weight and APOE mRNA gene expression (A,C,E,G). In the regression plots, black circles represent the beta weight of distinct brain regions, the black line indicates the linear regression fit, and the r2 and p-values from spatial correlation tests are presented in the bottom right. APOE mRNA gene expression values for each region for reference (I).
3. Discussion
Given that tau is more strongly associated with cortical atrophy and cognitive decline compared to Aβ pathology (53–55), understanding the mechanism underpinning tau pathology could be key to the prevention of AD. Furthermore, possession of the APOE ε4 allele has been identified as posing the greatest genetic risk factor for developing late-onset AD. Therefore, it is imperative to investigate whether there is a relationship between APOE ε4 and tau deposition and to quantify what influence this relationship has above that of Aβ. While a strong association between the APOE ε4 allele and Aβ pathology is well-known (10, 12, 14), the ε4 allele relationship with tauopathy is less established with inconsistent findings from postmortem, CSF, or PET studies (10, 17, 18, 20, 26–33). The goals of this study were to examine the influence of APOE ε4 carrier status on PET and CSF measures of tau pathology, test if there was an interaction between Aβ pathology and APOE ε4 carrier status, and to compare how the spatial impact of this pathology relates to regional levels of APOE mRNA gene expression.
There exists an overwhelming body of literature describing the widespread effect of APOE ε4 on Aβ in postmortem (10), neuroimaging (56), biofluid (13, 16) studies. In contrast, the relationship between APOE ε4 and tau has been less consistent in the literature. Several postmortem studies have found the presence of the APOE ε4 allele increased tauopathy (10, 57) revealing greater pathology in diffuse cortical areas in AD patients compared to APOE ε4 noncarriers (57), while other studies did not find this relationship (58). Studies using CSF measures of tau pathology reported no association with APOE ε4 carrier status (13, 14, 17, 59), as well as significantly elevated levels of tau (23, 33, 60, 61). There are also mixed results on the relationship within PET studies where many have shown greater tau load on APOE ε4 carriers compared to noncarriers (27, 28, 30–32, 62), while others have not (34).
In the current study, we found a main effect where participants carrying the APOE ε4 allele had elevated levels of both CSF ptau181 and tau PET relative to non-carriers. However, when a summary measure of Aβ PET was included as a covariate in the model, the main effect of APOE ε4 carrier status was greatly reduced, suggesting much of the influence that the APOE ε4 carrier status has on tau pathology is mediated through its regulation of Aβ. This suggests minimal additive influences of APOE and Aβ levels. When examining the interaction between APOE ε4 carrier status and Aβ, we found a significant effect for tau PET but not CSF ptau181 levels, where the presence of an APOE ε4 allele potentiated the degree of tauopathy above the effects that can be ascribed to Aβ alone. This result is consistent with animal models where P301S tau transgenic mice expressing human APOE ε4 exhibited greater tau burden as well as neurodegeneration as compared to mice expressing APOE ε2 or APOE ε3 with the absence of APOE being protective (25).
In the current analyses the three-way interaction suggests a greater influence of the ε4 allele in women. This result is consistent with prior work suggesting APOE genotype has a differential AD risk by sex (63, 64) and may have a sex-dependent effect on in vivo measures of tau pathology (62). Although the size of our population is robust (n=350) it is still modestly powered to estimate the three-way interaction between genotype, levels of Aβ, and sex. As a result, replication of this interaction between sex and genotype using additional cohorts in future studies is warranted. Although there was a modest effect of Aβ levels on medial temporal volumes, we found no evidence of potentiation by ε4 status. This is consistent with prior human work (20), but a difference from mouse models (25).
The discrepancy between CSF and PET is not entirely unexpected given the modest correlations seen between these modalities in literature (55, 65–67) suggesting they capture unique properties of the disease. The APOE ε4 carrier status may only exert its influence selectively in insoluble, aggregated, forms of tau. There is also emerging evidence that tau phosphorylated at different sites such as ptau217 and ptau231 changes quite early in the diseases (68–70), while tau PET is relatively late (71). Increased levels of ptau181, ptau205, ptau217, ptau231 may reflect a response to amyloidosis more than the aggregation of tau into neurofibrillary tangles per se. As a result the difference may not be due to the soluble and insoluble distinction, but instead an erroneous conceptualization in the field about how strongly ptau reflects tauopathy as defined neuropathological and with PET. Future analyses of other ptau phosphorylation sites, as well as other candidate markers such as microtubule binding region (MTBR)(72), may provide important insight into how APOE genotype influences biofluid measures beyond ptau181.
The observed genotype effects on Aβ were prominent in medial parietal and frontal areas, although elevation was observed across the cortex. Elevated tau was most prominently observed in temporal, hippocampus, and amygdala regions. These spatial signatures (Fig. 1) are highly consistent with the stereotypical patterns of these pathologies found in the literature (48, 55). While APOE is often viewed holistically, its mRNA expression levels vary across the brain and spatially resemble structural and functional networks (35, 73). To understand our findings in relation to genetic expression in the cortex, we utilized the AHBA APOE mRNA genetic expression data translated to FreeSurfer regions (50). We found that higher regional APOE mRNA gene expression levels in the brain are more associated with the APOE ε4 influence on regional tau, rather than regional Aβ. This suggests that the local levels of APOE expression in the tissue may regulate each region’s vulnerability to tauopathy.
The mechanism through which APOE influences tau pathology are unclear but there is building evidence that it has an immunomodulatory function (74, 75). ApoE plays a role in regulating microglial metabolism which is tied to microglial activation (76). Removal of astrocyte-derived apoE reduces tau-associated neurodegeneration (77), and overexpression of low-density lipoprotein receptor, an apoE metabolic receptor, alters markers of microglial suppression (76). The apoE protein may be an important therapeutic target, and lowering apoE ε4 levels with antisense oligonucleotides has been shown to reduce tauopathy and neurodegeneration in mouse models. (78). The APOE ε4 allele has also been shown to lead to blood-brain barrier dysfunction in the temporal lobe (79) which may also impact inflammation.
There are many strengths in this study. First, as there are discrepancies in literature on the APOE ε4 and tau relationship, we incorporated and compared both PET and CSF modalities. While there are suggestions that the effects of Aβ and APOE ε4 carrier status may be additive, we explicitly tested both additive and interactive effects. When performing these analyses Aβ was analyzed as a continuous variable rather than binarized into positive or negative. Such an approach avoids potential confounds such as genotype serving simply as a proxy of Aβ level or ε4 carriers on averaging simply having higher levels of Aβ. Our cohort contains over three hundred individuals, providing a robust sample for analysis. Finally, we integrated mRNA gene expression data from the AHBA to gain a more comprehensive understanding of the relationship. To our knowledge, this is the first study to compare the regional APOE mRNA gene expression patterns, tau PET, and Aβ PET effect to APOE.
There were also limitations. The current work focuses on the overall effect of the APOE ε4 allele without considering gene dosage as the analyses only had 18 ε4 homozygotes. The cohort is also primarily white, limiting the generalizability of APOE genotype effects to non-white cohorts. While the most comprehensive data of its kind, the AHBA mRNA dataset is derived from only a handful of individual brains that did not have AD. This means that it is only a rough approximation of mRNA expression and precludes the ability to examine whether mRNA expression varies as a function of demographic factors such as sex. There are also minimal samples available from the right hemisphere, resulting in the mRNA gene expression values being mirrored across hemispheres. This limits the ability to look at hemispheric specific effects. Additionally, AHBA data represents an aggregate expression in a bulk tissue sample. As APOE is primarily produced by astrocytes (80), the spatial association seen between APOE mRNA levels and tau pathology may be driven by the heterogeneity in cell distribution across the brain rather than the spatial distribution of APOE itself. The AHBA data has become an invaluable tool to the field. As comparing data from the AHBA to in vivo human imaging data is becoming common place (35–40), it would benefit the field if this data were expanded to provide opportunities to ask more in-depth questions. Given the strong animal work linking this gene to tau pathology (25, 74), we a priori looked at only APOE gene expression. Future research endeavors should expand analyses to consider additional genes or networks of gene expression. Finally, our cohort was comprised of individuals who were classified as either cognitively normal or who had mild dementia. Therefore, our results cannot directly assess how the influence of APOE may vary as dementia progressively worsens.
Conclusion
We found that presence of the APOE ε4 allele influences levels of Aβ PET, tau PET, and CSF ptau181. We additionally found that an interaction of APOE ε4 and Aβ PET is associated with elevated regional tau PET but not CSF ptau181 levels. The spatial pattern of the interaction effect on tau PET is mirrored by the levels of APOE mRNA gene expression in the cortex. Our results further elucidate the influence this prominent risk allele has on the pathogenesis of AD. Therefore, APOE ε4 carrier status needs to be considered for clinical trials targeting tau hyperphosphorylation or aggregation.
4. Materials and Methods
4.1. Participants
Participants were enrolled in the longitudinal studies of memory and aging at the Charles F. and Joanne Knight Alzheimer Disease Research Center (Knight ADRC) at Washington University in Saint Louis. Dementia severity was defined by the global Clinical Dementia Rating© (CDR©) (41), where CDR 0 indicates cognitive normality and CDR > 0 indicates cognitive impairment. For this study, three hundred and fifty participants were included who had a tau PET scan, an Aβ PET scan, a clinical assessment, and an APOE genotype assessment between years 2014 and 2018. APOE genotyping was performed as previously described (42). Participants who had one or more ε4 allele were assigned a positive APOE ε4 carrier status, while those with no ε4 allele were assigned a negative APOE carrier ε4 status. To fulfill the study criteria, each participant’s Aβ PET and clinical assessments were required to have been completed within one year of their tau PET visit. Of the three hundred and fifty participants, two hundred and seventy of the participants had CSF ptau181 within two years of Aβ PET. A summary table of demographic information for these individuals is provided in table 1. Data from the Knight ADRC can be freely requested (https://knightadrc.wustl.edu/professionals-clinicians/request-center-resources/).
4.2. Ethics Statement
All participants, or their caregivers, signed a standard informed consent document, and the Institutional Review Board at Washington University in St. Louis approved all procedures. ICMJE guidelines were followed in preparation of the manuscript.
4.3. Imaging Acquisition and Analysis
Structural T1-weighted scans were acquired on three Siemens MRI 3 Tesla scanners using a magnetization-prepared rapid gradient-echo sequence. The Siemens Biograph mMR (n = 346) and the MAGNETOM Vida (n = 3) T1 scans were acquired with a 1 × 1 × 1.2 mm resolution, 2300 ms repetition time, 2.95 ms echo time, 9 degree flip angle, 176 frames, and a 240 × 256 field of view in sagittal orientation. Structural T1 scans for the Siemens TIM Trio (n = 1) were acquired with a 1 × 1 × 1 mm resolution, 2400 ms repetition time, 3.16 echo time, 8 degree flip angle, 176 frames, and a 256 × 256 field of view in sagittal orientation. MRI images were processed through FreeSurfer v5.3-HCP (43) and were visually inspected. The FreeSurfer ROIs were subsequently utilized for PET processing. Tau PET imaging was performed on the Siemens Biograph PET CT using the 18F-flortaucipir tracer and Aβ PET imaging was performed on the Siemens PET/MR using 18F-florbetapir. All PET scans were processed through the PET Unified Pipeline (PUP, https://github.com/ysu001/PUP) using an 80-to-100-minute post-injection window for 18F-flortaucipir and a 50-to-70-minute post-injection window for the 8F-florbetapir tracer. The standardized uptake value ratio (SUVR) was calculated relative to the cerebellar cortex using the derived FreeSurfer ROIs (44, 45). Partial volume correction was performed using a geometric transfer matrix (44, 46). To calculate the Aβ summary measure, we averaged the left and right hemisphere partial volume corrected SUVRs from the lateral orbitofrontal, mesial orbitofrontal, rostral mesial frontal, superior frontal, superior temporal, mesial temporal, and precuneus regions as previously defined (45). Aβ positivity was defined using a cutoff of 1.22 for AV45 (Centiloid value 22.2(47)). As previously reported (48) tau PET positivity was defined as a value >1.22 from using an arithmetic mean of partial volume corrected SUVR values from the entorhinal cortex, amygdala, inferior temporal gyrus, and lateral occipital cortex.
4.4. CSF Assays
CSF (20 to 30 ml) was collected via lumbar puncture after overnight fasting as previously described (19). Samples were analyzed using the automated Lumipulse assay platform (LUMIPULSE G1200, Fujirebio, Malvern, PA) to determine levels of tau phosphorylated at threonine 181 (ptau181).
4.5. APOE Gene Expression
To analyze the genetic spatial relationship, we obtained the AHBA APOE gene expression data (49). The APOE gene expression data was summarized from 58,692 measurements of gene expression from postmortem brains of six cognitively normal individuals and transformed onto the Desikan-Killiany cortical atlas built into FreeSurfer (50). In brief, mean averaged gene expression values were calculated for each gene from multiple probes and were spatially mapped from MNI152 space to FreeSurfer cortical regions. As there was limited data for the right hemisphere, gene expression data converted into FreeSurfer regions only included the 34 left hemisphere cortical regions. For the six individual brains, the median gene expression value for each cortical region was determined, and a summary brain map was created by calculating the median from the six median gene expression values for each region (50).
4.6. Statistical Analysis
All statistical tests were done in R version 4.0.3 (51). Demographics were compared between ε4 carriers and non-carriers using t-tests and Chi Squared tests as appropriate. To answer the main questions of interest first, we performed linear models to predict regional Aβ and regional tau PET levels, regional volume derived from MRI, and CSF ptau181 from APOE ε4 carrier status (model 1).
Models 1 and 2 were run for each region independently with regional SUVRs or regional volumes or CSF ptau181 as the dependent variable. P-values were corrected for multiple comparisons using the Benjamini-Hochberg procedure at a false discovery rate of 0.05. Subsequently, we ran additional linear models to examine the additive and interaction effects between the Aβ summary measure and APOE ε4 carrier status on regional tau PET, regional volumes, or CSF ptau181 (models 3 and 4). The final model included a three-way interaction between sex, the Aβ summary measure, and APOE ε4 carrier status (model 5). For the regional tau PET interaction models, only the regions that were found to be statistically significant in model 1 predicting regional Aβ PET or regional tau PET respectively were analyzed in model 4 and 5. For all models age at tau PET visit and sex were included as covariates, and all continuous variables were scaled in all linear models to aid interpretation.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
To examine the relationship between spatial APOE mRNA expression, APOE ε4 carrier status, and Aβ summary measure, we performed a spatial rotational permutation as previously described (52). In brief, this method defines a set of null correlations by comparing the empirical correlation against two spatial maps by randomly rotating the spherical projections of one spatial map before projecting it back to the brain surface. The projection conserves spatial continuity of the empirical maps as well as hemispheric symmetry. Specifically, the p-value is derived by comparing the empirical Spearman’s ρ to a null distribution of 10,000 correlations between one empirical map and the randomly rotated spherical projections using the total APOE mRNA expression values from the AHBA and the beta weights from the interaction term in model 4. The code used to perform spatial permutation testing can be downloaded here: https://github.com/frantisekvasa/rotate_parcellation. The APOE mRNA expression data was only available for the left hemisphere, so the gene expression data was mirrored for the left and right hemisphere for these analyses.
Supplementary Material
Fig. S1. Comparison between the entorhinal tau PET SUVR (A), CSF ptau181 (B), and hippocampus volume (C) relationship relative to Aβ summary measure levels and APOE ε4 carrier status.
Fig. S2. Models predicting regional tau PET SUVRs. Values represent the non-significant and significant model estimates.
Fig. S3. Models predicting MRI regional volumes. Values represent the non-significant and significant model estimates.
Fig. S4. Relating regional weights from the models examining main effects of APOE genotype (A,B), the main effects of Aβ (C,D), the interaction between Aβ and genotype (E,F) and the three-way interaction additionally with sex (G,H) relative to APOE mRNA expression derived from the Allan Human Brain Atlas (I). Although there were minimal significant main effects of either Aβ or genotype on regional volumes, the relatively pattern of these non-significant effects was strongly related to the spatial pattern of APOE mRNA expression. This suggests a structured, albeit weak, effect on volumes.
Table S1. Effects of APOE ε4 status on Regional Aβ PET (Model 1).
Table S2. Individual effects of APOE ε4 status and Aβ summary measure on regional tau PET (Model 1 and 2, respectively).
Table S3. Concurrent effects of APOE ε4 status and Aβ summary measure on regional tau PET (Model 3).
Table S4. Interaction effects of APOE ε4 status, Aβ summary measure, and sex on regional tau PET (Model 4 and 5, respectively).
Table S5. Individual effects of APOE ε4 status and Aβ summary measure on regional volume (Model 1 and 2, respectively).
Table S6. Concurrent effects of APOE ε4 status and Aβ summary measure on regional volume (Model 3).
Table S7. Interaction effects of APOE ε4 status, Aβ summary measure, and sex on regional volume (Model 4 and 5, respectively).
Acknowledgements
We acknowledge the altruism of the participants and their families and contributions of the Knight ADRC research and support staff for their contributions to this work. Without the generous contribution and time the participants gave for these studies, this work would not be possible.
Funding
Support for these analyses was provided by NIH (P30AG066444, P01AG003991, P01AG026276, K01AG053474, RF1AG053303, RF1AG044546, UL1TR000448, P30NS098577, R01EB009352), Alzheimer’s Association (AARG-17-532945), Alzheimer Association International Research Program (AARFD-20-681815), and The National Science Foundation (DGE-1745038). Computations were performed using the facilities of the Washington University Center for High Performance Computing, which were partially funded by NIH grants 1S10RR022984-01A1 and 1S10OD018091-01. We also acknowledge support from the Hope Center for Neurological Disorders, the Barnes-Jewish Hospital Foundation Willman Scholar Award, and Avid Radiopharmaceuticals (a wholly owned subsidiary of Eli Lilly) which provided technology transfer and provided precursor for AV-1451.
Abbreviations:
- AD
Alzheimer disease
- APOE
apolipoprotein E
- Aβ
amyloid-β
- PET
positron emission tomography
- CSF
cerebrospinal fluid
- AHBA
Allen Human Brain Atlas
- Knight ADRC
Charles F. and Joanne Knight Alzheimer Disease Research Center
- CDR
Clinical Dementia Rating
- PUP
PET Unified Pipeline
- SUVR
standardized uptake value ratio
Footnotes
Declaration of Competing Interest
The following authors have no conflicts of interest regarding this study: AD, CDC, NSM, AM, LNK, SF, SJK, RLF, RCH, AMF, JCM, SES. SAS receives research support from The National Science Foundation. NJM is supported partly by the Alzheimer Association International Research Program. CC received grants from NIH and reports fees from GSK and Takeda. DMH is a Co-founder, C2N Diagnostics LLC serves on the Scientific advisory boards or consults for Genentech, Denali, C2N Diagnostics, Cajal Neurosciences, Takeda, Casma, and Eli Lilly. DMH is an inventor on a 1) a patent licensed by Washington University to C2N Diagnostics on the therapeutic use of anti-tau antibodies. This anti-tau antibody program has now been licensed to Abbvie; 2) a patent licensed by Washington University to Eli Lilly on a humanized anti-Aβ antibody. The lab of DMH receives research grants from the National Institutes of Health, Cure Alzheimer’s Fund, Tau Consortium, the JPB Foundation, Good Ventures, C2N Diagnostics, NextCure, and Denali. TLSB has investigator initiated research funding from the NIH, the Alzheimer’s Association, the Barnes-Jewish Hospital Foundation and Avid Radiopharmaceuticals (a wholly owned subsidiary of Eli Lilly) Dr. Benzinger participates as a site investigator in clinical trials sponsored by Avid Radiopharmaceuticals, Eli Lilly, Biogen, Eisai, Jaansen, and Roche. She serves as an unpaid consultant to Eisai and Siemens. She is on the Speaker’s Bureau for Biogen.
References
- 1.Karch CM, Goate AM, Alzheimer’s disease risk genes and mechanisms of disease pathogenesis, Biol. Psychiatry 77, 43–51 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA, Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families, Science (80-. ). 261, 921–923 (1993). [DOI] [PubMed] [Google Scholar]
- 3.Poirier J, Bertrand P, Poirier J, Kogan S, Gauthier S, Poirier J, Gauthier S, Davignon J, Bouthillier D, Davignon J, Apolipoprotein E polymorphism and Alzheimer’s disease, Lancet 342, 697–699 (1993). [DOI] [PubMed] [Google Scholar]
- 4.Holtzman DM, Herz J, Bu G, Apolipoprotein E and apolipoprotein E receptors: Normal biology and roles in Alzheimer disease, Cold Spring Harb. Perspect. Med 2, a006312–a006312 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Namba Y, Tomonaga M, Kawasaki H, Otomo E, Ikeda K, Apolipoprotein E immunoreactivity in cerebral amyloid deposits and neurofibrillary tangles in Alzheimer’s disease and kuru plaque amyloid in Creutzfeldt-Jakob disease, Brain Res. 541, 163–166 (1991). [DOI] [PubMed] [Google Scholar]
- 6.Wisniewski T, Frangione B, Apolipoprotein E: A pathological chaperone protein in patients with cerebral and systemic amyloid, Neurosci. Lett 135, 235–238 (1992). [DOI] [PubMed] [Google Scholar]
- 7.Schmechel DE, Saunders AM, Strittmatter WJ, Crain BJ, Hulette CM, Joo SH, Pericak-Vance MA, Goldgaber D, Roses AD, Increased amyloid β-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease, Proc. Natl. Acad. Sci. U. S. A 90, 9649–9653 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olichney JM, Hansen LA, Galasko D, Saitoh T, Hofstetter CR, Katzman R, Thal LJ, The apolipoprotein E ε4 allele is associated with increased neuritic plaques and cerebral amyloid angiopathy in Alzheimer’s disease and Lewy body variant, Neurology 47, 190–196 (1996). [DOI] [PubMed] [Google Scholar]
- 9.Polvikoski T, Sulkava R, Haltia M, Kainulainen K, Vuorio A, Verkkoniemi A, Niinistö L, Halonen P, Kontula K, Apolipoprotein E, Dementia, and Cortical Deposition of β-Amyloid Protein, N. Engl. J. Med 333, 1242–1248 (1995). [DOI] [PubMed] [Google Scholar]
- 10.Tiraboschi P, Hansen LA, Masliah E, Alford M, Thal LJ, Corey-Bloom J, Impact of APOE genotype on neuropathologic and neurochemical markers of Alzheimer disease, Neurology 62, 1977–1983 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Reiman EM, Chen K, Liu X, Bandy D, Yu M, Lee W, Ayutyanont N, Keppler J, Reeder SA, Langbaum JBS, Alexander GE, Klunk WE, Mathis CA, Price JC, Aizensteini HJ, DeKosky ST, Caselli RJ, Fibrillar amyloid-β burden in cognitively normal people at 3 levels of genetic risk for Alzheimer’s disease, Proc. Natl. Acad. Sci. U. S. A 106, 6820–6825 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vemuri P, Wiste HJ, Weigand SD, Knopman DS, Shaw LM, Trojanowski JQ, Aisen PS, Weiner M, Petersen RC, Jack CR, Effect of APOE on biomarkers of amyloid load and neuronal pathology in AD, Ann. Neurol 67, NA-NA (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lautner R, Palmqvist S, Mattsson N, Andreasson U, Wallin A, Pålsson E, Jakobsson J, Herukka SK, Owenius R, Olsson B, Hampel H, Rujescu D, Ewers M, Landén M, Minthon L, Blennow K, Zetterberg H, Hansson O, Apolipoprotein e genotype and the diagnostic accuracy of cerebrospinal fluid biomarkers for alzheimer disease, JAMA Psychiatry 71, 1183–1191 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Morris JC, Roe CM, Xiong C, Fagan AM, Goate AM, Holtzman DM, Mintun MA, APOE predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging, Ann. Neurol 67, 122–131 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galasko D, Chang L, Motter R, Clark CM, Kaye J, Knopman D, Thomas R, Kholodenko D, Schenk D, Lieberburg I, Miller B, Green R, Basherad R, Kertiles L, Boss MA, Seubert P, High cerebrospinal fluid tau and low amyloid β42 levels in the clinical diagnosis of Alzheimer disease and relation to apolipoprotein E genotype, Arch. Neurol 55, 937–945 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Fagan AM, Younkin LH, Morris JC, Fryer JD, Cole TG, Younkin SG, Holtzman DM, Differences in the Aβ40/Aβ42 ratio associated with cerebrospinal fluid lipoproteins as a function of apolipoprotein E genotype, Ann. Neurol 48, 201–210 (2000). [PubMed] [Google Scholar]
- 17.Sunderland T, Mirza N, Putnam KT, Linker G, Bhupali D, Durham R, Soares H, Kimmel L, Friedman D, Bergeson J, Csako G, Levy JA, Bartko JJ, Cohen RM, Cerebrospinal fluid β-amyloid 1–42 and tau in control subjects at risk for Alzheimer’s disease: The effect of APOE ε4 allele, Biol. Psychiatry 56, 670–676 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Risacher SL, Kim S, Nho K, Foroud T, Shen L, Petersen RC, Jack CR, Beckett LA, Aisen PS, Koeppe RA, Jagust WJ, Shaw LM, Trojanowski JQ, Weiner MW, Saykin AJ, Alzheimer’s Disease Neuroimaging Initiative (ADNI), APOE effect on Alzheimer’s disease biomarkers in older adults with significant memory concern, Alzheimer’s Dement. 11, 1417–1429 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fagan AM, Mintun MA, Mach RH, Lee SY, Dence CS, Shah AR, LaRossa GN, Spinner ML, Klunk WE, Mathis CA, DeKosky ST, Morris JC, Holtzman DM, Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Abeta;42 in humans, Ann. Neurol 59, 512–519 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Mishra S, Blazey TM, Holtzman DM, Cruchaga C, Su Y, Morris JC, Benzinger TLS, Gordon BA, Longitudinal brain imaging in preclinical Alzheimer disease: Impact of APOE ϵ4 genotype, Brain 141, 1828–1839 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farfel JM, Yu L, De Jager PL, Schneider JA, Bennett DA, Association of APOE with tau-tangle pathology with and without β-amyloid, Neurobiol. Aging 37, 19–25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crary JF, Trojanowski JQ, Schneider JA, Abisambra JF, Abner EL, Alafuzoff I, Arnold SE, Attems J, Beach TG, Bigio EH, Cairns NJ, Dickson DW, Gearing M, Grinberg LT, Hof PR, Hyman BT, Jellinger K, Jicha GA, Kovacs GG, Knopman DS, Kofler J, Kukull WA, Mackenzie IR, Masliah E, McKee A, Montine TJ, Murray ME, Neltner JH, Santa-Maria I, Seeley WW, Serrano-Pozo A, Shelanski ML, Stein T, Takao M, Thal DR, Toledo JB, Troncoso JC, Vonsattel JP, White CL, Wisniewski T, Woltjer RL, Yamada M, Nelson PT, Primary age-related tauopathy (PART): a common pathology associated with human aging, Acta Neuropathol. 128, 755–766 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Risacher SL, Kim S, Shen L, Nho K, Foroud T, Green RC, Petersen RC, Jack CR, Aisen PS, Koeppe RA, Jagust WJ, Shaw LM, Trojanowski JQ, Weiner MW, Saykin AJ, The role of apolipoprotein E (APOE) genotype in early mild cognitive impairment (E-MCI), Front. Aging Neurosci 5, 11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cruchaga C, Kauwe JSK, Harari O, Jin SC, Cai Y, Karch CM, Benitez BA, Jeng AT, Skorupa T, Carrell D, Bertelsen S, Bailey M, McKean D, Shulman JM, De Jager PL, Chibnik L, Bennett DA, Arnold SE, Harold D, Sims R, Gerrish A, Williams J, Van Deerlin VM, Lee VMY, Shaw LM, Trojanowski JQ, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD, Peskind ER, Galasko D, Fagan AM, Holtzman DM, Morris JC, Goate AM, GWAS of cerebrospinal fluid tau levels identifies risk variants for alzheimer’s disease, Neuron 78, 256–268 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Y, Yamada K, Liddelow SA, Smith ST, Zhao L, Luo W, Tsai RM, Spina S, Grinberg LT, Rojas JC, Gallardo G, Wang K, Roh J, Robinson G, Finn MB, Jiang H, Sullivan PM, Baufeld C, Wood MW, Sutphen C, McCue L, Xiong C, Del-Aguila JL, Morris JC, Cruchaga C, Fagan AM, Miller BL, Boxer AL, Seeley WW, Butovsky O, Barres BA, Paul SM, Holtzman DM, ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy, Nature 549, 523–527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weigand AJ, Thomas KR, Bangen KJ, Eglit GML, Delano-Wood L, Gilbert PE, Brickman AM, Bondi MW, APOE interacts with tau PET to influence memory independently of amyloid PET in older adults without dementia, Alzheimer’s Dement. 17, 61–69 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Therriault J, Benedet AL, Pascoal TA, Mathotaarachchi S, Chamoun M, Savard M, Thomas E, Kang MS, Lussier F, Tissot C, Parsons M, Qureshi MNI, Vitali P, Massarweh G, Soucy JP, Rej S, Saha-Chaudhuri P, Gauthier S, Rosa-Neto P, Association of Apolipoprotein e ϵ4 with Medial Temporal Tau Independent of Amyloid-β, JAMA Neurol. 77, 470–479 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baek MS, Cho H, Lee HS, Lee JH, Ryu YH, Lyoo CH, Effect of APOE ε4 genotype on amyloid-β and tau accumulation in Alzheimer’s disease, Alzheimer’s Res. Ther 12, 140 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jack CR, Wiste HJ, Weigand SD, Therneau TM, Lowe VJ, Knopman DS, Botha H, Graff-Radford J, Jones DT, Ferman TJ, Boeve BF, Kantarci K, Vemuri P, Mielke MM, Whitwell J, Josephs K, Schwarz CG, Senjem ML, Gunter JL, Petersen RC, Predicting future rates of tau accumulation on PET, Brain 143, 3136–3150 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.La Joie R, Visani AV, Lesman-Segev OH, Baker SL, Edwards L, Iaccarino L, Soleimani-Meigooni DN, Mellinger T, Janabi M, Miller ZA, Perry DC, Pham J, Strom A, Gorno-Tempini ML, Rosen HJ, Miller BL, Jagust WJ, Rabinovici GD, Association of APOE4 and Clinical Variability in Alzheimer Disease With the Pattern of Tau- and Amyloid-PET, Neurology 96, e650–e661 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salvadó G, Grothe MJ, Groot C, Moscoso A, Schöll M, Gispert JD, Ossenkoppele R, Differential associations of APOE-ε2 and APOE-ε4 alleles with PET-measured amyloid-β and tau deposition in older individuals without dementia, Eur. J. Nucl. Med. Mol. Imaging 2021 487 48, 2212–2224 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan S, Zheng C, Paranjpe MD, Li Y, Li W, Wang X, Benzinger TLS, Lu J, Zhou Y, for the ADN Initiative, Sex modifies APOE ε4 dose effect on brain tau deposition in cognitively impaired individuals, Brain (2021), doi: 10.1093/BRAIN/AWAB160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Therriault J, Benedet AL, Pascoal TA, Mathotaarachchi S, Savard M, Chamoun M, Thomas E, Kang MS, Lussier F, Tissot C, Soucy JP, Massarweh G, Rej S, Saha-Chaudhuri P, Poirier J, Gauthier S, Rosa-Neto P, APOEε4 potentiates the relationship between amyloid-β and tau pathologies, Mol. Psychiatry (2020), doi: 10.1038/s41380-020-0688-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mattsson N, Ossenkoppele R, Smith R, Strandberg O, Ohlsson T, Jögi J, Palmqvist S, Stomrud E, Hansson O, Greater tau load and reduced cortical thickness in APOE ε4-negative Alzheimer’s disease: a cohort study, Alzheimer’s Res. Ther. 10, 77 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burt JB, Demirtaş M, Eckner WJ, Navejar NM, Ji JL, Martin WJ, Bernacchia A, Anticevic A, Murray JD, Hierarchy of transcriptomic specialization across human cortex captured by structural neuroimaging topography, Nat. Neurosci 21, 1251–1259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gryglewski G, Seiger R, James GM, Godbersen GM, Komorowski A, Unterholzner J, Michenthaler P, Hahn A, Wadsak W, Mitterhauser M, Kasper S, Lanzenberger R, Spatial analysis and high resolution mapping of the human whole-brain transcriptome for integrative analysis in neuroimaging, Neuroimage 176, 259–267 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Hess A, Hinz R, Keliris GA, Boehm-Sturm P, On the Usage of Brain Atlases in Neuroimaging Research, Mol. Imaging Biol 20, 742–749 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Romero-Garcia R, Seidlitz J, Whitaker KJ, Morgan SE, Jones PB, Goodyer IM, Suckling J, Vértes PE, Bullmore ET, Fonagy P, Dolan RJ, Vaghi M, Moutoussis M, Hauser T, Neufeld S, St Clair M, Inkster B, Prabhu G, Ooi C, Toseeb U, Widmer B, Bhatti J, Villis L, Alrumaithi A, Birt S, Bowler A, Cleridou K, Dadabhoy H, Davies E, Firkins A, Granville S, Harding E, Hopkins A, Isaacs D, King J, Kokorikou D, Maurice C, McIntosh C, Memarzia J, Mills H, O’Donnell C, Pantaleone S, Scott J, Fearon P, van Harmelen AL, Kievit R, Schizotypy-Related Magnetization of Cortex in Healthy Adolescence Is Colocated With Expression of Schizophrenia-Related Genes, Biol. Psychiatry 88, 248–259 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neitzel J, Franzmeier N, Rubinski A, Dichgans M, Brendel M, Weiner M, Aisen P, Petersen R, Jack CR, Jagust W, Trojanowki JQ, Toga AW, Beckett L, Green RC, Saykin AJ, Morris J, Shaw LM, Liu E, Montine T, Thomas RG, Donohue M, Walter S, Gessert D, Sather T, Jiminez G, Harvey D, Bernstein M, Fox N, Thompson P, Schuff N, DeCArli C, Borowski B, Gunter J, Senjem M, Vemuri P, Jones D, Kantarci K, Ward C, Koeppe RA, Foster N, Reiman EM, Chen K, Mathis C, Landau S, Cairns NJ, Householder E, Reinwald LT, Lee V, Korecka M, Figurski M, Crawford K, Neu S, Foroud TM, Potkin S, Shen L, Kelley F, Kim S, Nho K, Kachaturian Z, Frank R, Snyder PJ, Molchan S, Kaye J, Quinn J, Lind B, Carter R, Dolen S, Schneider LS, Pawluczyk S, Beccera M, Teodoro L, Spann M, Brewer J, Vanderswag H, Fleisher A, Heidebrink JL, Lord JL, Mason SS, Albers S, Knopman D, Johnson K, Doody RS, Meyer JV, Chowdhury M, Rountree S, Dang M, Stern Y, Honig LS, Bell KL, Ances B, Morris JC, Carroll M, Leon S, Mintun MA, Schneider S, OliverNG A, Griffith R, Clark D, Geldmacher D, Brockington J, Roberson E, Grossman H, Mitsis E, deToledo-Morrell L, Shah RC, Duara R, Varon D, Greig MT, Roberts P, Albert M, Onyike C, D’Agostino D, Kielb S, Galvin JE, Pogorelec DM, Cerbone B, Michel CA, Rusinek H, de Leon MJ, Glodzik L, De Santi S, Doraiswamy PM, Petrella JR, Wong TZ, Arnold SE, Karlawish JH, Wolk D, Smith CD, Jicha G, Hardy P, Sinha P, Oates E, Conrad G, Lopez OL, Oakley MA, Simpson DM, Porsteinsson AP, Goldstein BS, Martin K, Makino KM, Ismail MS, Brand C, Mulnard RA, Thai G, Mc Adams Ortiz C, Womack K, Mathews D, Quiceno M, Arrastia RD, King R, Weiner M, Cook KM, DeVous M, Levey AI, Lah JJ, Cellar JS, Burns JM, Anderson HS, Swerdlow RH, Apostolova L, Tingus K, Woo E, Silverman DHS, Lu PH, Bartzokis G, Radford NRG, ParfittH F, Kendall T, Johnson H, Farlow MR, Hake AM, Matthews BR, Herring S, Hunt C, van Dyck CH, Carson RE, MacAvoy MG, Chertkow H, Bergman H, Hosein C, Black S, Stefanovic B, Caldwell C, Hsiung GYR, Feldman H, Mudge B, Past MA, Kertesz A, Rogers J, Trost D, Bernick C, Munic D, Kerwin D, Mesulam MM, Lipowski K, Wu CK, Johnson N, Sadowsky C, Martinez W, Villena T, Turner RS, Johnson K, Reynolds B, Sperling RA, Johnson KA, Marshall G, Frey M, Yesavage J, Taylor JL, Lane B, Rosen A, Tinklenberg J, Sabbagh MN, Belden CM, Jacobson SA, Sirrel SA, Kowall N, Killiany R, Budson AE, Norbash A, Johnson PL, Obisesan TO, Wolday S, Allard J, Lerner A, Ogrocki P, Hudson L, Fletcher E, Carmichael O, Olichney J, DeCarli C, Kittur S, Borrie M, Lee TY, Bartha R, Johnson S, Asthana S, Carlsson CM, Potkin SG, Preda A, Nguyen D, Tariot P, Reeder S, Bates V, Capote H, Rainka M, Scharre DW, Kataki M, Adeli A, Zimmerman EA, Celmins D, Brown AD, Pearlson GD, Blank K, Anderson K, Santulli RB, Kitzmiller TJ, Schwartz ES, SinkS KM, Williamson JD, Garg P, Watkins F, Ott BR, Querfurth H, Tremont G, Salloway S, Malloy P, Correia S, Rosen HJ, Miller BL, Mintzer J, Spicer K, Bachman D, Finger E, Pasternak S, Rachinsky I, Drost D, Pomara N, Hernando R, Sarrael A, Schultz SK, Ponto LLB, Shim H, Smith KE, Relkin N, Chaing G, Raudin L, Smith A, Fargher K, Raj BA, Malik R, Ewers M, KL-VS heterozygosity is associated with lower amyloid-dependent tau accumulation and memory impairment in Alzheimer’s disease, Nat. Commun. 2021 121 12, 1–12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mattsson N, Palmqvist S, Stomrud E, Vogel J, Hansson O, Staging β-Amyloid Pathology With Amyloid Positron Emission Tomography, JAMA Neurol. 76, 1319–1329 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morris JC, The clinical dementia rating (cdr): Current version and scoring rules, Neurology 43, 2412–2414 (1993). [DOI] [PubMed] [Google Scholar]
- 42.Talbot C, Lendon C, Craddock N, Shears S, Morris JC, Goate A, Protection against Alzheimer’s disease with apoE 2, Lancet 343, 1432–1433 (1994). [DOI] [PubMed] [Google Scholar]
- 43.Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, Van Der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM, Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain, Neuron 33, 341–355 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Su Y, Blazey TM, Snyder AZ, Raichle ME, Marcus DS, Ances BM, Bateman RJ, Cairns NJ, Aldea P, Cash L, Christensen JJ, Friedrichsen K, Hornbeck RC, Farrar AM, Owen CJ, Mayeux R, Brickman AM, Klunk W, Price JC, Thompson PM, Ghetti B, Saykin AJ, Sperling RA, Johnson KA, Schofield PR, Buckles V, Morris JC, Benzinger TLS, Partial volume correction in quantitative amyloid imaging, Neuroimage 107, 55–64 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su Y, D’Angelo GM, Vlassenko AG, Zhou G, Snyder AZ, Marcus DS, Blazey TM, Christensen JJ, Vora S, Morris JC, Mintun MA, Benzinger TLS, Quantitative analysis of PiB-PET with FreeSurfer ROIs, PLoS One 8, e73377–e73377 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rousset OG, Ma Y, Evans AC, Correction for partial volume effects in PET: Principle and validation, J. Nucl. Med 39, 904–911 (1998). [PubMed] [Google Scholar]
- 47.Su Y, Flores S, Hornbeck RC, Speidel B, Vlassenko AG, Gordon BA, Koeppe RA, Klunk WE, Xiong C, Morris JC, Benzinger TLS, Utilizing the Centiloid scale in cross-sectional and longitudinal PiB PET studies, NeuroImage Clin. 19, 406–416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mishra S, Gordon BA, Su Y, Christensen J, Friedrichsen K, Jackson K, Hornbeck R, Balota DA, Cairns NJ, Morris JC, Ances BM, Benzinger TLS, AV-1451 PET imaging of tau pathology in preclinical Alzheimer disease: Defining a summary measure, Neuroimage 161, 171–178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, Van De Lagemaat LN, Smith KA, Ebbert A, Riley ZL, Abajian C, Beckmann CF, Bernard A, Bertagnolli D, Boe AF, Cartagena PM, Mallar Chakravarty M, Chapin M, Chong J, Dalley RA, Daly BD, Dang C, Datta S, Dee N, Dolbeare TA, Faber V, Feng D, Fowler DR, Goldy J, Gregor BW, Haradon Z, Haynor DR, Hohmann JG, Horvath S, Howard RE, Jeromin A, Jochim JM, Kinnunen M, Lau C, Lazarz ET, Lee C, Lemon TA, Li L, Li Y, Morris JA, Overly CC, Parker PD, Parry SE, Reding M, Royall JJ, Schulkin J, Sequeira PA, Slaughterbeck CR, Smith SC, Sodt AJ, Sunkin SM, Swanson BE, Vawter MP, Williams D, Wohnoutka P, Ronald Zielke H, Geschwind DH, Hof PR, Smith SM, Koch C, Grant SGN, Jones AR, An anatomically comprehensive atlas of the adult human brain transcriptome, Nature 489, 391–399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.French L, Paus T, A FreeSurfer view of the cortical transcriptome generated from the Allen Human Brain Atlas, Front. Neurosci 9 (2015), doi: 10.3389/fnins.2015.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.R Core Team, R: A language and environment for statistical computing. R Foundation for Statistical Computing, R Found. Stat. Comput. Vienna, Austria: (2020). [Google Scholar]
- 52.Váša F, Seidlitz J, Romero-Garcia R, Whitaker KJ, Rosenthal G, Vértes PE, Shinn M, Alexander-Bloch A, Fonagy P, Dolan RJ, Jones PB, Goodyer IM, Sporns O, Bullmore ET, Adolescent tuning of association cortex in human structural brain networks, Cereb. Cortex 28, 281–294 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gordon BA, McCullough A, Mishra S, Blazey TM, Su Y, Christensen J, Dincer A, Jackson K, Hornbeck RC, Morris JC, Ances BM, Benzinger TLS, Cross-sectional and longitudinal atrophy is preferentially associated with tau rather than amyloid β positron emission tomography pathology, Alzheimer’s Dement. Diagnosis, Assess. Dis. Monit 10, 245–252 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aschenbrenner AJ, Gordon BA, Benzinger TLS, Morris JC, Hassenstab JJ, Influence of tau PET, amyloid PET, and hippocampal volume on cognition in Alzheimer disease, Neurology 91, e859–e866 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brier MR, Gordon B, Friedrichsen K, McCarthy J, Stern A, Christensen J, Owen C, Aldea P, Su Y, Hassenstab J, Cairns NJ, Holtzman DM, Fagan AM, Morris JC, Benzinger TLS, Ances BM, Tau and Aβ imaging, CSF measures, and cognition in Alzheimer’s disease, Sci. Transl. Med 8, 338ra66–338ra66 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bussy A, Snider BJ, Coble D, Xiong C, Fagan AM, Cruchaga C, Benzinger TLS, Gordon BA, Hassenstab J, Bateman RJ, Morris JC, Effect of apolipoprotein E4 on clinical, neuroimaging, and biomarker measures in noncarrier participants in the Dominantly Inherited Alzheimer Network, Neurobiol. Aging 75, 42–50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sabbagh MN, Malek-Ahmadi M, Dugger BN, Lee K, Sue LI, Serrano G, Walker DG, Davis K, Jacobson SA, Beach TG, The influence of Apolipoprotein E genotype on regional pathology in Alzheimer’s disease, BMC Neurol. 13 (2013), doi: 10.1186/1471-2377-13-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Landén M, Hesse C, Fredman P, Regland B, Wallin A, Blennow K, Apolipoprotein e in cerebrospinal fluid from patients with alzheimer’s disease and other forms of dementia is reduced but without any correlation to the apoe4 isoform, Dement. Geriatr. Cogn. Disord 7, 273–278 (1996). [DOI] [PubMed] [Google Scholar]
- 59.Buckley RF, Mormino EC, Chhatwal J, Schultz AP, Rabin JS, Rentz DM, Acar D, Properzi MJ, Dumurgier J, Jacobs H, Gomez-Isla T, Johnson KA, Sperling RA, Hanseeuw BJ, Associations between baseline amyloid, sex, and APOE on subsequent tau accumulation in cerebrospinal fluid, Neurobiol. Aging 78, 178–185 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Glodzik-Sobanska L, Pirraglia E, Brys M, de Santi S, Mosconi L, Rich KE, Switalski R, Saint Louis L, Sadowski MJ, Martiniuk F, Mehta P, Pratico D, Zinkowski RP, Blennow K, de Leon MJ, The effects of normal aging and ApoE genotype on the levels of CSF biomarkers for Alzheimer’s disease, Neurobiol. Aging 30, 672–681 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Slot RER, Kester MI, Van Harten AC, Jongbloed W, Bouwman FH, Teunissen CE, Scheltens P, van der Flier WM, Veerhuis R, ApoE and clusterin CSF levels influence associations between APOE genotype and changes in CSF tau, but not CSF Aβ42, levels in non-demented elderly, Neurobiol. Aging 79, 101–109 (2019). [DOI] [PubMed] [Google Scholar]
- 62.Wang Y-TT, Pascoal TA, Therriault J, Kang MS, Benedet AL, Savard M, Tissot C, Lussier FZ, Arias JF, Mathotaarachchi S, Rajah MN, Gauthier S, Rosa-Neto P, for the ADN Initiative, Interactive rather than independent effect of APOE and sex potentiates tau deposition in women, Brain Commun. 3 (2021), doi: 10.1093/BRAINCOMMS/FCAB126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altmann A, Tian L, Henderson VW, Greicius MD, Sex modifies the APOE-related risk of developing Alzheimer disease, Ann. Neurol 75, 563–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, Van Duijn CM, Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease: A meta-analysis, J. Am. Med. Assoc 278, 1349–1356 (1997). [PubMed] [Google Scholar]
- 65.De Wilde A, Reimand J, Teunissen CE, Zwan M, Windhorst AD, Boellaard R, Van Der Flier WM, Scheltens P, Van Berckel BNM, Bouwman F, Ossenkoppele R, Discordant amyloid-β PET and CSF biomarkers and its clinical consequences, Alzheimer’s Res. Ther 11, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gordon BA, Friedrichsen K, Brier M, Blazey T, Su Y, Christensen J, Aldea P, McConathy J, Holtzman DM, Cairns NJ, Morris JC, Fagan AM, Ances BM, Benzinger TLS, The relationship between cerebrospinal fluid markers of Alzheimer pathology and positron emission tomography tau imaging, Brain 139, 2249–2260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chhatwal JP, Schultz AP, Marshall GA, Boot B, Gomez-Isla T, Dumurgier J, La Point M, Scherzer C, Roe AD, Hyman BT, Sperling RA, Johnson KA, Temporal T807 binding correlates with CSF tau and phospho-tau in normal elderly, Neurology 87, 920–926 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barthélemy NR, Horie K, Sato C, Bateman RJ, Blood plasma phosphorylated-tau isoforms track CNS change in Alzheimer’s disease, J. Exp. Med 217 (2020), doi: 10.1084/JEM.20200861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mattsson-Carlgren N, Andersson E, Janelidze S, Ossenkoppele R, Insel P, Strandberg O, Zetterberg H, Rosen HJ, Rabinovici G, Chai X, Blennow K, Dage JL, Stomrud E, Smith R, Palmqvist S, Hansson O, Aβ deposition is associated with increases in soluble and phosphorylated tau that precede a positive Tau PET in Alzheimer’s disease, Sci. Adv 6, eaaz2387 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ashton NJ, Pascoal TA, Karikari TK, Benedet AL, Lantero-Rodriguez J, Brinkmalm G, Snellman A, Schöll M, Troakes C, Hye A, Gauthier S, Vanmechelen E, Zetterberg H, Rosa-Neto P, Blennow K, Plasma p-tau231: a new biomarker for incipient Alzheimer’s disease pathology, Acta Neuropathol. 141, 709–724 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jack CR, Wiste HJ, Botha H, Weigand SD, Therneau TM, Knopman DS, Graff-Radford J, Jones DT, Ferman TJ, Boeve BF, Kantarci K, Lowe VJ, Vemuri P, Mielke MM, Fields JA, Machulda MM, Schwarz CG, Senjem ML, Gunter JL, Petersen RC, The bivariate distribution of amyloid-β and tau: Relationship with established neurocognitive clinical syndromes, Brain 142, 3230–3242 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Horie K, Barthelemy NR, Sato C, Bateman RJ, CSF tau microtubule binding region identifies tau tangle and clinical stages of Alzheimer’s disease, Brain 144, 515–527 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Demirtaş M, Burt JB, Helmer M, Ji JL, Adkinson BD, Glasser MF, Van Essen DC, Sotiropoulos SN, Anticevic A, Murray JD, Hierarchical Heterogeneity across Human Cortex Shapes Large-Scale Neural Dynamics, Neuron 101, 1181–1194.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shi Y, Manis M, Long J, Wang K, Sullivan PM, Serrano JR, Hoyle R, Holtzman DM, Microglia drive APOE-dependent neurodegeneration in a tauopathy mouse model, J. Exp. Med 216, 2546–2561 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mancuso R, Fryatt G, Cleal M, Obst J, Pipi E, Monzón-Sandoval J, Ribe E, Winchester L, Webber C, Nevado A, Jacobs T, Austin N, Theunis C, Grauwen K, Ruiz ED, Mudher A, Vicente-Rodriguez M, Parker CA, Simmons C, Cash D, Richardson J, Jones DNC, Lovestone S, Gómez-Nicola D, Hugh Perry V, Bullmore ET, Bhatti J, Chamberlain SJ, Correia MM, Crofts AL, Dickinson A, Foster AC, Kitzbichler MG, Knight C, Lynall ME, Maurice C, O’Donnell C, Pointon LJ, St George Hyslop P, Turner L, Vertes P, Widmer B, Williams GB, Paul Morgan B, Leckey CA, Morgan AR, O’Hagan C, Touchard S, Cavanagh J, Deith C, Farmer S, McClean J, McColl A, McPherson A, Scouller P, Sutherland M, Boddeke HWGM, Richardson JC, Khan S, Murphy P, Parker CA, Patel J, Jones D, De Boer P, Kemp J, Drevets WC, Nye JS, Wittenberg G, Isaac J, Bhattacharya A, Carruthers N, Kolb H, Pariante CM, Turkheimer F, Barker GJ, Byrom H, Cash D, Cattaneo A, Gee A, Hastings C, Mariani N, McLaughlin A, Mondelli V, Nettis M, Nikkheslat N, Randall K, Sheridan H, Simmons C, Singh N, Van Loo V, Vicente-Rodriguez M, Wood TC, Worrell C, Zajkowska Z, Plath N, Egebjerg J, Eriksson H, Gastambide F, Adams KH, Jeggo R, Thomsen C, Pederson JT, Campbell B, Möller T, Nelson B, Zorn S, O’Connor J, Attenburrow MJ, Baird A, Benjamin J, Clare S, Cowen P, Huang IS, Hurley S, Jones H, Mada F, Nevado-Holgado A, Oladejo A, Ribe E, Smith K, Vyas A, Hughes Z, Balice-Gordon R, Duerr J, Piro JR, Sporn J, Hugh Perry V, Cleal M, Fryatt G, Gomez-Nicola D, Mancuso R, Reynolds R, Harrison NA, Cercignani M, Clarke CL, Hoskins E, Kohn C, Murray R, Wilcock L, Wlazly D, Mount H, CSF1R inhibitor JNJ-40346527 attenuates microglial proliferation and neurodegeneration in P301S mice, Brain 142, 3243–3264 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi Y, Andhey PS, Ising C, Wang K, Snipes LL, Boyer K, Lawson S, Yamada K, Qin W, Manis M, Serrano JR, Benitez BA, Schmidt RE, Artyomov M, Ulrich JD, Holtzman DM, Overexpressing low-density lipoprotein receptor reduces tau-associated neurodegeneration in relation to apoE-linked mechanisms, Neuron 109, 2413–2426.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang C, Xiong M, Gratuze M, Bao X, Shi Y, Andhey PS, Manis M, Schroeder C, Yin Z, Madore C, Butovsky O, Artyomov M, Ulrich JD, Holtzman DM, Selective removal of astrocytic APOE4 strongly protects against tau-mediated neurodegeneration and decreases synaptic phagocytosis by microglia, Neuron 109, 1657–1674.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Litvinchuk A, Huynh TPV, Shi Y, Jackson RJ, Finn MB, Manis M, Francis CM, Tran AC, Sullivan PM, Ulrich JD, Hyman BT, Cole T, Holtzman DM, Apolipoprotein E4 Reduction with Antisense Oligonucleotides Decreases Neurodegeneration in a Tauopathy Model, Ann. Neurol 89, 952–966 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Montagne A, Nation DA, Sagare AP, Barisano G, Sweeney MD, Chakhoyan A, Pachicano M, Joe E, Nelson AR, D’Orazio LM, Buennagel DP, Harrington MG, Benzinger TLS, Fagan AM, Ringman JM, Schneider LS, Morris JC, Reiman EM, Caselli RJ, Chui HC, Tcw J, Chen Y, Pa J, Conti PS, Law M, Toga AW, Zlokovic BV, APOE4 leads to blood–brain barrier dysfunction predicting cognitive decline, Nat. 2020 5817806 581, 71–76 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pitas RE, Boyles JK, Lee SH, Foss D, Mahley RW, Astrocytes synthesize apolipoprotein E and metabolize apolipoprotein E-containing lipoproteins, Biochim. Biophys. Acta (BBA)/Lipids Lipid Metab 917, 148–161 (1987). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Comparison between the entorhinal tau PET SUVR (A), CSF ptau181 (B), and hippocampus volume (C) relationship relative to Aβ summary measure levels and APOE ε4 carrier status.
Fig. S2. Models predicting regional tau PET SUVRs. Values represent the non-significant and significant model estimates.
Fig. S3. Models predicting MRI regional volumes. Values represent the non-significant and significant model estimates.
Fig. S4. Relating regional weights from the models examining main effects of APOE genotype (A,B), the main effects of Aβ (C,D), the interaction between Aβ and genotype (E,F) and the three-way interaction additionally with sex (G,H) relative to APOE mRNA expression derived from the Allan Human Brain Atlas (I). Although there were minimal significant main effects of either Aβ or genotype on regional volumes, the relatively pattern of these non-significant effects was strongly related to the spatial pattern of APOE mRNA expression. This suggests a structured, albeit weak, effect on volumes.
Table S1. Effects of APOE ε4 status on Regional Aβ PET (Model 1).
Table S2. Individual effects of APOE ε4 status and Aβ summary measure on regional tau PET (Model 1 and 2, respectively).
Table S3. Concurrent effects of APOE ε4 status and Aβ summary measure on regional tau PET (Model 3).
Table S4. Interaction effects of APOE ε4 status, Aβ summary measure, and sex on regional tau PET (Model 4 and 5, respectively).
Table S5. Individual effects of APOE ε4 status and Aβ summary measure on regional volume (Model 1 and 2, respectively).
Table S6. Concurrent effects of APOE ε4 status and Aβ summary measure on regional volume (Model 3).
Table S7. Interaction effects of APOE ε4 status, Aβ summary measure, and sex on regional volume (Model 4 and 5, respectively).