Abstract
Background:
Bacillus Calmette–Guérin (BCG) remains the only vaccine to prevent tuberculosis (TB) during childhood, with relatively low to no efficacy against pulmonary TB in adolescents and adults. BCG consists of close to 15 different substrains, where genetic variations among them might contribute to the variable protective efficacy afforded against pulmonary TB. We have shown that the vaccine candidate, BCGΔBCG1419c, which is based on BCG Pasteur, improved protection against chronic TB in murine models, as well as against pulmonary and extrapulmonary TB in guinea pigs. Here, to confirm deletion of the BCG1419c gene and to detect possible genetic variations occurring as a consequence of the spontaneous mutations that may arise during in vitro culture of mycobacteria, the genomes of BCG Pasteur ATCC 35734 and its isogenic derivative, BCGΔBCG1419c, were sequenced and subjected to a comparative analysis between them and against BCG Pasteur 1173P2.
Results:
The complete catalog of variants in genes relative to the reference genome BCG Pasteur 1173P2 (GenBank NC008769) showed that the parental strain BCG Pasteur ATCC 35734, from which the mutant BCGΔBCG1419c originated, showed five synonymous mutations, three missense mutations, and five codon insertions, whereas the BCGΔBCG1419c mutant reported the same changes. When BCG Pasteur ATCC 35734 and BCGΔBCG1419c were compared, we confirmed that the latter was devoid of the BCG1419c gene, with only one unanticipated SNP at position 2, 828, 791 which we consider has no role in vaccine properties reported thus far.
Conclusion:
We provide evidence that the mutagenesis performed to remove BCG1419c from BCG Pasteur ATCC 35734 solely deleted this gene, and that compared with the reference strain BCG Pasteur 1173P2, few changes were present confirming that they are BCG Pasteur strains, and that changes in immunogenicity or efficacy observed thus far in BCGΔBCG1419c are most likely derived solely from the elimination of the BCG1419c gene.
Keywords: BCG, Vaccine, Pasteur, Genomic analysis, BCGΔBCG1419c
Background
Bacille Calmette-Guérin (BCG) is an attenuated strain of Mycobacterium bovis and is the only available vaccine against tuberculosis (TB). Since its introduction 100 years ago, it is estimated that more than 3 billion individuals have received BCG and over 100 million doses of BCG are administered annually to reduce TB burden worldwide. BCG is generally safe and can protect children against disseminated disease, including meningitis; in fact, a very recent meta-analysis suggests that BCG vaccination at birth is effective at preventing TB in young children, but as previously thought, is ineffective in adolescents and adults [1].
BCG typically refers to several substrains, each having genomic differences concerning reference strains [2]. Close to 50 production substrains have been used at one time or another in various parts of the world [3], including the major BCG vaccines in current use (BCG-Danish, -Glaxo, -Russia, and -Japan), which have recently been shown to differ in their viability, RNA content and capacity to induce ex vivo immune responses [4].
Considering that the relative protective efficacy of BCG substrains is a matter of debate [5], coupled with the inefficacy of BCG to protect adolescents and adults against pulmonary TB, there is an urgent need for novel and improved vaccines that could replace or boost the protective effect produced upon immunization with BCG.
In this regard, we developed the BCGΔBCG1419c vaccine candidate based on BCG substrain Pasteur. The second-generation version of BCGΔBCG1419c, devoid of antibiotic markers and based on Pasteur ATCC 35734 was recently shown to improve protection of C57BL/6 mice against the Haarlem strain M. tuberculosis M2 in reducing lung pathology compared with BCG Pasteur ATCC 35734 [6]. Also, BCGΔBCG1419c improved protection in guinea pigs against pulmonary and extrapulmonary TB better than parental BCG [7], and it showed variations in its cellular and secreted proteome compared with parental BCG [8].
Here, to identify potential genomic polymorphisms in BCGΔBCG1419c compared with its parental BCG Pasteur ATCC 35734 substrain and the reference genome of BCG Pasteur 1173P2, as well as to evaluate whether additional genetic events (insertion/deletion) other than the targeted deletion of the BCG1419c gene, we have obtained the whole genome sequences (WGS) of the BCG ATCC 35734 and that of the BCGΔBCG1419c strains. Obtained results were assembled and compared with the genome of the reference strain BCG Pasteur 1173P2 to identify eventual major genomic rearrangements. A mapping strategy was also used to evaluate SNPs/InDels variability acquired.
Thus, we confirmed that BCGΔBCG1419c has a single deletion of the BCG1419c gene and identified novel genomic polymorphisms of both BCG Pasteur ATCC 35734 and BCGΔBCG1419c compared with BCG Pasteur 1173P2.
Results
A total of 7,772,967 and 7,577,201 paired ends raw reads have been obtained from the BCG Pasteur ATCC 35734 and BCGΔBCG1419c strains, respectively. The two genome reads datasets have been mapped against the reference genome M. tuberculosis BCG str. Pasteur 1173P2, obtaining an average coverage of 388x and 379x, respectively.
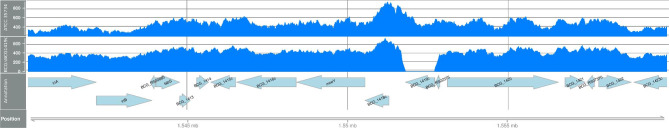
The parental strain BCG Pasteur ATCC 35734, from which the mutant BCGΔBCG1419c originated, belonged to the BCG str. Pasteur substrain . The sequenced BCG Pasteur ATCC 35734 strain showed five synonymous mutations, three missense mutations, and four codon insertions compared with the BCG str. Pasteur 1173P2 (Table 1). The BCGΔBCG1419c mutant, on the other hand, had the same mutations as BCG Pasteur ATCC 35734 (Table 1) plus the deletion of BCG1419c, which is annotated as cyclic diguanylate phosphodiesterase. Further to this, we found an unanticipated SNP at position 2, 828, 791. In Table 1, the column “Evidence” indicates the frequencies (sequences) of the nucleotides in the reference (REF) genome with respect to its alternative (ALT, mutation) genome. Figure 1 shows the region surrounding the deletion.
Table 1.
Summary of changes detected in the genomes of BCG Pasteur ATCC 35734 and its isogenic derivative BCGΔBCG1419c compared with BCG Pasteur 1173P2
| BCG Pasteur ATCC 35734 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CHROM | POSITION | TYPE | REF | ALT | EVIDENCE | FTYPE | STRAND | NT_POS | AA_POS | EFFECT | LOCUS_TAG | BCG 1172P2 GENE | H37Rv GENE | PRODUCT |
| NC_008769 | 705623 | complex | GTGG | ATGC | ATGC:125 GTGG:0 | CDS | - | 1176/3912 | 391/1303 | synonymous_variant c.1173_1176delCCACinsGCAT p.393 | BCG_RS03180 | BCG_0623c | PE_PGRS7 | PE family protein |
| NC_008769 | 1344671 | mnp | CG | GC | GC:119 CG:0 | |||||||||
| NC_008769 | 1661323 | snp | G | C | C:12 G:0 | CDS | - | 1926/2223 | 642/740 | synonymous_variant c.1926C > G p.Gly642Gly | BCG_RS07850 | BCG_1513c | PE_PGRS28 | PE family protein |
| NC_008769 | 2002385 | complex | G | CGGC | CGGC:61 G:3 | CDS | - | 666/2690 | 222/895 | disruptive_inframe_insertion&synonymous_variant c.666delCinsGCCG p.Thr222_Val223insPro | BCG_RS09290 | BCG_1799c | PE-PGRS family protein wag22b | PE family protein |
| NC_008769 | 2764157 | snp | T | A | A:260 T:0 | CDS | - | 459/3342 | 153/1113 | synonymous_variant c.459A > T p.Thr153Thr | BCG_RS12955 | BCG_2507c | Rv2488c | LuxR family transcriptional regulator |
| NC_008769 | 2766103 | complex | G | CGGC | CGGC:103 G:0 | CDS | - | 3930/4972 | 1310/1656 | missense_variant&disruptive_inframe_insertion c.3930delCinsGCCG p.Cys1310delinsTrpPro | BCG_RS12965 | BCG_2509c | PE_PGRS43b | PE family protein |
| NC_008769 | 3833488 | ins | G | GGCC | GGCC:141 G:9 | CDS | - | 1298/1419 | 433/472 | disruptive_inframe_insertion c.1298_1299insGGC p.Ala433dup | BCG_RS17970 | BCG_3499c | Rv3433c | Bifunctional ADP-dependent NAD(P)H-hydrate dehydratase/NAD(P)H-hydrate epimerase |
| NC_008769 | 3854969 | ins | G | GGTC | GGTC:289 G:0 | CDS | + | 783/786 | 261/261 | disruptive_inframe_insertion c.782_783insTCG p.Gly261_Ter262insArg | BCG_RS18060 | BCG_3517 | Cut3 | Cutinase family protein |
| NC_008769 | 3907860 | snp | A | G | G:81 A:0 | CDS | + | 1795/4119 | 599/1372 | missense_variant c.1795A > G p.Asn599Asp | BCG_RS18320 | BCG_3571 | PE_PGRS53 | PE family protein |
| NC_008769 | 3908180 | snp | C | T | T:70 C:0 | CDS | + | 2115/4119 | 705/1372 | synonymous_variant c.2115C > T p.Gly705Gly | BCG_RS18320 | BCG_3571 | PE_PGRS53 | PE family protein |
| NC_008769 | 3927720 | snp | A | G | G:13 A:0 | CDS | + | 1792/3228 | 598/1075 | missense_variant c.1792A > G p.Thr598Ala | BCG_RS18350 | BCG_3577 | PE_PGRS57 | PE family protein |
| BCGΔBCG1419c | ||||||||||||||
| CHROM | POSITION | TYPE | REF | ALT | EVIDENCE | FTYPE | STRAND | NT_POS | AA_POS | EFFECT | LOCUS_TAG | BCG 1172P2 GENE | H37Rv GENE | PRODUCT |
| NC_008769 | 705623 | complex | GTGG | ATGC | ATGC:137 GTGG:0 | CDS | - | 1176/3912 | 391/1303 | synonymous_variant c.1173_1176delCCACinsGCAT p.393 | BCG_RS03180 | BCG_0623c | PE_PGRS7 | PE family protein |
| NC_008769 | 1344671 | mnp | CG | GC | GC:91 CG:0 | |||||||||
| NC_008769 | 1661323 | snp | G | C | C:16 G:0 | CDS | - | 1926/2223 | 642/740 | synonymous_variant c.1926C > G p.Gly642Gly | BCG_RS07850 | BCG_1513c | PE_PGRS28 | PE family protein |
| NC_008769 | 2002385 | complex | G | CGGC | CGGC:61 G:0 | CDS | - | 666/2690 | 222/895 | disruptive_inframe_insertion&synonymous_variant c.666delCinsGCCG p.Thr222_Val223insPro | BCG_RS09290 | BCG_1799c | PE-PGRS family protein wag22b | PE family protein |
| NC_008769 | 2764157 | snp | T | A | A:266 T:0 | CDS | - | 459/3342 | 153/1113 | synonymous_variant c.459A > T p.Thr153Thr | BCG_RS12955 | BCG_2507c | Rv2488c | LuxR family transcriptional regulator |
| NC_008769 | 2766103 | complex | G | CGGC | CGGC:110 G:0 | CDS | - | 3930/4972 | 1310/1656 | missense_variant&disruptive_inframe_insertion c.3930delCinsGCCG p.Cys1310delinsTrpPro | BCG_RS12965 | BCG_2509c | PE_PGRS43b | PE family protein |
| NC_008769 | 2828791 | snp | G | A | A:155 G:0 | |||||||||
| NC_008769 | 3833488 | ins | G | GGCC | GGCC:159 G:6 | CDS | - | 1298/1419 | 433/472 | disruptive_inframe_insertion c.1298_1299insGGC p.Ala433dup | BCG_RS17970 | BCG_3499c | Rv3433c | Bifunctional ADP-dependent NAD(P)H-hydrate dehydratase/NAD(P)H-hydrate epimerase |
| NC_008769 | 3854969 | ins | G | GGTC | GGTC:279 G:0 | CDS | + | 783/786 | 261/261 | disruptive_inframe_insertion c.782_783insTCG p.Gly261_Ter262insArg | BCG_RS18060 | BCG_3517 | Cut3 | Cutinase family protein |
| NC_008769 | 3907860 | snp | A | G | G:37 A:0 | CDS | + | 1795/4119 | 599/1372 | missense_variant c.1795A > G p.Asn599Asp | BCG_RS18320 | BCG_3571 | PE_PGRS53 | PE family protein |
| NC_008769 | 3908180 | snp | C | T | T:99 C:1 | CDS | + | 2115/4119 | 705/1372 | synonymous_variant c.2115C > T p.Gly705Gly | BCG_RS18320 | BCG_3571 | PE_PGRS53 | PE family protein |
| NC_008769 | 3927720 | snp | A | G | G:11 A:0 | CDS | + | 1792/3228 | 598/1075 | missense_variant c.1792A > G p.Thr598Ala | BCG_RS18350 | BCG_3577 | PE_PGRS57 | PE family protein |
The column “Evidence” indicates the frequencies (sequences) of the nucleotides in the reference (REF) with respect to its alternative (ALT, mutation) genome
Fig. 1.
Schematic representarion showing the genomic region upstream and downstream the deletion of the BCG1419c gene. The upper panels shows the region present in wild type BCG Pasteur ATCC 35734, the middle panel, the region present in BCG?BCG1419c, and the bottom panel shows gene names when they have an annotation available.
Specifically, synonymous changes were found for PE_PGRS7, PE_PGRS28, and PE_PGRS53, whereas changes possibly affecting function were found for PE-PGRS family protein Wag22b, PE_PGRS43b, PE_PGRS53, and PE_PGRS57 (Table 1). Regarding non-PE_PGRS family genes, we found a synonymous change in BCG_2507c, which encodes for a LuxR-family transcriptional regulator, and we found two disruptive in-frame insertions, one in BCG_3499c and another in BCG_3517 (Table 1).
Discussion
Previously, spontaneous heterogeneity of BCG seed lots and commercial vaccines used during vaccine production was demonstrated in the BCG Tokyo-172 vaccine strain as determined by deep-sequencing [9]. Because of this reason that may impact on immunogenicity and/or efficacy of protection of TB vaccines in general, we decided to determine the WGS of our BCGΔBCG1419c vaccine candidate and its parental strain, BCG Pasteur ATCC 35734, a passage “zero” strain as obtained from ATCC. BCG Pasteur 35734 was passaged 3 times in our lab, and BCGΔBCG1419c was passaged 9 times by the time genomic DNA was obtained from them for WGS. We cannot rule out the fact that spontaneous mutations could arise during subsequent passages of our BCGΔBCG1419c vaccine candidate, which could lead to changes affecting efficacy of protection, as we have hypothesized to occur for other vaccine strains where the global regulator gene phoP is affected [10]. With the current availability of WGS, it would be convenient to monitor these possible changes over time to make sure that the vaccine strain maintains or not its reported properties.
In our study, most changes detected in both BCG Pasteur ATCC 35734 and its isogenic derivative BCGΔBCG1419c compared with BCG Pasteur 1173P2 were found in PE family genes , including PE_PGRS7, PE_PGRS28, PE-PGRS family Wag22b, PE_PGRS43b, PE_PGRS53, and PE_PGRS57. From these, PE_PGRS43, PE_PGRS53, and PE_PGRS57 have been found in infected guinea pig lungs, and overall, this family has been suggested to play important roles in virulence [11].
Rv2488c (mclx3), homologous to BCG_2507c, showed a synonymous variant among our strains and BCG Pasteur 1173P2 (Table 1). Rv2488c presented a higher tendency for pseudogenization among isolates from patients born on the Western Pacific area, and from isolates causing extra-pulmonary infections [12]. Rv3433c, homologous to BCG_3499c, presented a disruptive in-frame insertion among our strains and BCG Pasteur 1173P2 (Table 1). Rv3433c was identified by mass spectrometry in M. tuberculosis H37Rv-infected guinea pig lungs at 90 days but not 30 days [13].
As for BCG_3517, this gene also showed a disruptive in-frame insertion among our strains and BCG Pasteur 1173P2 (Table 1). Transcripts from its homologous gene cut3 (Rv3451) were increased in a mce1 mutant, along with transcripts of mmpL3, fas, kasA, kasB and acpM, involved in mycolic acids transport and metabolism [14].
Overall, considering that other than deletion of BCG1419c, BCGΔBCG1419c differs from BCG Pasteur ATCC 35734 only in a SNP at position 2,828,791, this support the notion that the improved efficacy and changes to the proteome we have reported for the BCG ATCC 35734-derived version of BCGΔBCG1419c [6–8] are most likely the sole consequence of the gene deletion we created. The SNP at position 2,828,791 is located in the intergenic region of what would be homologous to BCG_2563 (annotated as hypothetical alanine rich protein) and BCG_2564 (a conserved hypothetical protein with an α/β hydrolase 8 family protein). Considering its location and the predicted functions of these genes, we hypothesize that this SNP plays no role in vaccine efficacy reporte thus far.
Conclusion
Recently, comparative studies of genomic variations in BCG strains at their different stages of production and utilization (production strains, their seeds, administered vaccine lots) was suggested to potentially provide data to better understand the bases of vaccine efficacy and adverse reactions of present and future BCG-based vaccines [15]. Here, we provide the WGS of our vaccine candidate, BCGΔBG1419c and its parental strain, BCG Pasteur ATCC 35734. Our analysis show that BCGΔBCG1419c differs from BCG Pasteur ATCC 35734 only in a SNP at position 2,828,791, hereby supporting the notion that the improved efficacy we have observed for BCGΔBG1419c in preclinical models are most likely the sole consequence of the gene deletion of BCG1419c we created and support further development of this vaccine candidate.
Methods
Construction of the BCGΔBCG1419c mutant
BCG Pasteur ATCC 35734 was used as parental strain to promote homologous recombination to create the antibiotic-less version of BCGΔBCG1419c as already described in detail [8]. Succinctly, sequences upstream and downstream of BCG1419c were amplified by PCR and cloned into pUCHyg (a kind gift from Dr. Yi-.
Cheng Sun), sequences were verified, and this plasmid was transformed by electroporation into BCG Pasteur ATCC 35734 harboring pJV53 (a kind gift from Dr. Graham Hatfull). Recombination and successful mutagenesis was verified as described [16].
Genomic DNA extraction
BCG Pasteur ATCC 35734 and its isogenic derivative, BCGΔBG1419c, were cultured in Middlebrook 7H9 broth, supplemented with 10% OADC, at 37ºC, 100 rpm, until OD600nm 0.8. Then, cell pellets were obtained by centrifugation at 3,200 x g for 10 min. The bacterial pellets were resuspended in SET buffer (0.25 M sucrose, 0.05 M EDTA, 0.03 M Tris) and lysozyme (50 mg/mL) was added followed by incubation overnight at 37ºC. RNAse A was added (10 mg/mL) and incubated at 37ºC for 30 min, followed by the addition of proteinase K (1 mg/mL) to incubate at 55ºC for 2 h. A phenol-chloroform–isoamyl alcohol extraction step was performed, to separate the aqueous phase and add 0.1 V of 3 M sodium acetate (pH 5.2), and 0.7 volume of isopropanol for precipitation performed by centrifugation at 16,000 x g, 4ºC for 30 min. The DNA pellets were washed with 7% ethanol, the supernatant discarded, and the pellets were air-dried to resuspend in molecular biology-grade water finally. This protocol was adapted from that described by van Soolingen et al. [17].
Library Preparation and sequencing
The genomic DNA was randomly sheared into short fragments using enzymes provided in the Nextera XT DNA Library Preparation Kit (Illumina, USA) following the manufacturer’s protocol to achieve equimolar pools of each library sample. The obtained fragments were end-repaired, A-tailed, and further ligated to Illumina adapters by “tagmentation”. The fragments with adapters were PCR amplified, size selected using MPure XP Beads (Beckman Coulter, USA), and purified. The size distribution of fragments was checked using an Agilent DNA High Sensitivity chip (2100 Bionanalyzer, Agilent, USA). An Illumina library was prepared with the Nextera DNA Flex kit and Nextera DNA CD indexes, and 2 × 150-bp sequencing was performed on a MiSeq sequencer using the MiSeq reagent kit v.3 (Illumina, USA).
Sequencing quality assessment
Illumina output files in FASTQ format were loaded into Geneious Prime software (v.2021.2.2) and trimmed with the BBDuk plugin (v.1.0, https://sourceforge.net/projects/bbmap/). Adapters on the right and low-quality ends (quality below 20%) were trimmed, while reads shorter than 200 bp were discarded. Then, the reads were subjected to preprocessing (https://www.geneious.com/tutorials/map-to-reference/). The genomes were assembled by using the “Map to reference” tool. BCG Pasteur 1173P2 (GenBank accession no. NC008769) was used as a reference genome. A consensus sequence from aligned reads was extracted. We visually confirmed the circular genomes of the M. bovis BCG strains by assessing the reads spanning the junction between the two linearized ends and overlapping with them. Annotation was generated with the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (v.4.13) [18–20].
The sequencing data belonging to the BCG Pasteur ATCC 35734 (NCBI Locus tag: CP109681; https://www.ncbi.nlm.nih.gov/nuccore/CP109681) strain and its isogenic derivative, BCGΔBCG1419c (NCBI Locus tag: CP110223; https://www.ncbi.nlm.nih.gov/nuccore/CP110223), were mapped against the reference genome Mycobacterium bovis BCG Pasteur 1173P2 (NCBI Locus tag: NC_008769; https://www.ncbi.nlm.nih.gov/nuccore/NC_008769). The mapping process was carried out using the Snippy pipeline (V 4.6.0; Seemann 2015, https://github.com/tseemann/snippy). Genomes alignment was visualized using Mauve program [21] (snapshot 2015-02-13).
Acknowledgements
M.J.A.S. received a Ph.D. fellowship from CONACYT number 745841.
Abbreviations
- BCG
Bacille Calmette-Guérin
- TB
Tuberculosis
- SNP
Single nucleotide polymorphisms
- WGS
Whole-genome sequencing
- CDS
Coding DNA sequences
Author contribution
M.J.A.S., M.AF.V., and Y.H. performed the experiments, Y.H. and G.d.A analyzed WGS data, G.d.A., Y.H., and M.A.F.V. wrote the draft of manuscript. M.A. F.V. conceptualized the study, M.A.F.V., A.M, and S.P. edited the manuscript. All authors read and approved the final version of manuscript.
Funding
This work received no specific funding.
Data availability
The raw sequence reads can be found as CP110223.1 - Mycobacterium tuberculosis variant bovis strain BCG delta BCG1419c mutant chromosome.
https://www.ncbi.nlm.nih.gov/nuccore/CP110223, and CP109681.1 - Mycobacterium tuberculosis variant bovis strain BCG Pasteur ATCC 35,734 chromosome https://www.ncbi.nlm.nih.gov/nuccore/CP109681. All data generated or analyzed during this study are included in this article and its supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
M.A.F.V. and M.J.A.S are inventors in the patent 363576 issued in Mexico for the BCGΔBG1419c vaccine candidate. All other authors have no interests to declare.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Giuseppe D’Auria and Yordan Hodzhev contributed equally and should be considered joint first authors.
Contributor Information
Stefan Panaiotov, Email: spanaiotov@ncipd.org.
Mario Alberto Flores-Valdez, Email: floresv@ciatej.mx.
References
- 1.Martinez L, Cords O, Liu Q, Acuna-Villaorduna C, Bonnet M, Fox GJ, Carvalho ACC, Chan PC, Croda J, Hill PC, et al. Infant BCG vaccination and risk of pulmonary and extrapulmonary tuberculosis throughout the life course: a systematic review and individual participant data meta-analysis. Lancet Glob Health. 2022;10(9):e1307–16. doi: 10.1016/S2214-109X(22)00283-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brosch R, Gordon SV, Garnier T, Eiglmeier K, Frigui W, Valenti P, Dos Santos S, Duthoy S, Lacroix C, Garcia-Pelayo C, et al. Genome plasticity of BCG and impact on vaccine efficacy. Proc Natl Acad Sci U S A. 2007;104(13):5596–601. doi: 10.1073/pnas.0700869104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corbel MJ, Fruth U, Griffiths E, Knezevic I. Report on a WHO consultation on the characterisation of BCG strains, Imperial College, London 15–16 December 2003. Vaccine. 2004;22(21–22):2675–80. doi: 10.1016/j.vaccine.2004.01.050. [DOI] [PubMed] [Google Scholar]
- 4.Angelidou A, Conti MG, Diray-Arce J, Benn CS, Shann F, Netea MG, Liu M, Potluri LP, Sanchez-Schmitz G, Husson R, et al. Licensed Bacille Calmette-Guerin (BCG) formulations differ markedly in bacterial viability, RNA content and innate immune activation. Vaccine. 2020;38(9):2229–40. doi: 10.1016/j.vaccine.2019.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antas PRZ, Flores-Valdez M, Shann F. An opportunity to compare the effects of BCG-Moreau and BCG-Russia in Brazil. Int J Tuberc Lung Dis. 2018;22(9):1108–9. doi: 10.5588/ijtld.18.0271. [DOI] [PubMed] [Google Scholar]
- 6.Kwon KW, Aceves-Sanchez MJ, Segura-Cerda CA, Choi E, Bielefeldt-Ohmann H, Shin SJ, Flores-Valdez MA. BCGDeltaBCG1419c increased memory CD8(+) T cell-associated immunogenicity and mitigated pulmonary inflammation compared with BCG in a model of chronic tuberculosis. Sci Rep. 2022;12(1):15824. doi: 10.1038/s41598-022-20017-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aceves-Sanchez MJ, Flores-Valdez MA, Pedroza-Roldan C, Creissen E, Izzo L, Silva-Angulo F, Dawson C, Izzo A, Bielefeldt-Ohmann H, Segura-Cerda CA, et al. Vaccination with BCGDeltaBCG1419c protects against pulmonary and extrapulmonary TB and is safer than BCG. Sci Rep. 2021;11(1):12417. doi: 10.1038/s41598-021-91993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Velazquez-Fernandez JB, Ferreira-Souza GHM, Rodriguez-Campos J, Aceves-Sanchez MJ, Bravo-Madrigal J, Vallejo-Cardona AA, Flores-Valdez MA. Proteomic characterization of a second-generation version of the BCGDeltaBCG1419c vaccine candidate by means of electrospray-ionization quadrupole time-of-flight mass spectrometry.Pathog Dis2021, 79(1). [DOI] [PubMed]
- 9.Wada T, Maruyama F, Iwamoto T, Maeda S, Yamamoto T, Nakagawa I, Yamamoto S, Ohara N. Deep sequencing analysis of the heterogeneity of seed and commercial lots of the bacillus Calmette-Guerin (BCG) tuberculosis vaccine substrain Tokyo-172. Sci Rep. 2015;5:17827. doi: 10.1038/srep17827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flores-Valdez MAS-C. Cristian Alfredo; Vallejo-Cardona, Alba Adriana; Velázquez-Fernández, Jesús Bernardino: Understanding mycobacterial lipid metabolism and employing it as a tool to produce attenuated TB vaccine candidates. In: Biology of Mycobacterial Lipids Edited by Zeeshan Fatima SC, 1st edn: Elsevier; 2022: 221–233.
- 11.Fishbein S, van Wyk N, Warren RM, Sampson SL. Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Mol Microbiol. 2015;96(5):901–16. doi: 10.1111/mmi.12981. [DOI] [PubMed] [Google Scholar]
- 12.Lopes Santos C, Nebenzahl-Guimaraes H, Vaz Mendes M, van Soolingen D, Correia-Neves M. To be or not to be a pseudogene: a Molecular Epidemiological Approach to the mclx genes and its impact in tuberculosis. PLoS ONE. 2015;10(6):e0128983. doi: 10.1371/journal.pone.0128983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kruh NA, Troudt J, Izzo A, Prenni J, Dobos KM. Portrait of a pathogen: the Mycobacterium tuberculosis proteome in vivo. PLoS ONE. 2010;5(11):e13938. doi: 10.1371/journal.pone.0013938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Queiroz A, Medina-Cleghorn D, Marjanovic O, Nomura DK, Riley LW. Comparative metabolic profiling of mce1 operon mutant vs wild-type Mycobacterium tuberculosis strains. Pathog Dis. 2015;73(8):ftv066. doi: 10.1093/femspd/ftv066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narvskaya O, Starkova D, Levi D, Alexandrova N, Molchanov V, Chernyaeva E, Vyazovaya A, Mushkin A, Zhuravlev V, Solovieva N, et al. First insight into the whole-genome sequence variations in Mycobacterium bovis BCG-1 (Russia) vaccine seed lots and their progeny clinical isolates from children with BCG-induced adverse events. BMC Genomics. 2020;21(1):567. doi: 10.1186/s12864-020-06973-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Kessel JC, Hatfull GF. Recombineering in Mycobacterium tuberculosis. Nat Methods. 2007;4(2):147–52. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 17.van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29(11):2578–86. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44(14):6614–24. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haft DH, DiCuccio M, Badretdin A, Brover V, Chetvernin V, O’Neill K, Li W, Chitsaz F, Derbyshire MK, Gonzales NR, et al. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018;46(D1):D851–60. doi: 10.1093/nar/gkx1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W, O’Neill KR, Haft DH, DiCuccio M, Chetvernin V, Badretdin A, Coulouris G, Chitsaz F, Derbyshire MK, Durkin AS, et al. RefSeq: expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res. 2021;49(D1):D1020–8. doi: 10.1093/nar/gkaa1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14(7):1394–403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw sequence reads can be found as CP110223.1 - Mycobacterium tuberculosis variant bovis strain BCG delta BCG1419c mutant chromosome.
https://www.ncbi.nlm.nih.gov/nuccore/CP110223, and CP109681.1 - Mycobacterium tuberculosis variant bovis strain BCG Pasteur ATCC 35,734 chromosome https://www.ncbi.nlm.nih.gov/nuccore/CP109681. All data generated or analyzed during this study are included in this article and its supplementary information files.