Abstract
JSNP is a repository of Japanese Single Nucleotide Polymorphism (SNP) data, begun in 2000 and developed through the Prime Minister’s Millennium Project. The aim of this undertaking is to identify and collate up to 150 000 SNPs from the Japanese population, located in genes or in adjacent regions that might influence the coding sequence of the genes. The project has been carried out by a collaboration between the Human Genome Center (HGC) in the Institute of Medical Science (IMS) at the University of Tokyo and the Japan Science and Technology Corporation (JST). JSNP serves as both a storage site for the Japanese SNPs obtained from the ongoing project and as a facility for public dissemination to allow researchers access to high quality SNP data. A primary motivation of the project is the construction of a basic data set to identify relationships between polymorphisms and common diseases or the reaction to drugs. As such, emphasis has been placed on the identification of SNPs that lie in candidate regions which may affect phenotype but which would not necessarily directly cause disease. Unrestricted access to JSNP and any associated files is available at http://snp.ims.u-tokyo.ac.jp/.
INTRODUCTION
Moving on from the completion of the human genome sequence, studies on DNA variation between individuals will contribute further to the understanding of human genetics, influencing medical treatment and drug discoveries. In Japan, through the Prime Minister’s Millennium Project, collaborative research between the HGC in the University of Tokyo and the JST has led to a genome-wide screen to find common single nucleotide polymorphisms (SNPs), within the Japanese gene pool. With biological and medical interests in mind, screening targets suggestive of phenotypic impact are selected from annotated genes, gene predictions and the adjacent sequence regions. The objective of this project is to identify up to 150 000 common DNA variants within the Japanese population and submit them to a database, JSNP, which will provide the basic data for genotyping and allele frequencies.
JSNP has been developed as a database for the SNP discovery project. The project spans the selection of screening targets, to establishing links with public databases, and to the presentation of data on the web site. The database itself is in two parts: the data management system and the data publication system. This ensures that the data management system has a minimal operational load and that the data publication system can include up-to-date information from public human genomic databases and other related efforts towards SNP identification in Japan. The JSNP provides an access to non-redundant SNP data and other polymorphisms resulting from the project with any relevant maps. As many genes as possible are added computationally, even if that data appear inconsistent, so that the end user may accurately assess the data available.
In the first year, 77 141 records were entered into the database. By the first quarter of 2002, the identification of new SNPs will cease. Parallel projects such as genotyping, by JBiC, and SNP development in drug metabolizing genes, by RIKEN, will exploit the SNPs available in JSNP, as well as return further data for verification. JSNP will continue to be refined by introducing novel data from any related research.
DATA SOURCE
The purpose of the project is to discover common DNA sequence variants amongst the Japanese population, especially those that might affect phenotype, because they reside within the regulatory domain of a gene, an intron or an exon.
The target regions are selected from genomic DNA sequences annotated as genes in GenBank, homologous to known gene sequences in UniGene, from transcripts from the NCBI Reference Sequences (1,2) and from GenScan predictions (3). The sequences of the target regions are extracted from DNA sequences in GenBank and the genomic contigs of the NCBI database. Primers are designed using Primer3.0 (4) on the extracted sequences to cover the exons, introns and promoter regions, excluding any repetitive regions.
Twenty-four unrelated anonymous Japanese DNA samples are used as the template. These samples are pooled into batches of three. After PCR amplification of the target region from the DNA pools, sequencing of the PCR products is performed using ABI3700 capillary-based sequencers. Almost 10 000 samples are loaded per day. Polyphred (5) is used to detect any SNPs in the assembled sequences. The positions of the SNP candidates and possible insertions or deletions are verified by manual inspection using Consed (6) and the web interfaces of JSNP, which can show the evidence and statistics required for a decision and the results are loaded into the data management system.
DATA FEATURES
The density of SNPs discovered among the regions surveyed was one SNP per 960 bp, based upon the number of SNPs from the JSNP release 7 and the number of bases sequenced for the screening.
The SNPs have been categorized according to the nucleotide substitution observed (Table 1). The results show that the transition rate is 70% and the transversion rate is 30%, corresponding with previously published data. In comparison with the ratio of the SNPs on chromosome 22, discovered in overlapping clones used for sequencing (7), and the gene targeted cSNPs on chromosome 21 (8), we find very similar results. These ratios differ somewhat from those derived from the whole genome (Celera PFP) (9). However, C/T substitutions tend to occur at CpG dinucleotides and there is a higher frequency of CpGs associated with the first exon (9). Chromosome 22 is one of the most gene-rich chromosomes and the cSNP map for chromosome 21 and JSNP are both targeted at regions known to contain genes, thus these transition and transversion ratios are probably associated with the skew towards SNPs chosen from gene rich regions in the human genome.
Table 1. Ratios of different types of substitutions.
| Type of substitution | Percentage transitions | Percentage transversions | |||||||
|---|---|---|---|---|---|---|---|---|---|
| C/T | G/A | C/G | T/A | T/G | C/A | ||||
| JSNP | 34.80 | 35.20 | 10.20 | 4.60 | 7.30 | 7.60 | |||
| CSNP map for chromosome 21a | – | 70.00 | – | – | – | 30.00 | – | – | |
| SNP resource chromosome 22b | – | 70.40 | – | 9.44 | 4.94 | – | 15.38 | – | |
| Celera PFPc | 30.70 | 30.70 | 9.20 | 8.60 | 10.30 | 10.30 | |||
aFrom table 2 in Deutsch et al. (8).
bFrom table 3 in Dawson et al. (7).
cFrom table 16 in Venter et al. (9).
We also examined the overlapping data between JSNP and dbSNP, comparing the JSNP data with the mapped SNPs on the NCBI contigs (Table 2). The locations of the SNPs against NCBI contigs were found by electronic (e-)PCR (10) using primers used to amplify the screened regions and by using BLAST with the flanking sequences of the SNPs. The conditions for identifying data in dbSNP and JSNP are the same as is required for the identification of a Reference SNP in dbSNP (11), by comparing the flanking sequences of the SNP, derived from the NCBI contigs. 24 246 SNPs from the dbSNP build96 overlapped with JSNP data and the results, excluding any data registered by JSNP in dbSNP, show 20.9% identity between the two databases.
Table 2. Overlapping data between dbSNP build96 and JSNP Release 6.
| Chromosome | JSNP | % Overlapping dbSNPs |
|---|---|---|
| 1 | 5799 | 22.7 |
| 2 | 4321 | 19.1 |
| 3 | 3227 | 21.1 |
| 4 | 2227 | 18.7 |
| 5 | 2840 | 28.7 |
| 6 | 4834 | 21.2 |
| 7 | 5478 | 20.3 |
| 8 | 2040 | 18.9 |
| 9 | 2090 | 19.2 |
| 10 | 2255 | 19.4 |
| 11 | 3332 | 27.8 |
| 12 | 3860 | 21.1 |
| 13 | 1044 | 24.7 |
| 14 | 3069 | 19.0 |
| 15 | 1933 | 19.3 |
| 16 | 2818 | 20.9 |
| 17 | 2805 | 19.3 |
| 18 | 929 | 28.0 |
| 19 | 3571 | 21.2 |
| 20 | 2763 | 18.5 |
| 21 | 2203 | 18.5 |
| 22 | 4035 | 21.3 |
| X | 2018 | 14.3 |
| Y | 50 | 14.0 |
| Multi | 1172 | 29.3 |
| Unknown | 3592 | 15.8 |
| Total | 74305 | 20.9 |
The JSNP data are those SNPs found to map to an NCBI contig. The ratio is based on the number of SNPs found in both databases, excluding any data from dbSNP derived directly from JSNP submissions. Those SNPs whose flanking sequences match more than one chromosome are included in Multi and those with no match to NCBI contigs are shown in Unknown.
These are preliminary analyses and any candidate polymorphisms developed by JSNP should be user verified by further studies, including allele frequencies and with genotyping.
DATABASE STRUCTURE
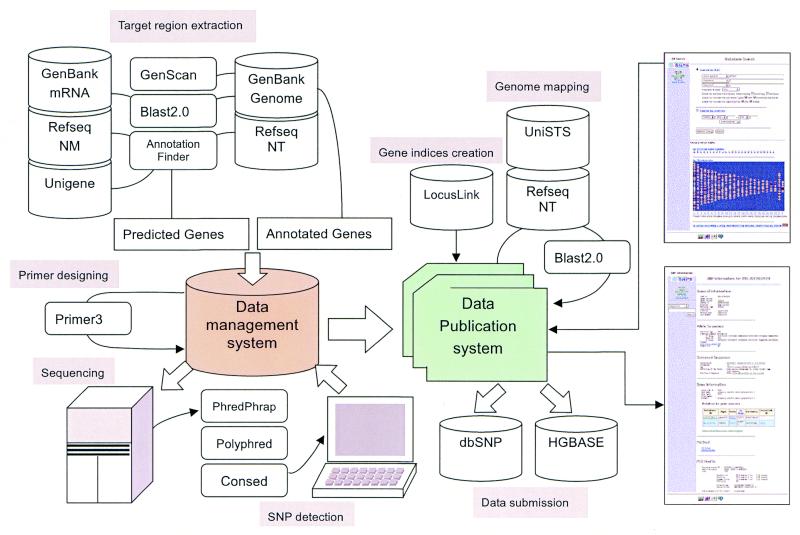
To develop JSNP, we separated the data management system, which stores all the output from the experimental processes, from the data publication system, which serves as a user interface to retrieve and display the JSNP data using the Internet (Fig. 1 shows an overview of JSNP dataflow). The data management system uses Oracle 8 and controls the applications that are used to extract the target sequences from the public sequence databases, design primers for amplification of the target regions and perform the sequencing and verification of any detected SNP candidates.
Figure 1.
Overview of JSNP dataflow.
Because large amounts of data have to be processed but only a small part of this information needs to be accessed by the user, we constructed a text based data publication system, which is distinct from the Oracle based system. As the primary focus of this report is the publicly accessible section of JSNP, the data management system will not be described any further.
The data publication system maintains the verified SNP data and any computed subsidiary information. The SNP data include the allelic bases, any insertions or deletions, and 60 bases of flanking sequences both 5′ and 3′. The sequence trace around the detected SNP is converted to GIF files and is available on the web site. The information used to design primers, which defines the target region and the associated phenotype, keeps the SNP data linked with the current public databases.
The most interesting feature of a SNP is the gene with which it is associated and how that SNP might affect the gene. We employ multiple methods to identify the target sequences, such as public database annotation, homology searches and prediction programs. Even though much evidence supports the location of the gene, biological information such as a gene name or product is often not available and its original entry has a limited description. Thus, we designed gene indices to combine as much information as possible in order to derive the gene names. In short, the gene indices contain relationships between the accession numbers or gene symbols derived from the target sequences and the gene names in the public databases, such as UniGene and LocusLink. Every target region, except the GenScan predictions, has at least one gene name defined when it was entered into the database. Thus, the gene indices work to unify any database descriptions into the format defined by LocusLink. Although the SNP location within a gene can be deduced from their primer positions, mapping to current gene sequences from UniGene clusters is performed to obtain links between the SNP entry and other database information.
Another important feature of a SNP is its location within the human genome. While most of the target regions are derived from genomic contigs from the NCBI Reference Sequences, the location of the primers are often lost because the contig sequences are updated so frequently. Therefore, the location of the SNPs are mapped to the current contig sequences, masked by MaskerAid (12), using BLAST2.0 (13) with the SNP flanking sequence, after e-PCR preformed with the primers for amplification to extract their screened regions. The criteria for this analysis, are the same as for identifying SNPs as Reference SNP in dbSNPs (11). The SNPs that map to multiple locations in the genome will not be included on the map, unless they are on the same chromosome.
Redundant SNPs are removed from the database, except those that map to two or more locations or to repetitive sequences. These SNPs are detected by the ID of the source sequences, and by the similarity of the primers and flanking sequences of the SNPs.
DATABASE USAGE
JSNP can be searched directly via the Internet (Table 3). Searching by text with Gene Symbol or Gene Name looks for a match with the GenBank annotation, UniGene ID/Gene and cross-linked information to LocusLink for each JSNP record. Searching by Accession ID identifies any DDBJ/EMBL/GenBank accession number (14–16) in the supporting information of the SNPs. Searching by dbSNP (17), JSNP and HGVbase (18) finds each SNP record by its ID directly. Options can be chosen which will limit the results returned. If the user checks the homology and GenScan boxes, the results will show any SNP found in sequences annotated as genes in GenBank. Searching by location allows the user to specify the cytogenetic location of the SNP. The ‘Contained in’ option narrows the result to within one or two specific cytogenetic bands. Lists, which have been sorted alphabetically (using the gene symbol) or by chromosome, are available for browsing. The targets for these searches are the data extracted from the public databases used to design the screening primers.
Table 3. URLs for JSNP services.
| Description | URL |
|---|---|
| JSNP home | http://snp.ims.u-tokyo.ac.jp/ |
| BLAST search for JSNP | http://www-scc.jst.go.jp/cgi-bin/sankichi/Homology_Blast2/submission_new.cgi?PROGRAM=blastn&DATABASE=SNP |
| JSNP FTP site | ftp://blue3.ims.u-tokyo.ac.jp/pub/snp/ |
| JSNP search through HOWDY | http://www-alis.tokyo.jst.go.jp/HOWDY/ |
The map view of the relationship between the SNP and other sequences such as contigs, genes and clones is also available by chromosome. These maps are created by the method previously described, so that the SNP has to be linked to a currently available sequence. The search fields seen on the map view allow the user to retrieve the SNPs by computationally associated database data.
JSNP can be searched using the standard BLAST algorithm that will compare a user-submitted sequence against all the flanking sequence records in JSNP. The flanking sequences are also available as FASTA format files from the JSNP FTP site.
DATA REPRESENTATION
The results are initially presented as a list sorted by JSNP identifier, with a hit word, such as the gene symbol, and the location of the SNP within the gene. The detailed information on each SNP includes General Information, Allele Sequence, Screened Sequence, Gene Information, Methods and PCR Profile. General Information gives the JSNP and public database (dbSNP and HGVbase) identifiers and its release date. Allele Sequence shows the allelic bases and the 60 bases of 5′ and 3′ flanking sequence. Graphics of the sequence traces and the number of valid sequence loads to determine the SNP are also shown. Screened Sequence contains the information derived from the sequence record used to define the target region. The actual position of the SNP on the sequence is shown in the sequence view of the record. The map shows all the SNPs relevant to that entry. Gene Information includes information from the gene indices and the annotations corresponding to the SNP position. These data may contain inconsistencies due to differences in derivation, because of the current state of the gene definitions.
The calculated maps consist of the Navigation Map and the Access Map. The Navigation Map, which shows density of genes, markers and SNPs, allows the user to center the map upon a point on the chromosome of interest and expand to obtain a more detailed view. The Access Map, which shows the SNP positions in detail on the chromosome, is used to obtain all the SNP data seen on the map, with a selectable scale.
FUTURE DEVELOPMENT
The identification of any new SNPs will end in early 2002. The database will continue to be updated, as human sequences and the annotated genes are improved, to maintain its effectiveness for further research. The SNPs in JSNP will be the baseline data for Japanese medical and pharmaceutical research and further development of the database will be continued. Under the Prime Minister’s Millennium Project, genotyping and feasibility studies for custom-made medical treatments are ongoing and these will give the SNP data confidence. JSNP will be improved by cross-linking the data from the latest research. The collaborations are continuing in the hope of developing tools to perform linkage analysis and to simulate polygenic disorder traits.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr Yozo Ohnishi, Dr Ryo Yamada and the members of the SNP team for discovering SNPs. We also wish to thank Hiroshi Fukagawa, Yuichi Kumaki, Hidekazu Nakamura and Yutaka Nakamura and other staff in the Bio Informatics group, INTEC Web & Genome Informatics Corporation for the database construction and Takeaki Taniguchi, Jiro Araki and Takehiko Itoh from Mitsubishi Research Institute Inc. for the primer designing and providing interfaces for the maps. This work has been supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and its predecessor, the Science and Technology Agency (STA).
REFERENCES
- 1.Wheeler D.L., Deanna,M., Church,D.M., Lash,A.E., Leipe,D.D., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Tatusova,T.A., Wagner,L. and Rapp,B.A. (2001) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 29, 11–16. Updated article in this issue: Nucleic Acids Res. (2002), 30, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pruitt K. and Maglott,D. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burge C.B. (1998) Modeling dependencies in pre-mRNA splicing signals. In Salzberg,S., Searls,D. and Kasif,S. (eds), Computational Methods in Molecular Biology. Elsevier Science, Amsterdam, The Netherlands, pp. 127–163.
- 4.Rozen S. and Skaletsky,H. (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol., 132, 365–386. [DOI] [PubMed] [Google Scholar]
- 5.Nickerson D.A., Tobe,V.O. and Taylor,S.L. (1997) Polyphred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res., 25, 2745–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon D., Abajian,C. and Green,P. (1998) Consed: a graphical tool for sequence finishing. Genome Res., 8, 195–202. [DOI] [PubMed] [Google Scholar]
- 7.Dawson E., Chen,Y., Hunt,S., Smink,L.J., Hunt,A., Rice,K., Livingston,S., Bumpstead,S., Bruskiewich,R., Sham,P., Ganske,R., Adams,M., Kawasaki,K., Shimizu,N., Minoshima,S., Roe,B., Bentley,D. and Dunham,I. (2001) A SNP resource for human chromosome 22: extracting dense clusters of SNPs from the genomic sequence. Genome Res., 11, 170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deutsch S., Iseli,C., Bucher,P., Antonarakis,S.E. and Scott,H.S. (2001) A cSNP map and database for human chromosome 21. Genome Res., 11, 300–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 10.Schuler G.D. (1998) Electronic PCR: bridging the gap between genome mapping and genome sequencing. Trends Biotechnol., 11, 456–459. [DOI] [PubMed] [Google Scholar]
- 11.Sherry S.T., Ward,M. and Sirotkin,K. (1999) dbSNP—database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res., 9, 677–679. [PubMed] [Google Scholar]
- 12.Bedell J.A., Korf,I. and Gish,W. (2000) MaskerAid: a performance enhancement to RepeatMasker. Bioinformatics, 11, 1040–1041. [DOI] [PubMed] [Google Scholar]
- 13.Gish W. and States,D.J. (1993) Identification of protein coding regions by database similarity search. Nature Genet., 3, 266–272. [DOI] [PubMed] [Google Scholar]
- 14.Tateno Y., Miyazaki,S., Ota,M., Sugawara,H. and Gojobori,T. (2000) DNA Data Bank of Japan (DDBJ) in collaboration with mass sequencing teams. Nucleic Acids Res., 28, 24–26. Updated article in this issue: Nucleic Acids Res. (2002), 30, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoesser G., Baker,W., van den Broek,A., Camon,E., Garcia-Pastor,M., Kanz,C., Kulikova,T., Lombard,V., Lopez,R., Parkinson,H., Redaschi,N., Sterk,P., Stoehr,P. and Tuli,M.A. (2001) The EMBL Nucleotide Sequence Database. Nucleic Acids Res., 29, 17–21. Updated article in this issue: Nucleic Acids Res. (2002), 30, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J., Rapp,B.A. and Wheeler,D.L. (2000) GenBank. Nucleic Acids Res., 28, 15–18. Updated article in this issue: Nucleic Acids Res. (2002), 30, 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherry S.T., Ward,M.-H., Kholodov,M., Baker,J., Phan,L., Smigielski,E.M. and Sirotkin,K. (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res., 29, 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brookes A.J., Lehvaslaiho,H., Siegfried,M., Boehm,J.G., Yuan,Y.P., Sarkar,C.M., Bork,P. and Ortigao,F. (2000) HGBASE: a database of SNPs and other variations in and around human genes. Nucleic Acids Res., 28, 356–360. Updated article in this issue: Nucleic Acids Res. (2002), 30, 387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]