Abstract
In recent dog and cat experiments, a novel milk oligosaccharide biosimilar (GNU100) positively modulated fecal microbiota and metabolite profiles, suggesting benefits to gastrointestinal health. The objective of this study was to investigate the effects of GNU100 on the fecal characteristics, microbiota, and bile acid (BA) concentrations of healthy adult dogs treated with antibiotics. Twelve healthy adult female dogs (mean age: 3.74 ± 2.4 yr) were used in an 8-wk crossover design study (dogs underwent both treatments). All dogs were fed a control diet during a 2-wk baseline, then randomly allotted to 1 of 2 treatments (diet only or diet + 1% GNU100) for another 6 wk. From weeks 2 to 4, dogs were orally administered metronidazole (20 mg/kg BW) twice daily. Fecal scores were recorded daily and fresh fecal samples were collected at weeks 2, 4, 5, 6, and 8 for measurement of pH, dry matter, microbiota populations, and BA, immunoglobulin A, and calprotectin concentrations. On weeks 0, 4, and 8, blood samples were collected for serum chemistry and hematology analysis. All data were analyzed as repeated measures using the Mixed Models procedure of SAS version 9.4, with significance considered P < 0.05. Metronidazole increased (P < 0.0001) fecal scores (looser stools) and modified (P < 0.05) fecal microbiota and BA profiles. Using qPCR, metronidazole reduced fecal Blautia, Fusobacterium, Turicibacter, Clostridium hiranonis, and Faecalibacterium abundances, and increased fecal Streptococcus and Escherichia coli abundances. DNA sequencing analysis demonstrated that metronidazole reduced microbial alpha diversity and influenced the relative abundance of 20 bacterial genera and families. Metronidazole also increased primary BA and reduced secondary BA concentrations. Most antibiotic-induced changes returned to baseline by week 8. Fecal scores were more stable (P = 0.01) in GNU100-fed dogs than controls after antibiotic administration. GNU100 also influenced fecal microbiota and BA profiles, reducing (P < 0.05) the influence of metronidazole on microbial alpha diversity and returning some fecal microbiota and secondary BA to baseline levels at a quicker (P < 0.05) rate than controls. In conclusion, our results suggest that GNU100 supplementation provides benefits to dogs treated with antibiotics, providing more stable fecal scores, maintaining microbial diversity, and allowing for quicker recovery of microbiota and secondary BA profiles which play an essential role in gut health.
Keywords: canine microbiome, canine nutrition, gastrointestinal health
This study shows that feeding a supplemental novel milk oligosaccharide biosimilar during antibiotic administration has the potential for maintaining normal stool consistency and facilitating a quicker recovery of microbiota and bile acid concentrations.
Introduction
The gastrointestinal (GI) microbiome is a complex and diverse community of bacteria and other microorganisms that are important in maintaining host health. Commensal bacteria are important producers of short-chain fatty acids (SCFA), which serve as a source of energy by the host, metabolize and alter bile acids (BA), and protect the host from invading pathogens. Maintaining host health and a stable GI tract over time, in part, depends on the level of microbiome resistance (ability to withstand change when challenged) and resilience (rate a community returns to initial composition; Shade et al., 2012; Fassarella et al., 2021). In times of distress (e.g., alterations to GI stability via stress, diet change, and/or antibiotic use), the GI microbial community may become unbalanced (i.e., dysbiosis) and have negative implications on the host.
Antibiotics are a common method to relieve GI distress and clinical signs of disease (e.g., diarrhea, loss of appetite) and function by killing or inhibiting bacterial growth. Metronidazole is one of the most commonly prescribed medications for acute diarrhea due to Giardia or bacterial infections in dogs (Singleton et al., 2019). In most cases, however, diarrhea may resolve independent of medical intervention after a week (Fenimore et al., 2017; Langlois et al., 2019) so it may be overprescribed. Despite being a popular treatment, frequent use of antibiotics may increase host susceptibility to GI disease, relapse of bacterial infection, and antibiotic resistance (Ungaro et al., 2014; Stravroulaki et al., 2021). Previous reports have demonstrated decreased bacterial species richness and dysbiosis and altered fecal metabolite profiles during antibiotic administration (Chaitman et al., 2020; Pilla et al., 2020). The antibiotic-induced dysbiosis that occurs negatively impacts stool quality and leads to the dysmetabolism of BA that are important for maintaining GI health (Manchester et al., 2019; Pilla et al., 2020).
A variety of functional ingredients may be used in the diet or supplement form to combat the negative effects of antibiotics and other factors leading to an altered GI microbiota and loose stools. Dietary fibers, probiotics, and prebiotics are known to support GI health by promoting the growth and activity of beneficial commensal bacteria that work to reduce pathogen growth, improve epithelial barrier function, and beneficially influence immunological responses (Swanson et al., 2002; Apanavicius et al., 2007; Rivière et al., 2016; Parada Venegas et al., 2019; Schmitz, 2021). Traditional plant-based prebiotics such as fructooligosaccharides and galactooligosaccharides are linear compounds that are relatively small (degree of polymerization for most is less than 10) and rapidly fermentable. The benefits of these traditional prebiotics are well-documented in companion animals (Vester and Fahey, 2009).
The search for other GI microbiome and health modulators continues, with recent interest being on milk oligosaccharides (MO) from natural and synthetic sources. Compared with plant-based oligosaccharides, MO are much more structurally diverse and complex carbohydrates. MO are nondigestible glycans composed of a variety of monosaccharides (e.g., glucose, galactose, N-acetylglucosamine, fucose, sialic acid) that vary in composition across species. In humans and animals, they are known to influence neonate development by enhancing bacterial colonization and promoting beneficial bacterial growth, aiding in the development and maturation of the gut-associated immune system, and increasing SCFA production that improves epithelial barrier function (Bode et al. 2012, 2020; McKeen et al., 2019).
In recent studies, over 55 MO from dogs and cats have been described (Macias Rostami et al., 2014; Wrigglesworth et al., 2020). With increased research and improvements to technology, human MO products are now available to support GI health for humans (e.g., 2ʹ-fucosyllactose, lacto-N-neotetraose). A similar product named GNU100 was concluded to be generally recognized as safe (GRAS) for use in the diet of cats and dogs by an independent expert panel in July 2021, and is currently undergoing FDA review. This status was obtained through safety and tolerance testing in animals and evaluation of elements such as the production process. GNU100 is a highly complex mixture of oligosaccharides and peptides derived from hydrolyzed porcine intestinal mucosa, with the structures of the oligosaccharides linked to the peptides sharing many similarities with those in dog milk, including N-acetylneuraminic acid, N-acetylgalactosamine, N-acetylglucosamine, 2ʹ-fucosyllactose, and 3ʹ-sialyllactose (Wrigglesworth et al., 2020). GNU100 was previously shown to be well-tolerated at doses up to 1.5% of the diet on an as-is basis (1.6% on a dry matter [DM] basis), not affecting total tract macronutrient digestibility, and modulating the fecal microbiota and metabolites of healthy adult dogs and cats (Lee et al., 2021; Oba et al., 2021).
In this study, the objective was to determine the effects of GNU100 supplementation on fecal characteristics, fecal microbiota, and BA concentrations of healthy adult dogs treated with antibiotics. We hypothesized that GNU100 would lead to fewer changes to fecal characteristics and BA concentrations during and after antibiotic treatment, along with fewer changes to the microbiota when compared with dogs treated with antibiotics only. Additionally, we hypothesized that GNU100-supplemented dogs would have higher concentrations of fecal immunoglobulin A (IgA), but lower fecal calprotectin concentrations than dogs treated with antibiotics alone.
Materials and Methods
All animal procedures were approved by the University of Illinois Institutional Animal Care and Use Committee prior to experimentation (IACUC #21052).
Experimental design
Twelve healthy adult female beagle dogs (11 spayed females, 1 intact female; mean age = 3.74 ± 2.40 yr; mean body weight [BW] = 8.74 ± 1.02 kg) were used in a crossover design study. All dogs were housed individually in an environmentally-controlled facility at the University of Illinois at Urbana-Champaign. Despite individual housing, dogs were socialized at least twice per week, which provided them with the ability to socialize with humans and other dogs, and had constant access to toys.
Dogs had free access to fresh water at all times and were fed twice daily (9 a.m.; 4 p.m.) at a rate to maintain BW. Dogs were fed a commercial extruded dry diet formulated to meet all nutrient recommendations for adult dogs at maintenance provided by the Association of American Feed Control Officials (AAFCO, 2021), was free of prebiotics and probiotics, and contained a low level of fermentable fiber (Purina ONE Smartblend Chicken & Rice Formula; Nestlé Purina PetCare Company, St. Louis, MO). Food offered and refused was measured each day to calculate intake and any observations of vomiting or negative reactions were recorded.
The study consisted of two 8-wk periods. Dogs were given a 2-wk baseline to consume the diet only. After baseline, dogs were allotted to two groups: diet alone or the diet + GNU100 (top dressed to the diet at each feeding at a rate to provide 1% on an as-is basis [1.1% on a DM basis] of the dietary intake). Dogs then received metronidazole (Avet Pharmaceuticals, Inc., East Brunswick, NJ) at a dosage of 20 mg/kg orally at meal times for 2 wk, which is a common treatment for bacterial intestinal infections. After antibiotic administration ceased, all dogs remained on their respective treatments for 4 wk. At the end of period 1, dogs received the opposite treatment in period 2. Dogs were weighed and body condition scores were assessed using a 9-point scale (Laflamme, 1997) once a week prior to morning feeding throughout the study. Daily fecal scores were recorded, with more frequent checks occurring during antibiotic administration because an increased number of defecations were anticipated.
Chemical analysis of diet
The diet was subsampled (100 g from 11 bags) and ground through a 2-mm screen using a Wiley mill (model 4, Thomas Scientific, Swedesboro, NJ). Procedures from the Association of Official Analytic Chemists (AOAC) were used to determine DM and ash, with organic matter (OM) being calculated (AOAC, 2006; method 934.1 for DM; method 942.05 for OM). Crude protein content was calculated from Leco total N values (TruMac N, Leco Corporation, St. Joseph, MI: AOAC, 2006; method 922.15). Acid-hydrolyzed fat content was determined according to the methods of the American Association of Cereal Chemists (AACC, 1983) and Budde (1952). Gross energy determination was calculated using an oxygen bomb calorimeter (model 6200, Parr Instruments, Moline, IL). Total dietary fiber (TDF) content was measured using methods according to Prosky et al. (1985).
Blood sample collection and analyses
At baseline and weeks 4 and 8, fasted (12 h overnight) blood samples were collected via jugular or cephalic venipuncture for serum chemistry and complete blood count (CBC). Blood was placed into BD Vacutainer Venous Blood Collection Tubes: SST Serum Separation Tubes (Becton Dickinson, Franklin Lakes, NJ) for serum metabolite concentrations and into BD Microtainer MAP Microtubes with K2EDTA and BD Microgard Closure Tubes (Becton Dickinson) for CBC. Blood for serum isolation was centrifuged 1,300 × g at 4 °C for 15 min (Beckman GS-6R centrifuge; Beckman Coulter Inc., Brea, CA). Fresh extracted serum was transported to the University of Illinois Veterinary Medicine Diagnostics Laboratory for serum chemistry analysis. Samples in K2EDTA tubes were cooled (but not frozen) and submitted to the University of Illinois Veterinary Medicine Diagnostics Laboratory for CBC analysis.
Fecal scores
During all observation and collection phases, daily fecal scores were recorded based on the following scale: 1 = hard, dry pellets, small hard mass; 2 = hard, formed, dry stool; remains firm and soft; 3 = soft, formed, and moist stool, retains shape; 4 = soft, unformed stool, assumes shape of container; and 5 = watery, liquid that can be poured.
Fecal sample collection
At weeks 2, 4, 5, 6, and 8, fresh fecal samples were collected for measurement of pH, DM percentage, microbiota populations, and BA, IgA, and calprotectin concentrations. Fecal pH was measured immediately using an AP10 pH meter (Denver Instrument, Bohemia, NY) with a Beckman Electrode (Beckman Instruments Inc., Fullerton, CA). Two aliquots were used for DM determination in accordance to the Association of Official Analytical Chemists (AOAC, 2006) using a 105 °C oven. Four additional aliquots were collected into sterile cryogenic vials (Corning Inc., Corning, NY), immediately frozen in dry ice, then stored at −80 °C until analyses.
Fecal protein analysis
Fecal proteins were extracted in accordance with Vilson et al. (2016) prior to IgA and calprotectin analysis. Fecal samples were weighed (500 mg) into conical tubes and vortexed in 1.5 mL of extraction buffer consisting of 100-mL ethylenediaminetetraacetic acid (ThermoFisher, Waltham, MA), 100-μg trypsin inhibitor (Sigma, St. Louis, MO), 20-mg bovine serum albumin (Tocris Bioscience, Bristol, UK) and 900 mL of phosphate buffered saline. Phenylmethanesulphonyl fluoride (20.1 μL, 350 mg/L; Sigma, St. Louis, MO) was added to each tube and centrifuged for 10 min at 2,500 × g. Supernatants were collected and fecal protein measurements were collected using commercial enzyme-linked immunosorbent assay kits for IgA (MBS018650; MyBioSource, Inc., San Diego, CA) and calprotectin (MBS030023; MyBioSource, Inc., San Diego, CA).
Fecal bile acid analysis
The protocol for quantifying BA was adapted and modified from the methods previously described by Batta et al. (2002). Briefly, an aliquot of 10- to 15-mg lyophilized stool was added to 200 µL of 1-butanol containing internal standards (cholic acid-d4 and lithocholic acid-d4) followed by adding 20 µl of hydrochloric acid. Samples were incubated for 4 h at 65 °C. Following incubation, samples were completely evaporated at 65 °C under nitrogen gas, 200-µL trimethylsilylation derivatization agent was added and samples were incubated for 30 min. The samples were then evaporated under nitrogen gas and resuspended in 200-µL hexane, vortexed, and centrifuged at 4 °C for 10 min at 3,000 × g. The supernatant was then analyzed by gas chromatography and mass spectrometry according to methods described by Blake et al. (2019). Cholic acid (CA), chenodeoxycholic acid (CDCA), lithocholic acid (LCA), deoxycholic acid (DCA), and ursodeoxycholic acid (UDCA) were measured.
Fecal DNA extraction
Total DNA was extracted from approximately 200 mg of fecal samples using the DNeasy PowerLyzer PowerSoil Kit (MoBio Laboratories, Carlsbad, CA) according to the manufacturer’s protocol. Samples underwent bead beating using a vortex, followed by further centrifugation to purify DNA, then quantified using a Qubit 3.0 Fluorometer (Life Technologies, Grand Island, NY). DNA quality was determined using an E-Gel Power Snap Electrophoresis Device (Invitrogen, Waltham, MA) on E-Gel EX 1% agarose gels. The DNA samples were stored at −20 °C until analysis.
Quantitative polymerase chain reaction analysis
qPCR analysis of selected bacterial groups commonly present in the GI tract of dogs was performed with specific primers targeting Faecalibacterium, Fusobacterium, Blautia, universal bacteria, Turicibacter, Escherichia coli, Clostridium hiranonis, and Streptococcus as described in AlShawaqfeh et al. (2017). Briefly, the conditions for qPCR were as follows: initial denaturation at 98 °C for 2 min, then 40 cycles with denaturation at 98 °C for 3 s, and annealing for 3 s (see Supplementary Table S1 for specific annealing temperatures for the final qPCR panel). Melt curve analysis was performed to validate the specific generation of the qPCR product using these conditions: 95 °C for 1 min, 55 °C for 1 min, and increasing incremental steps of 0.5 °C for 80 cycles for 5 s each. Each reaction was run in duplicate. The qPCR data were expressed as the log amount of DNA (fg) for each particular bacterial group/10 ng of isolated total DNA as reported previously (Suchodolski et al. 2012a; Panasevich et al. 2015).
Microbiota profiling with 16S rRNA gene amplicon sequencing and analysis
The 16S rRNA gene amplicon sequencing was performed using the Illumina MiSeq sequencing platform and paired-end technology (Microsynth AG, Balgach, Switzerland). The V3-V4 hypervariable region of bacterial 16S rRNA gene was amplified using the primer sets 341F_ill (5ʹ-CCTACGGGNGGCWGCAG-3ʹ) and 802R_ill (5ʹ-GACTACHVGGGTATCTAATCC-3ʹ) to generate an amplicon size of ~ 460 bp (Klindworth et al., 2012). The 16S rRNA gene PCR reactions, library preparation, quality control measures, and sequencing were performed by Microsynth. The Nextera XT Sample Preparation Kit (Illumina) was used for amplicon library preparation according to the manufacturer’s instructions. Equimolar amounts of each of the amplicon libraries were pooled and sequenced in multiplex on the MiSeq machine with a V2 reagent kit for 2 × 250 bp paired-end Nextera chemistry. Sequence demultiplexing was performed automatically by MiSeq Reporter software version 2.5. The raw Fastq files were trimmed for the presence of the Illumina adapter and primer sequences using Cutadapt v3.0 (Martin, 2011).
Quantitative Insights Into Microbial Ecology 2 (QIIME 2) version 2020.8 was used for the analysis of the sequence data (Bolyen et al., 2019). The sequences were imported into QIIME 2 using a Casava 1.8 single-end demultiplexed format. DADA2, a pooled-sample chimera filtering method, was used to denoise the sequences (Callahan et al., 2016). VSEARCH was also used to identify non16S rRNA genes, chimeric sequences, and open reference clustering of amplicon sequence variants (ASV; Rognes et al., 2016). All ASV were aligned de novo using MAFFT and used to construct a phylogenetic tree with FastTree 2 (via q2phylogeny; Price et al., 2010; Katoh and Standley, 2013). Taxonomy was assigned to ASV using a pretrained scikitlearn Naive Bayes classifier referencing SILVA database (v. 138) with a 99% identity threshold. Feature tables that represent the ASV counts for each sample were made in the HDF5based biological observation matrix format version. The QIIME2 q2-diversity plugin was used to perform alpha and beta diversity analysis to a depth of 12,465, based on the lowest number of sequences per sample, allowing for maximal retention of sequences per sample (Navas-Molina et al., 2013). The DESeq2 method was used to test significant differences in taxa abundance between microbiota of treatment and control microbiota (Love et al., 2014).
Statistical analysis
All data were analyzed as repeated measures using the Mixed Models procedure of SAS version 9.4 (SAS Institute Inc., Cary, NC), with treatment considered a fixed effect and dog a random effect. Data were tested for normality conditions using the UNIVARIATE procedure of SAS. If data did not meet normality, a logarithmic transformation was applied. If transformation failed, data were analyzed using npar1way procedures and Wilcoxson statistics were used to determine significance. Mean change from baseline differences due to diet, time, and diet*time were determined using a Fisher-protected least significant difference with a Tukey adjustment to control for experiment-wise error. To determine significance, a probability of P < 0.05 was accepted. Principal component analysis and nonmetric multidimensional scaling were used to evaluate the differences among microbial communities during treatment and control (P < 0.05) using QIIME2 and DeSeq2 methods.
Results
All 12 dogs finished period 1, but one dog was removed during period 2 due to an issue unrelated to treatments. This resulted in N = 12 for the GNU100 treatment and N = 11 for the control treatment. Mean baseline (after 2 wk of consuming diet only) data are presented in Supplementary Tables S2–S9. All animals were confirmed to be healthy prior to the study by a physical examination by a veterinarian, serum chemistry, and hematology. All serum chemistry and hematology measures were within reference ranges other than slightly lower serum globulin, slightly elevated albumin:globulin ratio, and slightly elevated monocyte counts (Supplementary Tables S3 and S4). All baseline measures were similar between treatment groups, except for red blood cell counts and platelet volume (Supplementary Table S4), fecal Turicibacter abundance (Supplementary Table S5), and fecal Butyricicoccaceae (uncultured), Coriobacteriaceae_UCG-002, and Peptostreptococcus relative abundances (Supplementary Table S6) that were different (P < 0.05).
Food intake, body weight, and body condition scores
Baseline food intake, BW, and body condition scores are presented in Supplementary Table S2. Change from baseline measurements were not affected by diet or time (Table 1). Change from baseline food intake, body condition scores, or BW were not affected by treatment or metronidazole administration (Table 1).
Table 1.
Mean change from baseline (week 2) food intake, body condition scores, and body weights of healthy adult dogs supplemented with GNU100 during and after metronidazole treatment
| Food intake, g/day (as-is) | Body condition score2 | Body weight, kg | |||||
|---|---|---|---|---|---|---|---|
| Item | CTRL1 | TRT | CTRL | TRT | CTRL | TRT | |
| ΔBL3-3 | −0.4 | −0.1 | 0.1 | 0.1 | 0.1 | 0.1 | |
| ΔBL-4 | −0.4 | −0.6 | 0.1 | 0.0 | 0.1 | 0.1 | |
| ΔBL-5 | −0.4 | −0.7 | 0.0 | 0.0 | 0.1 | 0.1 | |
| ΔBL-6 | −0.5 | −0.6 | 0.0 | 0.0 | 0.1 | 0.0 | |
| ΔBL-7 | −0.6 | −0.5 | 0.0 | 0.0 | 0.2 | 0.0 | |
| ΔBL-8 | −0.6 | −0.6 | −0.1 | 0.0 | 0.2 | 0.0 | |
| SEM4 | 0.28 | 0.07 | 0.07 | ||||
| P-value | Treatment | 0.7178 | 0.6481 | 0.9630 | |||
| Time | 0.5184 | 0.4458 | 0.0074 | ||||
| Treatment*Time | NA5 | NA | 0.5091 | ||||
1CTRL, kibble diet only; TRT, kibble diet top dressed with GNU100.
29-point scale (LaFlamme, 1997).
3∆BL, change from baseline.
4SEM, pooled standard errors of the means.
5NA, data not normal, requiring nonparametric statistical analysis.
Fecal characteristics
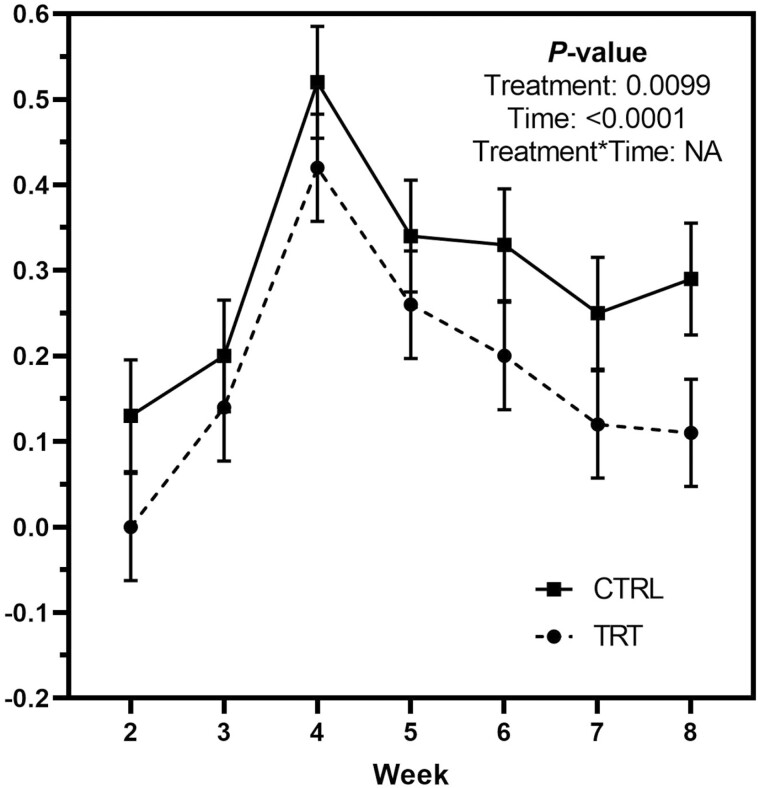
Change from baseline fecal scores were increased (P < 0.0001; looser stools) by metronidazole administration and were greater (P < 0.01; looser stools) in control dogs than those fed GNU100 (Figure 1). Change from baseline fecal pH and DM percentage were not affected by treatment or metronidazole administration (Table 2).
Figure 1.
Mean weekly change from baseline fecal scores of healthy adult dogs supplemented with GNU100 (TRT) compared with control (CTRL) dogs during and after metronidazole treatment.
Table 2.
Mean change from baseline (week 2) fecal dry matter (DM) and pH of adult dogs supplemented with GNU100 during and after metronidazole treatment
| ΔBL1-4 | ΔBL-5 | ΔBL-6 | ΔBL-8 | P-value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Item | CTRL2 | TRT | CTRL | TRT | CTRL | TRT | CTRL | TRT | SEM3 | Treatment | Time | Treatment*Time |
| Fecal DM, % | −2.09 | −2.06 | −2.04 | −1.4 | −1.71 | −1.55 | −1.54 | −0.61 | 0.73 | 0.9782 | 0.6223 | NA4 |
| Fecal pH | −0.36 | −0.62 | −0.31 | −0.34 | −0.44 | −0.23 | −0.3 | −0.23 | 0.23 | 0.9933 | 0.7997 | 0.7761 |
1∆BL, change from baseline.
2CTRL, kibble diet only; TRT, kibble diet top dressed with GNU100.
3SEM, pooled standard errors of the means.
4NA, data not normal, requiring nonparametric statistical analysis.
Serum chemistry and complete blood count
A few change from baseline serum chemistry measures were altered by treatment and metronidazole administration, but no interactions were observed (Supplementary Table S10). Change from baseline serum total protein, alkaline phosphatase, and corticosteroid-induced alkaline phosphatase were affected (P < 0.05) by treatment, while change from baseline serum Cl, glucose, bilirubin, creatine phosphokinase, and anion gap were affected (P < 0.05) by metronidazole administration. A few change from baseline CBC measures were altered by treatment, time, and treatment*time interaction (Supplementary Table S11). Change from baseline red blood cell counts, hemoglobin, hematocrit, mean cell volume, and mean platelet volume were affected (P < 0.05) by treatment, while change from baseline platelet counts were affected (P < 0.05) by metronidazole administration. Change from baseline monocyte percentage was affected (P < 0.05) by a treatment*metronidazole interaction, with all values being reduced except for baseline-week 8 values in the GNU100 group that were increased. Because all of the serum chemistry and CBC changes were small and dogs stayed within reference ranges, they are unlikely to be physiologically relevant.
Fecal microbial diversity and abundance
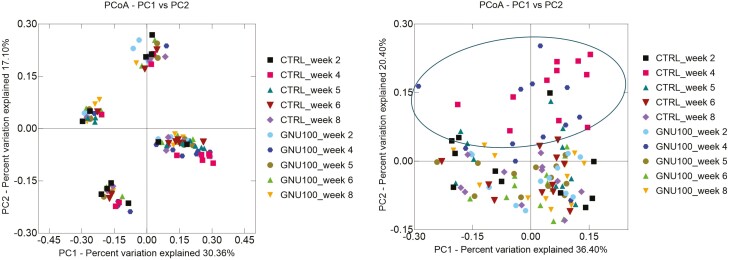
Change from baseline alpha diversity measures coming from DNA sequencing were dramatically altered (P < 0.001) by metronidazole administration. Faith’s PD, Shannon Index, and observed ASV all showed that diversity was initially reduced and then increased toward the baseline level over time (Table 3). GNU100 supplementation impacted Faith’s PD and observed ASV, with GNU100-fed dogs having smaller (P < 0.05) changes and a more stable microbiota than controls (Table 3). Microbial beta diversity, representing species richness among samples, is represented as principal coordinate analysis plots. The plot based on unweighted UniFrac distances did not show a shift over time, but the plot based on weighted UniFrac distances shows a strong effect of metronidazole treatment with week 4 samples clustering away from the other time points (Figure 2).
Table 3.
Mean change from baseline (week 2) fecal bacterial alpha diversity indices of adult dogs supplemented with GNU100 during and after metronidazole treatment
| ΔBL1-4 | ΔBL-5 | ΔBL-6 | ΔBL-8 | P-value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Item | CTRL2 | TRT | CTRL | TRT | CTRL | TRT | CTRL | TRT | SEM3 | Treatment | Time | Treatment*Time |
| Faith’s PD | −2.16 | −1.73 | −0.78 | 0.15 | −0.88 | 0.09 | −0.58 | 0.17 | 0.53 | 0.0244 | 0.0009 | 0.9380 |
| Shannon Index | −0.77 | −0.79 | −0.38 | −0.28 | −0.17 | −0.22 | −0.13 | −0.08 | 0.13 | 0.7692 | <0.0001 | 0.8564 |
| Pielou evenness | −0.03 | −0.04 | −0.02 | −0.02 | −0.01 | −0.02 | 0.00 | 0.00 | 0.02 | 0.2637 | 0.0982 | 0.9693 |
| Observed ASV | −72.04 | −69.92 | −37.58 | −14.08 | −20.04 | −5.92 | −22.76 | −10.25 | 10.35 | 0.0446 | <0.0001 | 0.6951 |
1∆BL, change from baseline.
2CTRL, kibble diet only; TRT, kibble diet top dressed with GNU100.
3SEM, pooled standard errors of the means.
Figure 2.
Beta diversity of fecal samples collected from healthy adult dogs supplemented with GNU100 during and after metronidazole treatment. Although the principal coordinate analysis plot of unweighted UniFrac distances of fecal microbial communities (left panel) showed clustering patterns based on individual animals, the principal coordinate analysis plot of weighted UniFrac distances of fecal microbial communities (right panel) demonstrated clustering patterns that separated samples immediately following metronidazole administration (week 4) from those collected at other time points.
Change from baseline fecal microbiota abundances analyzed by qPCR were altered (P < 0.05) over time due to metronidazole administration (Table 4). Change from baseline abundances of fecal total bacteria, Blautia, Fusobacterium, Turicibacter, C. hiranonis, and Faecalibacterium were initially decreased by metronidazole, but returned to baseline abundances over time. In contrast, change from baseline abundances of fecal Streptococcus and E. coli were initially increased by metronidazole, with abundances decreasing over time and approaching baseline values by week 8. Change from baseline abundances of fecal Fusobacterium, Turicibacter, and Streptococcus were affected by GNU100. Fecal Fusobacterium abundance was altered at a lower (P < 0.0001) rate and returned to baseline sooner in dogs fed GNU100 than controls. The opposite was true of fecal Turicibacter and Streptococcus, who were altered at a greater (P < 0.05) rate and returned to baseline values later in dogs fed GNU100 than controls.
Table 4.
Mean change from baseline (week 2) fecal bacterial abundance (log DNA/gram feces) of adult dogs supplemented with GNU100 during and after metronidazole treatment
| ΔBL1-4 | ΔBL-5 | ΔBL-6 | ΔBL-8 | P-value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Item | CTRL2 | TRT | CTRL | TRT | CTRL | TRT | CTRL | TRT | SEM3 | Treatment | Time | Trt*Time |
| Total bacteria | −0.21 | −0.27 | −0.05 | −0.13 | −0.04 | −0.08 | −0.05 | −0.02 | 0.05 | 0.1452 | <0.0001 | 0.3990 |
| Blautia | −1.38 | −1.12 | 0.05 | 0.16 | 0.01 | 0.17 | 0.10 | 0.09 | 0.17 | 0.2799 | <0.0001 | 0.8811 |
| Fusobacterium | −1.32 | −1.03 | −0.63 | 0.07 | −0.34 | 0.28 | −0.66 | 0.21 | 0.21 | <0.0001 | <0.0001 | 0.4236 |
| Turicibacter | −0.29 | −0.53 | 0.23 | −0.11 | 0.24 | −0.03 | 0.23 | −0.13 | 0.12 | 0.0007 | <0.0001 | 0.9541 |
| Streptococcus | 1.16 | 1.85 | −0.20 | 0.93 | −0.64 | 0.62 | −0.45 | −0.21 | 0.61 | 0.0312 | 0.0052 | 0.7608 |
| Clostridium hiranonis | −4.72 | −5.17 | −0.56 | −1.89 | −0.07 | 0.14 | 0.03 | 0.07 | 0.43 | 0.5896 | <0.0001 | NA4 |
| Escherichia coli | 2.39 | 2.39 | 1.31 | 1.35 | −0.49 | 0.96 | 0.45 | −0.27 | 0.61 | 0.5924 | 0.0002 | NA |
| Faecalibacterium | −0.96 | −0.96 | 0.32 | 0.22 | 0.14 | 0.19 | −0.14 | −0.06 | 0.23 | 0.9501 | <0.0001 | NA |
1∆BL, change from baseline.
2CTRL, kibble diet only; TRT, kibble diet top dressed with GNU100.
3SEM, pooled standard errors of the means.
4NA, not normal data, requiring nonparametric statistical analysis.
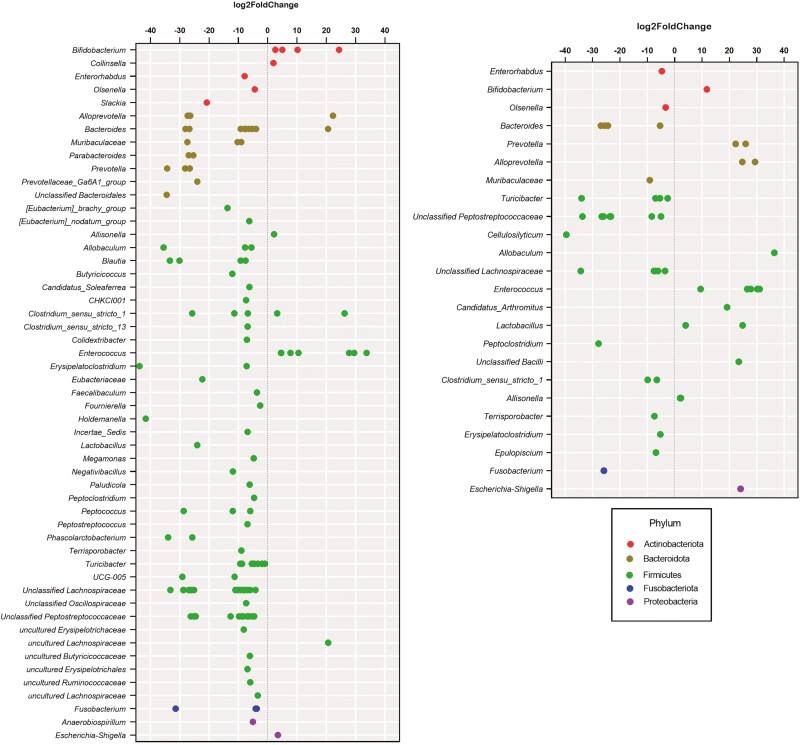
Change from baseline fecal microbiota relative abundances analyzed by DNA sequencing were drastically impacted by metronidazole administration, with 20 bacterial genera and families being changed (P < 0.05) over time (Table 5). Relative abundances of fecal Allisonella, Bifidobacterium, Catenibacterium, Collinsella, Enterococcus, Erysipelotrichaceae, Escherichia-Shigella, Faecalibacterium, Lactobacillus, and Streptococcus increased (P < 0.05) after metronidazole administration and returned to baseline over time. In contrast, relative abundances of fecal Blautia, Butyricicoccaceae (uncultured), Clostridium_sensu_stricto_1, Eggerthellaceae, Fusobacterium, Negativibacillus, Peptoclostridium, Peptococcus, Peptostreptococcaceae, and Peptostreptococcus decreased (P < 0.01) after metronidazole administration and returned to baseline over time. The metronidazole-induced changes to Blautia (reduced), Fusobacterium (reduced), Turicibacter (reduced), E. coli (increased), and Streptococcus (increased) measured by DNA sequencing were similar to that measured by qPCR. Relative abundances of fecal Allobaculum, Alloprevotella, Bacteroides, Bifidobacterium, Erysipelotrichaceae, Eubacteriaceae, Peptostreptococcus, and Turicibacter were altered by treatment, with most of these microbial taxa having smaller shifts in GNU100-supplemented dogs (Table 5). DESeq2 analysis of week 4 vs. week 2 data showed similar results, with GNU100-supplemented dogs having fewer microbial taxa changed (at least log2 fold-change; P < 0.05) by metronidazole than controls (Figure 3).
Table 5.
Mean change from baseline (week 2) fecal bacterial genera and family relative abundances (% of sequences) of adult dogs supplemented with GNU100 during and after metronidazole treatment
| ΔBL1-4 | ΔBL-5 | ΔBL-6 | ΔBL-8 | P-value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Item | CTRL2 | TRT | CTRL | TRT | CTRL | TRT | CTRL | TRT | SEM3 | Trt | Time | Trt* Time |
| Adlercreutzia | −0.06 | −0.11 | −0.02 | −0.06 | −0.05 | −0.08 | −0.09 | −0.05 | 0.04 | 0.4228 | 0.5367 | 0.5090 |
| Allisonella | 0.61 | 0.62 | 0.22 | 0.21 | 0.13 | 0.06 | 0.08 | 0.04 | 0.07 | 0.3209 | <0.0001 | NA4 |
| Allobaculum | −4.26 | −2.13 | −1.43 | −0.99 | −2.09 | −1.14 | −5.80 | 0.41 | 1.66 | 0.0125 | 0.4086 | 0.1284 |
| Alloprevotella | −1.27 | 0.81 | −1.39 | 1.07 | −1.23 | 0.41 | −1.48 | 0.65 | 0.64 | <0.0001 | 0.9470 | 0.8877 |
| Anaerobio-spirillum | 0.35 | 0.39 | 0.04 | 0.05 | 0.08 | 0.09 | 0.25 | 0.00 | 0.13 | 0.4860 | 0.2635 | NA |
| Bacteroides | −3.59 | −1.43 | −1.55 | 0.17 | −0.64 | −0.56 | −1.35 | 0.13 | 1.09 | 0.0423 | 0.1078 | 0.6965 |
| Bifidobacterium | 20.16 | 13.76 | 5.23 | −0.33 | 1.62 | 0.30 | 0.31 | 0.16 | 1.93 | 0.0113 | <0.0001 | 0.2317 |
| Blautia | −6.78 | −3.54 | 2.65 | 1.85 | 1.96 | 2.51 | 2.22 | 0.76 | 1.76 | 0.7485 | <0.0001 | 0.5097 |
| Butyricicoccaceae (uncultured) | −0.01 | −0.03 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.00 | 0.01 | 0.0755 | <0.0001 | NA |
| Catenibacterium | 0.85 | 0.13 | −0.15 | −0.21 | −0.15 | −0.04 | −0.12 | −0.38 | 0.34 | 0.8943 | 0.0427 | NA |
| Cellulosilyticum | −0.21 | −0.30 | −0.16 | −0.26 | −0.14 | −0.04 | −0.20 | −0.06 | 0.13 | 0.5470 | 0.3279 | NA |
| Clostridium_sensu_stricto_1 | −0.26 | −0.77 | 0.57 | 0.82 | 0.71 | 0.29 | 1.01 | 0.33 | 0.62 | 0.5951 | 0.0031 | NA |
| Collinsella | 1.46 | 0.53 | −0.42 | 0.05 | −0.11 | −0.08 | 0.40 | −0.13 | 0.51 | 0.4037 | 0.0169 | 0.3293 |
| Coriobacteriaceae_UCG−002 | −0.50 | −0.88 | 0.44 | −0.45 | 0.57 | −0.25 | 0.01 | −0.21 | 0.79 | 0.0627 | 0.1291 | NA |
| Dubosiella | 0.25 | 0.02 | 0.45 | 0.09 | 0.14 | 0.01 | 0.21 | 0.20 | 0.20 | 0.3813 | 0.7869 | NA |
| Eggerthellaceae | −0.09 | −0.14 | −0.04 | 0.02 | 0.00 | −0.03 | −0.01 | 0.00 | 0.04 | 0.8705 | 0.0038 | 0.3438 |
| Enterococcus | 6.35 | 4.60 | 0.24 | 0.18 | 0.27 | 0.04 | 1.77 | 0.20 | 1.09 | 0.1417 | <0.0001 | NA |
| (Eubacterium)_brachy_group | −1.00 | −1.44 | −1.00 | −1.34 | −1.00 | −1.27 | −0.96 | −0.93 | 0.27 | 0.0940 | 0.5590 | 0.6998 |
| Erysipelatoclostridium | −0.34 | −0.27 | −0.34 | −0.18 | −0.12 | −0.02 | −0.11 | 0.08 | 0.10 | 0.3209 | 0.1611 | NA |
| Erysipelotrichaceae | 1.00 | 0.02 | 0.00 | 0.01 | 0.00 | 0.00 | −0.01 | 0.00 | 0.25 | 0.0240 | 0.0006 | 0.8789 |
| Erysipelotrichaceae (uncultured) | −0.63 | −1.23 | −0.22 | 0.12 | −0.25 | −0.01 | −0.73 | 0.60 | 0.47 | 0.2838 | 0.1212 | 0.7180 |
| Erysipelotrichaceae_UCG-003 | −0.31 | −0.24 | −0.38 | −0.25 | −0.36 | −0.03 | −0.31 | −0.20 | 0.23 | 0.2410 | 0.9577 | NA |
| Escherichia-Shigella | 3.85 | 4.41 | 0.06 | 0.28 | −0.05 | 0.11 | 0.91 | 0.00 | 0.50 | 0.2442 | <0.0001 | NA |
| Eubacteriaceae | −0.05 | −0.07 | −0.03 | −0.07 | −0.01 | −0.09 | 0.03 | −0.01 | 0.03 | 0.0322 | 0.0735 | 0.7693 |
| Faecalibacterium | 0.31 | 1.81 | 1.99 | 2.90 | 1.43 | 1.26 | 0.08 | −0.73 | 1.32 | 0.7904 | 0.0358 | NA |
| Faecalibaculum | 0.08 | −0.12 | −0.09 | −0.01 | −0.04 | 0.25 | −0.16 | 0.09 | 0.13 | 0.1664 | 0.6569 | NA |
| Fournierella | −0.19 | −0.27 | −0.13 | −0.12 | 0.02 | −0.11 | −0.15 | −0.26 | 0.15 | 0.7133 | 0.4536 | NA |
| Fusobacterium | −4.17 | −3.73 | −1.28 | −0.54 | −1.00 | −0.50 | −2.15 | −0.99 | 0.97 | 0.2732 | 0.0019 | 0.9776 |
| Holdemanella | 1.45 | 0.45 | −0.30 | −0.27 | 0.09 | −0.07 | 0.62 | −0.26 | 0.62 | 0.2026 | 0.4798 | NA |
| Lachnospiraceae | 4.09 | 3.70 | 1.23 | 7.52 | 2.11 | 2.97 | 1.72 | 1.06 | 1.70 | 0.1771 | 0.2216 | 0.0994 |
| Lactobacillus | 2.60 | 3.97 | −0.25 | −0.60 | −1.29 | −1.09 | −1.76 | 1.31 | 1.33 | 0.5525 | 0.0263 | NA |
| Megamonas | −0.50 | −0.71 | −0.47 | −0.54 | −0.10 | 0.14 | 0.03 | 0.07 | 0.40 | 0.5950 | 0.2732 | NA |
| Muribaculaceae | −2.33 | −1.85 | −1.43 | −0.96 | −1.17 | −1.19 | −1.97 | −0.18 | 1.13 | 0.7427 | 0.1842 | NA |
| Negativibacillus | −0.19 | −0.24 | −0.18 | −0.18 | −0.10 | −0.10 | −0.02 | 0.05 | 0.07 | 0.9222 | <0.0001 | 0.6119 |
| Olsenella | −0.41 | −0.51 | −0.04 | −0.22 | −0.02 | −0.26 | −0.23 | 0.31 | 0.24 | 0.5114 | 0.1646 | NA |
| Parasutterella | −0.70 | 0.16 | −0.62 | −0.39 | −0.67 | −0.85 | −1.14 | −0.03 | 0.63 | 0.1703 | 0.8350 | 0.5971 |
| Peptoclostridium | −0.17 | −0.33 | 0.14 | 0.09 | 0.41 | 0.91 | 0.46 | 0.48 | 0.14 | 0.4303 | <0.0001 | 0.0801 |
| Peptococcus | −0.49 | −0.36 | −0.44 | −0.28 | −0.42 | −0.41 | 0.01 | −0.05 | 0.13 | 0.7724 | 0.0079 | NA |
| Peptostreptococcaceae | −12.37 | −12.93 | −0.25 | −5.65 | 0.81 | 2.10 | 2.80 | 0.66 | 1.82 | 0.1034 | <0.0001 | 0.1272 |
| Peptostreptococcus | −0.16 | −0.73 | −0.02 | −0.35 | 1.51 | 0.16 | 0.84 | 1.08 | 0.40 | 0.0016 | 0.0010 | NA |
| Phascolarctobacterium | −0.61 | −0.25 | −0.13 | 0.02 | −0.23 | −0.11 | −0.10 | −0.09 | 0.22 | 0.2681 | 0.2516 | 0.8441 |
| Prevotella | −3.38 | −0.67 | −1.05 | −0.04 | −1.71 | −1.27 | −1.63 | −1.20 | 1.38 | 0.5015 | 0.1966 | NA |
| Prevotellaceae_Ga6A1_group | −0.34 | −0.32 | −0.21 | −0.28 | −0.19 | −0.32 | −0.11 | −0.18 | 0.11 | 0.0673 | 0.3430 | NA |
| Slackia | −0.05 | −0.01 | 0.11 | 0.13 | 0.08 | 0.09 | 0.13 | 0.09 | 0.07 | 0.8512 | 0.1063 | NA |
| Streptococcus | 2.58 | 4.08 | −0.07 | 0.33 | 0.11 | 0.12 | 4.37 | −0.02 | 1.42 | 0.5683 | 0.0106 | NA |
| Succinivibrionaceae | 0.27 | 0.61 | −0.02 | −0.02 | −0.03 | 0.01 | 0.30 | −0.08 | 0.17 | 0.8389 | 0.0614 | NA |
| Sutterella | 0.19 | 0.64 | 0.01 | 0.16 | 0.04 | 0.07 | 0.26 | 0.00 | 0.17 | 0.3483 | 0.0871 | NA |
| Turicibacter | −0.72 | −4.55 | 0.69 | −1.10 | 1.06 | −1.85 | 1.89 | −2.42 | 1.67 | 0.0018 | 0.2341 | 0.8094 |
1∆BL, change from baseline.
2CTRL, kibble diet only; TRT, kibble diet top dressed with GNU100.
3SEM, pooled standard errors of the means.
4NA, data not normal, requiring nonparametric statistical analysis.
Figure 3.
DESeq2 analysis of week 4 vs. week 2 data demonstrates that fewer microbial taxa were affected (at least log2 fold-change; P < 0.05) by metronidazole administration in dogs supplemented with GNU100 (right panel) compared with controls (left panel). Each ASV affected is indicated by a dot, with dots colored according to the phylum of which they are a member.
Fecal bile acid
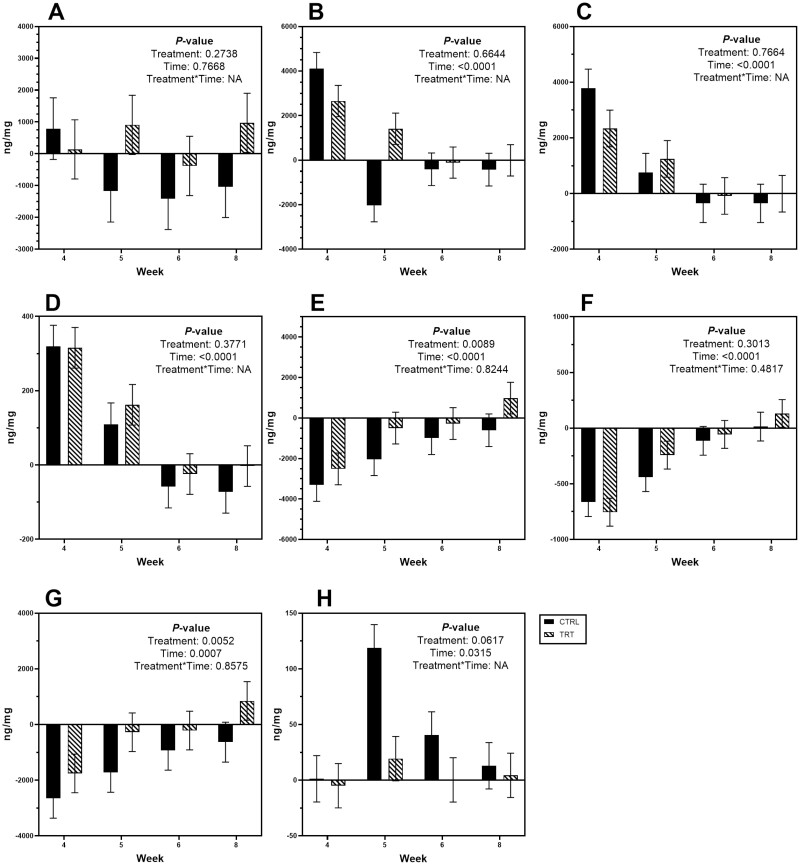
Total BA concentrations were unchanged over time, but change from baseline concentrations and percentages of individual BA were drastically altered (P < 0.05) by metronidazole administration (Table 6; Figure 4). Following metronidazole administration, fecal CA, CDCA, and total primary BA concentrations and percentages initially increased (P < 0.0001) and then returned closer to baseline over time. In contrast, change from baseline fecal LCA, DCA, and total secondary BA concentrations and percentages decreased (P < 0.001) after antibiotic administration and returned closer to baseline over time. Change from baseline fecal UDCA concentrations were increased (P < 0.05) by metronidazole, while fecal UDCA percentages were variable over time. GNU100 affected the change from baseline concentrations of DCA and total secondary BA as well as UDCA percentage. In general, dogs fed GNU100 had a smaller (P < 0.01) decrease in fecal DCA and total BA concentrations following metronidazole administration than those fed the control. Dogs fed GNU100 also had smaller (P < 0.01) changes to fecal UDCA percentage than those fed the control.
Table 6.
Mean change from baseline (week 2) fecal bile acid (BA) percentages of adult dogs supplemented with GNU100 during and after metronidazole treatment
| ΔBL1-4 | ΔBL-5 | ΔBL-6 | ΔBL-8 | P-value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Item | CTRL2 | TRT | CTRL | TRT | CTRL | TRT | CTRL | TRT | SEM3 | Treatment | Time | Trt*Time |
| Primary BA | 59.0 | 65.2 | 27.7 | 31.1 | −2.9 | −2.9 | −3.9 | −2.8 | 6.66 | 0.5593 | <0.0001 | 0.9640 |
| CA4 | 52.0 | 56.1 | 23.9 | 27.2 | −2.7 | −2.1 | −3.5 | −2.3 | 5.76 | 0.5529 | <0.0001 | 0.9871 |
| CDCA | 7.2 | 9.1 | 3.8 | 4.0 | −0.1 | −0.7 | −0.5 | −0.7 | 1.19 | 0.6902 | <0.0001 | 0.7473 |
| Secondary BA | −59.0 | −65.2 | −27.7 | −31.1 | 2.9 | 2.9 | 3.9 | 2.8 | 6.66 | 0.5593 | <0.0001 | 0.9640 |
| LCA | −14.8 | −20.8 | −9.2 | −8.5 | 0.3 | 2.5 | 1.6 | 0.1 | 3.24 | 0.7427 | <0.0001 | NA5 |
| DCA | −43.6 | −44.3 | −21.9 | −22.8 | 0.6 | −0.3 | 1.9 | 3.3 | 5.16 | 0.9383 | <0.0001 | 0.9943 |
| UDCA | −0.3 | −0.3 | 3.4 | 0.1 | 2.0 | 0.4 | 0.8 | −0.6 | 0.71 | 0.0032 | 0.0276 | 0.2516 |
1∆BL, change from baseline.
2CTRL, kibble diet only; TRT, kibble diet top dressed with GNU100.
3SEM, pooled standard errors of the means.
4CA, cholic acid; CDCA, chenodeoxycholic acid; LCA, lithocholic acid; DCA, deoxycholic acid; UDCA, ursodeoxycholic acid.
5NA, not normal data, requiring nonparametric statistical analysis.
Figure 4.
Mean change from baseline fecal (A) total bile acid, (B) total primary bile acid, (C) cholic acid, (D) chenodeoxycholic acid, (E) total secondary bile acid, (F) lithocholic acid, (G) deoxycholic acid, and (H) ursodeoxycholic acid concentrations (ng/mg feces) of healthy adult dogs supplemented with GNU100 (TRT) compared with control (CTRL) dogs during and after metronidazole treatment. Data are presented least square means ± standard errors of the means.
Fecal calprotectin and fecal IgA
All fecal calprotectin concentrations were low throughout the study. Change from baseline fecal calprotectin concentrations were affected by metronidazole administration, with concentrations decreasing (P < 0.01) after antibiotic administration (Table 7). Change from baseline fecal calprotectin concentrations were also affected by GNU100, with concentrations decreasing (P < 0.0001) to a greater extent in control dogs than those fed GNU100. Change from baseline fecal IgA concentrations were not affected by GNU100 or metronidazole administration.
Table 7.
Mean change from baseline (week 2) fecal calprotectin (μg/g feces, dry matter basis) and immunoglobulin A (mg/g feces, dry matter basis) concentrations of healthy adult dogs supplemented with GNU100 during and after metronidazole treatment
| ΔBL1-4 | ΔBL-5 | ΔBL-6 | ΔBL-8 | P-value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Item | CTRL2 | TRT | CTRL | TRT | CTRL | TRT | CTRL | TRT | SEM3 | Treatment | Time | Treatment*Time |
| Calprotectin | −0.55 | −0.15 | −0.44 | −0.12 | −0.34 | 0.05 | −0.05 | −0.01 | 0.11 | <0.0001 | 0.0085 | 0.2177 |
| Immunoglobulin A | 0.92 | 1.60 | 2.17 | 1.29 | 0.49 | 1.24 | 0.87 | 1.60 | 0.78 | 0.4849 | 0.6012 | 0.5004 |
1∆BL, change from baseline.
2CTRL, kibble diet only; TRT, kibble diet top dressed with GNU100.
3SEM, pooled standard errors of the means.
Discussion
Previous research has reported that MO stimulate the growth of beneficial bacteria, have anti-adhesive effects against pathogens, aid in the maturation of the gut immune system, and increase barrier function (Kunz et al., 2000; Boehm and Stahl, 2007; Oliveira et al., 2015; McKeen et al., 2019; Bode, 2020; Wiciński et al., 2020). Because MO may act as decoys and/or be selectively utilized substrates by beneficial bacteria, they may promote host health by enhancing GI microbial resistance and resilience. In comparison to what has been done in humans and animal models, little MO research has been conducted in dogs and cats, with only a couple reports on dog and cat MO profiles being reported (Macias Rostami et al., 2014; Wrigglesworth et al., 2020). Moreover, because the extraction of MO from dog and cat milk is not a viable option commercially, ingredient sources of other origins but having similar properties are of interest.
GNU100 is a functional ingredient option with the potential to play a nutritional role in supporting normal digestive function, as it is a highly complex mixture of oligosaccharides and peptides, with many of the oligosaccharide structures being similar to those in dog milk (Wrigglesworth et al., 2020). In vitro fermentation testing of GNU100 demonstrated that it was fermentable by commensal microbiota, increasing SCFA production (Oba et al., 2020), suggesting it should also influence microbiota in vivo. Consequent testing in canines and felines showed that GNU100 modified fecal microbiota and metabolites without negatively impacting apparent total tract macronutrient digestibility or fecal characteristics (Lee et al., 2021; Oba et al., 2021). Although fecal metabolites were not measured in the current study, SCFA production was shown to be increased with GNU100 fermentation in vitro and would be expected herein (Oba et al., 2020). In agreement with previous reports (Lee et al., 2021; Oba et al., 2021), few blood parameters were impacted by GNU100 supplementation in the current study, with all remaining within normal ranges.
In the current study, dogs consuming GNU100 demonstrated higher microbial resistance to antibiotic treatment and higher resilience following its cessation, with several microbial taxa and BA returning to baseline concentrations quicker than control dogs. Although the physiological significance of GNU100 providing greater stability of certain taxa (e.g., Allobaculum, Alloprevotella, Bacteroides, Bifidobacterium, Erysipelotrichaceae, and Fusobacterium) may not be apparent, Fusobacterium is negatively associated with GI disease and is thought to play a role in microbiome canine health (Suchodolski et al. 2012a, 2012b; AlShawaqfeh et al., 2017). Therefore, the smaller shift and quicker recovery in fecal Fusobacterium abundance of GNU100-supplemented dogs would also appear to be beneficial. For a few microbial taxa, such as fecal Turicibacter and Streptococcus abundances and fecal Peptostreptococcus relative abundance, GNU100 was not successful in promoting their stability or increasing the rate at which they returned to baseline. Because these taxa have not been associated with health or disease in dogs, the relevance of these changes are not easy to interpret. However, the greater overall stability demonstrated by DESeq2 analysis and greater resilience demonstrated by alpha diversity measures would be viewed as being beneficial.
Antibiotic administration is known to not only influence GI tract microbiota abundances (e.g., dysbiosis), but also influences microbial activity and stool quality and increases potential for relapse of disease. Such changes to the microbiota often impact fecal metabolite profiles, including SCFA, protein catabolites, and BA. Fecal SCFA and protein catabolites were not measured in the current study, but the changes to the BA concentrations and profiles were quite interesting. Primary BA are synthesized from cholesterol into CA and CDCA and are actively transported into the gut to aid in the absorption of lipids, fat-soluble vitamins, and cholesterol. Once they reach the ileum, they can be reabsorbed; however, any that escape active transport are exposed to bacteria in the large intestine and may be deconjugated and biotransformed, creating secondary BA (DCA, LCA, UDCA; Ciaula et al., 2017). Some species in the Clostridium genus produce bile salt hydrolases to deconjugate BA (e.g., C. hiranonis, C. scindens). C. hiranonis is the primary BA converter in dogs, while C. scindens is the primary taxa involved with BA dehydroxylation reactions in humans (Ridlon et al. 2006, 2016; Guzior and Quinn, 2021). Similar to the current study, those researchers reported a significant increase in primary BA with the decline in C. hiranonis and a sequential decline in secondary BA concentrations without the activity of the bile salt hydrolases. An inability to recover C. hiranonis and secondary BA profiles are known to increase the likelihood of relapse in diseased dogs. In a recent study, the actions of C. scindens were demonstrated to help a mouse model resist C. difficile infection by increasing secondary BA concentrations, suggesting that manipulation of BA-metabolizing bacteria may be a plausible therapeutic for individuals at risk of such infections (Buffie et al., 2015). In humans, some GI diseases (e.g., BA diarrhea, inflammatory bowel disease) have been correlated with increased primary BA loss in the feces (Vijayvargiya et al., 2019; Lavvelle and Sokol, 2020). In humans, low secondary BA concentrations and large microbial diversity shifts have been reported in patients diagnosed with cirrhosis or dysbiosis-induced secondary BA deficiency (Kakiyama et al., 2013; Sinha et al., 2020). While GNU100 supplementation did not appear to affect C. hiranonis recovery in the current study, it did quicken the return of secondary BA. This differential response could be due to the actions of other microbial taxa capable of BA metabolism or an increased metabolic capability of the C. hiranonis present in GNU100-supplemented dogs. In any case, this result was deemed to be a beneficial outcome of GNU100 supplementation.
In the present study, antibiotic administration did not influence total BA concentrations, but affected all specific BA concentrations and ratios. The influence of GNU100 was more targeted, with the metabolism and percentage of DCA and UDCA being impacted. During antibiotic administration, primary BA were dramatically increased, while secondary BA dramatically decreased. Because total BA were not influenced, it suggests a decreased conversion of primary to secondary BA by GI microbiota rather than changes in re-absorption. With GNU100 supplementation, BA were able to return to baseline concentrations at a quicker rate. These results, along with microbiota data, implies that GNU100 aids in a faster recovery of normal BA profiles by maintaining stability and aiding in a quicker recovery of the GI microbiome.
The current study had several strengths and limitations. Strengths included the number of relevant physiologic and microbiota-based measurements and the crossover experimental design that allowed for strong comparisons between treatments. Administering metronidazole for a full 2 wk mimicked that of a traditional clinical prescription. Finally, the high frequency of sample collections, fecal scoring, and behavioral observations allowed for longitudinal analysis and potential identification of treatment*time interactions. A potential limitation to the study included the timing of GNU100 supplementation, which was started at the time of metronidazole administration. Although this strategy is similar to what is often done in the clinic, with probiotics given to animals undergoing antibiotic therapy, this timing could have limited GNU100’s ability in stabilizing GI tract microbiota. In the future, dosing of GNU100 prior to antibiotic administration may allow it to provide GI and immunological benefits that would enhance its ability to resist change. Also, the protocol did not include the testing on clinically-symptomatic animals. Future studies in client-owned animals is warranted so that the effects of GNU100 can be tested in a more clinically relevant population.
In conclusion, oral metronidazole administration dramatically impacted the fecal microbiota populations of healthy adult dogs, reducing alpha diversity measures, altering beta diversity, and affecting the relative abundances of 20 bacterial genera and families. Metronidazole administration also increased fecal scores (looser stools) and influenced fecal BA profiles, increasing primary BA concentrations and reducing secondary BA concentrations. Some of the outcomes measured were attenuated by GNU100 supplementation. Based on alpha diversity measures (Shannon Index and observed ASV) and DESeq2 analysis, GNU100 appeared to lessen metronidazole’s effects on the overall diversity of the GI microbiota. GNU100-supplemented dogs also maintained fecal scores better than controls and had a quicker return of some fecal microbial taxa and secondary BA concentrations after antibiotic administration ceased. While more research is warranted, the current study suggests that GNU100 supplementation may provide benefits to dogs by providing gastrointestinal microbial stability and allowing for a quicker recovery of microbiota populations and metabolism following antibiotic administration.
Supplementary Material
Acknowledgments
The funding for this study was provided by Gnubiotics Sciences SA, Epalinges, Switzerland.
Glossary
Abbreviations
- ASV
amplicon sequence variants
- BA
bile acids
- BW
body weight
- CA
cholic acid
- CDCA
chenodeoxycholic acid
- DCA
deoxycholic acid
- DM
dry matter
- GI
gastrointestinal
- GRAS
generally recognized as safe
- IgA
immunoglobulin A
- LCA
lithocholic acid
- MO
milk oligosaccharides
- qPCR
quantitative polymerase chain reaction
- SCFA
short-chain fatty acids
- UDCA
ursodeoxycholic acid
Contributor Information
Sara E Belchik, Department of Animal Sciences, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.
Patricia M Oba, Department of Animal Sciences, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.
Romain Wyss, Gnubiotics Sciences, Route de la Corniche 6, Epalinges, Switzerland.
Paul T Asare, Gnubiotics Sciences, Route de la Corniche 6, Epalinges, Switzerland.
Sara Vidal, Gnubiotics Sciences, Route de la Corniche 6, Epalinges, Switzerland.
Yong Miao, Gnubiotics Sciences, Route de la Corniche 6, Epalinges, Switzerland.
Yemi Adesokan, Gnubiotics Sciences, Route de la Corniche 6, Epalinges, Switzerland.
Jan S Suchodolski, Gastrointestinal Laboratory, Department of Small Animal Clinical Sciences, Texas A&M University, College Station, TX, USA.
Kelly S Swanson, Department of Animal Sciences, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA; Division of Nutritional Sciences, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA; Department of Veterinary Clinical Medicine, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.
Conflict of Interest Statement
S.V., R.W., P.T.A., Y.M., and Y.A. are employed by Gnubiotics Sciences SA. K.S.S. has served as a paid consultant for Gnubiotics Sciences SA. The remaining authors have no conflicts of interest.
Literature Cited
- AlShawaqfeh, M. K., B. Wajid, Y. Minamoto, M. Markel, J. A. Lidbury, J. M. Steiner, E. Serpedin, and J. S. Suchodolski. . 2017. A dysbiosis index to assess microbial changes in fecal samples of dogs with chronic inflammatory enteropathy. FEMS Microbiol. Ecol. 93:fix136. doi: 10.1093/femsec/fix136 [DOI] [PubMed] [Google Scholar]
- American Association of Cereal Chemists (AACC). 1983. Approved methods. 8th ed. St Paul (MN): American Association of Cereal Chemists. [Google Scholar]
- Apanavicius, C. J., K. L. Powell, B. M. Vester, L. K. Karr-Lilienthal, L. L. Pope, N. D. Fastinger, M. A. Wallig, K. A. Tappenden, and K. S. Swanson. . 2007. Fructan supplementation and infection affect food intake, fever, and epithelial sloughing from Salmonella challenge in weanling puppies. J. Nutr. 137:1923–1930. doi: 10.1093/jn/137.8.1923 [DOI] [PubMed] [Google Scholar]
- Association of American Feed Control Officials (AAFCO). 2021. Official Publication 2021, Oxford (IN): AAFCO [Google Scholar]
- Association of Official Analytical Chemists (AOAC). 2006. Official methods of analysis. 17th ed. Gaithersburg (MD): Association of Official Analytical Chemists. [Google Scholar]
- Batta, A. K., G. Salen, P. Batta, G. S. Tint, D. S. Alberts, and D. L. Earnest. . 2002. Simultaneous quantitation of fatty acids, sterols and bile acids in human stool by capillary gas-liquid chromatography. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 775:153–161. doi: 10.1016/s1570-0232(02)00289-1 [DOI] [PubMed] [Google Scholar]
- Blake, A. B., B. C. Guard, J. B. Honneffer, J. A. Lidbury, J. M. Steiner, and J. S. Suchodolski. . 2019. Altered microbiota, fecal lactate, and fecal bile acids in dogs with gastrointestinal disease. PLoS One 14:e0224454. doi: 10.1371/journal.pone.0224454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode, L. 2012. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology 22:1147–1162. doi: 10.1093/glycob/cws074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode, L. 2020. Human milk oligosaccharides: structure and functions. Nestlé Nutr. Inst. Workshop Ser. Basel, Karger. 94:115–123. doi: 10.1159/000505339 [DOI] [PubMed] [Google Scholar]
- Boehm, G., and B. Stahl. . 2007. Oligosaccharides from milk. J. Nutr. 137:847S–849S. doi: 10.1093/jn/137.3.847s [DOI] [PubMed] [Google Scholar]
- Bolyen, E., J. R. Rideout, M. R. Dillon, N. A. Bokulich, C. C. Abnet, G. A. Al-Ghalith, H. Alexander, E. J. Alm, M. Arumugam, F. Asnicar, . et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37:852–857. doi: 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde, E. F. 1952. The determination of fat in baked biscuit type of dog foods. J. Assoc. Off. Agric. Chem. 35:799–805. doi: 10.1093/jaoac/35.3.799 [DOI] [Google Scholar]
- Buffie, C. C., V. Bucci, R. R. Stein, P. T. McKenney, L. Ling, A. Gobourne, D. No, H. Lui, M. Kinnebrew, A. Viale, . et al. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan, B. J., P. J. McMurdie, M. J. Rosen, A. W. Han, A. J. A. Johnson, and S. P. Holmes. . 2016. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13:581–583. doi: 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaitman, J., A. Ziese, R. Pilla, Y. Minamoto, A. B. Blake, B. C. Guard, A. Isaiah, J. A. Lidbury, J. M. Steiner, S. Unterer, . et al. 2020. Fecal microbial and metabolic profiles in dogs receiving either fecal microbiota transplantation or oral metronidazole. Front. Vet. Sci. 7:192. doi: 10.3389/fvets.2020.00192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciaula, A. D., G. Garruti, R. L. Baccetto, E. Molina-Molina, L. Bonfrate, D. Q. Wang, and P. Portincasa. . 2017. Bile acid physiology. Ann. Hepatol. 16:s4–s14. doi: 10.5604/01.3001.0010.5493 [DOI] [PubMed] [Google Scholar]
- Fassarella, M., E. E. Blaak, J. Penders, A. Nauta, H. Smidt, and E. G. Zoetendal. . 2021. Gut microbiome stability and resilience: elucidating the response to perturbations in order to modulate gut health. Gut 70:595–605. doi: 10.1136/gutjnl-2020-321747 [DOI] [PubMed] [Google Scholar]
- Fenimore, A., L. Martin, and M. R. Lappin. . 2017. Evaluation of metronidazole with and without Enterococcus faecium SF68 in shelter dogs with diarrhea. Top. Comp. Anim. Med. 32:100–103. doi: 10.1053/j.tcam.2017.11.001 [DOI] [PubMed] [Google Scholar]
- Guzior, D. V., and R. A. Quinn. . 2021. Review: microbial transformations of human bile acids. Microbiome. 9:140. doi: 10.1186/s40168-021-01101-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakiyama, G., W. M. Pandak, P. M. Gillevet, P. B. Hylemon, D. M. Heuman, K. Daita, H. Takei, A. M. Muto, H. Nittono, J. M. Ridlon, . et al. 2013. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J. Hepatol. 58:949–955. doi: 10.1016/j.jhep.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K., and D. M. Standley. . 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30:772–780. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klindworth, A., E. Pruesse, T. Schweer, J. Peplies, C. Quast, M. Horn, and F. O. Glöckner. . 2012. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1. doi: 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz, C., S. Rudloff, W. Baie, N. Klein, and S. Strobel. . 2000. Oligosaccharides in human milk: Structural, functional and metabolic aspects. Annu. Rev. Nutr. 20:699–722. doi: 10.1146/annurev.nutr.20.1.699 [DOI] [PubMed] [Google Scholar]
- Laflamme, D. P. 1997. Development and validation of a body condition score system for cats: a clinical tool. Feline Pract. 25:13–18. [Google Scholar]
- Langlois, D. K., A. M. Koenigshof, and R. Mani. . 2019. Metronidazole treatment of acute diarrhea in dogs: A randomized double blinded placebo-controlled clinical trial. J. Vet. Intern. Med. 34:98–104. doi: 10.1111/jvim.15664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavelle, A., and H. Sokol. . 2020. Gut microbiota-derived metabolites as key factors in inflammatory bowel diseases. Nat. Rev. Gastroenterol. Hepatol. 17:223–237. doi: 10.1038/s41575-019-0258-z [DOI] [PubMed] [Google Scholar]
- Lee, A. H., S. Vidal, P. M. Oba, R. Wyss, Y. Miao, Y. Adesokan, and K. S. Swanson. . 2021. Evaluation of a novel animal milk oligosaccharide biosimilar: macronutrient digestibility and gastrointestinal tolerance, fecal metabolites, and fecal microbiota of healthy adult dogs and in vitro genotoxicity assays. J. Anim. Sci. 99:1–14. doi: 10.1093/jas/skab014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M. I., W. Huber, and S. Anders. . 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias-Rostami, S., T. Bénet, J. Spears, A. Reynolds, E. Satyaraj, N. Sprenger, and S. Austin. . 2014. Milk oligosaccharides over time of lactation from different dog breeds. PLoS One 9:e99824. doi: 10.1371/journal.pone.0099824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchester, A. C., C. B. Webb, A. B. Blake, F. Sarwar, J. A. Lidbury, J. M. Steiner, and J. S. Suchodolski. . 2019. Long-term impact of tylosin on fecal microbiota and fecal bile acids of healthy dogs. J. Vet. Intern. Med. 33:2605–2617. doi: 10.1111/jvim.15635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:101. doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- McKeen, S., W. Young, K. Fraser, N. C. Roy, and W. C. McNabb. . 2019. Glycan utilisation and function in the microbiome of weaning infants. Microorganisms. 7:190. doi: 10.3390/microorganisms7070190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas-Molina, J. A., J. M. Peralta-Sánchez, A. González, P. J. McMurdie, Y. Vázquez-Baeza, Z. Xu, L. K. Ursell, C. Lauber, H. Zhou, S. J. Song, . et al. 2013. Advancing our understanding of the human microbiome using QIIME. Methods Enzymol. 531:371–444. doi: 10.1016/B978-0-12-407863-5.00019-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oba, P. M., S. Vidal, R. Wyss, Y. Miao, Y. Adesokan, and K. S. Swanson. . 2020. Effect of a novel animal milk oligosaccharide biosimilar on the gut microbial communities and metabolites of in vitro incubations using feline and canine fecal inocula. J. Anim. Sci. 98:1–11. doi: 10.1093/jas/skaa273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oba, P. M., A. H. Lee, S. Vidal, R. Wyss, Y. Miao, Y. Adesokan, and K. S. Swanson. . 2021. Effect of novel animal milk oligosaccharide biosimilar on macronutrient digestibility and gastrointestinal tolerance, fecal metabolites, and fecal microbiota of healthy adult cats. J. Anim. Sci. 99:1–15. doi: 10.1093/jas/skaa399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira, D. L., R. A. Wilbey, A. S. Grandison, and L. B. Roseiro. . 2015. Milk oligosaccharides: a review. Int. J. Dairy Tech. 68:305–321. doi: 10.1111/1471-0307.12209 [DOI] [Google Scholar]
- Panasevich, M. R., K. R. Kerr, R. N. Dilger, G. C. Fahey, Jr, L. Guérin-Deremaux, G. L. Lynch, D. Wils, J. S. Suchodolski, J. M. Steiner, S. E. Dowd, . et al. 2015. Modulation of the faecal microbiome of healthy adult dogs by inclusion of potato fibre in the diet. Brit. J. Nutr. 113:125–133. doi: 10.1017/S0007114514003274 [DOI] [PubMed] [Google Scholar]
- Parada Venegas, D., M. K. De la Fuente, G. Lanskron, M. J. González, R. Quera, G. Dijkstra, H. J. M. Harmsen, K. N. Faber, and M. A. Hermoso. . 2019. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10:277. doi: 10.3389/fimmu.2019.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilla, R., F. P. Gaschen, J. W. Barr, E. Olson, J. Honneffer, G. C. Guard, A. B. Blake, D. Villanueva, M. R. Khattab, M. K. AlShawaqfeh, . et al. 2020. Effects of metronidazole on the fecal microbiome and metabolome in healthy dogs. J. Vet. Intern. Med. 34:1853–1866. doi: 10.1111/jvim.15871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, M. N., P. S. Dehal, and A. P. Arkin. . 2010. FastTree 2 - Approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosky, L., N. G. Asp, I. Furda, J. W. DeVries, T. F. Schweizer, and B. F. Harland. . 1985. Determination of total dietary fiber in foods and food products: collaborative study. J. AOAC Int. 68:677–679. doi: 10.1093/jaoac/68.4.677 [DOI] [PubMed] [Google Scholar]
- Ridlon, J. M., D. -J. Kang, and P. B. Hylemon. . 2006. Bile salt transformations by human intestinal bacteria. J. Lipid Res. 47:241–259. doi: 10.1194/jlr.r500013-jlr200 [DOI] [PubMed] [Google Scholar]
- Ridlon, J. M., S. C. Harris, S. Bhowmik, D. -J. Kang, P. B. Hylemon, and P. B. Hylemon. . 2016. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes. 7:22–39. doi: 10.1080/19490976.2015.1127483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière, A., M. Selak, D. Lantin, F. Leroy, and L. de Vuyst. . 2016. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front. Microbiol. 7:979. doi: 10.3389/fmicb.2016.00979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognes, T., T. Flouri, B. Nichols, C. Quince, and F. Mahé. . 2016. VSEARCH: A versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz, S. S. 2021. Value of probiotics in canine and feline gastroenterology. Vet. Clin. Small Anim. 51:171–217. doi: 10.1016/j.cvsm.2020.09.011 [DOI] [PubMed] [Google Scholar]
- Shade, A., H. Peter, S. D. Allison, D. L. Baho, M. Berga, H. Bürgmann, D. H. Huber, S. Langenheder, J. T. Lennon, J. B. H. Martiny, . et al. 2012. Fundamentals of microbial community resistance and resilience. Front. Microbiol. 3:217. doi: 10.3389/fmicb.2012.00417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton, D. A., P. J. M. Noble, F. Sánchez-Vizcaíno, S. Dawson, G. L. Pinchbeck, N. J. Williams, A. D. Radford, and P. H. Jones. . 2019. Pharmaceutical prescription in canine acute diarrhoea: A longitudinal electronic health record analysis of first opinion veterinary practices. Front. Vet. Sci. 6:218. doi: 10.3389/fvets.2019.00218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, S. R., Y. Haileselassie, L. P. Nguyen, C. Tropini, M. Wang, L. S. Becker, D. Sim, K. Jarr, E. T. Spear, G. Singh, . et al. 2020. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe. 27:659–670. doi: 10.1016/j.chom.2020.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavroulaki, E. M., J. S. Suchodolski, R. Pilla, G. T. Fosgate, C. -H. Sung, J. A. Lidbury, J. M. Steiner, and P. G. Xenoulis. . 2021. Short- and long-term effects of amoxicillin/clavulanic acid or doxycycline on the gastrointestinal microbiome of growing cats. PLoS One 16:e0253031. doi: 10.1371/journal.pone.0253031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchodolski, J. S., M. E. Markel, J. F. Garcia-Mazcorro, S. Unterer, R. M. Heilmann, S. E. Dowd, P. Kachroo, I. Ivanov, Y. Minamoto, E. M. Dillman, . et al. 2012a. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS One 7:e51907. doi: 10.1371/journal.pone.0051907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchodolski, J. S., S. E. Dowd, V. Wilke, J. M. Steiner, and A. E. Jergens. . 2012b. 16S rRNA gene pyrosequencing reveals bacterial dysbiosis in the duodenum of dogs with idiopathic inflammatory bowel disease. PLoS One 7:e39333. doi: 10.1371/journal.pone.0039333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, K. S., C. M. Grieshop, E. A. Flickinger, L. L. Bauer, J. Chow, B. W. Wolf, K. A. Garleb, and G. C. Fahey, Jr. 2002. Fructooligosaccharides and Lactobacillus acidophilus modify gut microbial populations, total tract nutrient digestibilities, and fecal protein catabolite concentrations in healthy adult dogs. J. Nutr. 132:3721–3731. doi: 10.1093/jn/132.12.3721 [DOI] [PubMed] [Google Scholar]
- Ungaro, R., C. N. Bernstein, R. Gearry, A. Hviid, K. -L. Kolho, M. P. Kronman, S. Shaw, H. Van Kruinigen, J. -F. Colombel, and A. Atreja. . 2014. Antibiotics associated with increased risk of new-onset Crohn’s disease but not ulcerative colitis: a meta-analysis. Am. J. Gastroenterol. 109:1728–1738. doi: 10.1038/ajg.2014.246 [DOI] [PubMed] [Google Scholar]
- Vester, B. M., and G. C. Fahey, Jr. 2009. Prebiotics and probiotics in companion animal nutrition. In: Cho, S. S., and T. Finocchiaro (eds.). Handbook of Prebiotics and Probiotics: Health Benefits and Food Applications. Boca Raton (FL): CRC Press. [Google Scholar]
- Vijayvargiya, P., M. Camilleri, V. Chedid, P. Carlson, I. Busciglio, D. Burton, and L. Donato. . 2019. Analysis of fecal primary bile acids detects increased stool weight and colonic transit in patients with chronic functional diarrhea. Clin. Gastroenterol. Hepatol. 17:922–929. doi: 10.1016/j.cgh.2018.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilson, A., A. Hedhammar, A. Reynolds, J. Spears, E. Satyaraj, R. Pelker, C. Rottman, B. Björkstén, and H. Hansson-Hamlin. . 2016. Immunoglobulins in dogs: correspondence and maturation in 15 litters of German shepherd dogs and their dams. Vet. Rec. Open 3:e000173. doi: 10.1136/vetreco-2016-000173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiciński, M., E. Sawicka, J. Gębalski, F. Kubiak, and B. Malinowski. . 2020. Human milk oligosaccharides: health benefits, potential applications in infant formulas, and pharmacology. Nutrients 12:266. doi: 10.3390/nu12010266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrigglesworth, D. J., E. Goonatilleke, R. Haydock, K. R. Hughes, C. B. Lebrilla, K. S. Swanson, P. Jones, and P. Watson. . 2020. High-throughput glycomic analyses reveal unique oligosaccharide profiles of canine and feline milk samples. PLoS One 15:e0243323. doi: 10.1371/journal.pone.0243323 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.