Abstract
The Structural Classification of RNA (SCOR) database provides a survey of the three-dimensional motifs contained in 259 NMR and X-ray RNA structures. In one classification, the structures are grouped according to function. The RNA motifs, including internal and external loops, are also organized in a hierarchical classification. The 259 database entries contain 223 internal and 203 external loops; 52 entries consist of fully complementary duplexes. A classification of the well-characterized tertiary interactions found in the larger RNA structures is also included along with examples. The SCOR database is accessible at http://scor.lbl.gov.
INTRODUCTION
The number of RNA structures whose coordinates are available in the Protein Data Bank (PDB) (1) and the Nucleic Acid Database (NDB) (2), though small compared with the number of protein structures available, is substantial and rapidly growing. Although the great majority of the structures in the databases are made up of only one helical stack, the recent determination of structures containing two or more helical stacks, such as the hammerhead ribozyme (3), the P4–P6 domain of the Tetrahymena group I intron (4), and recently the 5S, 16S and 23S RNAs of the ribosome has greatly increased our knowledge of RNA folds (5–7). Collectively, these structures provide a large amount of information about RNA structural motifs. These motifs have also been studied extensively (8).
In order to organize this information and make it available to the non-specialist, to discover new features of RNA structure and relationships to sequence and function, and to enumerate and classify substructures for model building and RNA engineering, we are developing a database for the Structural Classification of RNA (SCOR).
As the first stage of a classification of RNA structures, we have examined, cataloged and classified all of the internal and external loops in a comprehensive collection of RNA structures contained in the PDB and NDB. A total of 259 PDB entries were examined; this includes all PDB entries, unique by structure or primary reference, containing more than one RNA residue and solved by NMR spectroscopy or X-ray crystallography, whose coordinates were released before October 4, 2000 with the exception of the 30S (9,10) and 50S (11) ribosomal subunit structures. We have initiated an analysis of domain and subdomain structure in ribosomal and other large RNAs that will be incorporated into an upcoming version of the SCOR database. Of the 259 entries currently classified, 184 contain at least one internal or external loop.
DATABASE ORGANIZATION AND INTERFACE
Although the SCOR database has much in common with the widely used SCOP database of protein structure (12), fundamental differences between RNA and proteins have prompted a somewhat different database design. RNA structure is considered to be modular at the motif level, while protein structure is modular at the domain level; RNA sequence within helical regions can easily co-vary without affecting structure. We therefore emphasize conserved motifs found in RNA secondary and tertiary structure and how these motifs combine to build functional RNA molecules.
Our goal is an interconnected database in which RNAs are dissected into structural elements and, conversely, from which all examples of structures containing specific structural elements may be easily accessed. From the SCOR home page one can choose a classification according to function, three-dimensional motif or tertiary interaction. The classification pages are primarily hierarchical, but have some aspects of the directed acyclic graph structure found in the Gene Ontology (13). Proceeding along these hierarchies provides a more detailed classification and ultimately leads to a specific structural example with PDB identification code. Selecting the PDB ID code yields a page containing the following information about the structure: a link to its PDB summary information, its NDB ID, its functional classification, the authors, title and journal reference of its primary reference, and a list of its motifs with their classification and sequence. Alternatively, a PDB code can be entered at the home page and a page is immediately displayed showing the above information.
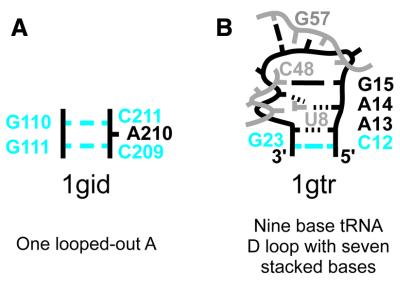
Sketches of representative loops are provided at each level of the classification to supplement the class, subclass and motif definitions. Two sample sketches, which illustrate the range of complexity of the loops in this classification, are shown in Figure 1. The first, from PDB entry 1gid (4), consists of a single looped-out A base; the second, from PDB entry 1gtr (14), is a tRNA D loop. Transfer RNA D loops are remarkable in that tertiary interactions, particularly with the T loop, play a greater role in determining their structure than secondary interactions. In addition, an option for RasMol (15) interactive viewing is available for every entry at both the structure and loop level of the classification; this viewer highlights the selected motif in the overall structure. A machine parseable version of the database is available upon request.
Figure 1.
Sample loop sketches, illustrating the range in complexity of the loops that have been classified, with the loops’ classifications. In these sample images, Watson–Crick base pairs are shown as short lines, non-Watson–Crick base pairs as dotted lines; the closing Watson–Crick base pairs are shown in blue. RNA residues which are not part of the loop but interact with it are shown in gray. (A) The loop from PDB entry 1gid (4) is a very simple internal loop. (B) The loop from PDB entry 1gtr (14) exhibits base pairing—both canonical and noncanonical—with residues outside the loop, base triples, unpaired, stacked bases and looped-out bases.
FUNCTIONAL CLASSIFICATION
As an aid in accessing the loop information, the 259 database entries have been classified by function. These categories include transfer RNAs, ribosomal RNAs, ribozymes, small nuclear RNAs (snRNAs), signal recognition particle (SRP) RNAs, genetic control elements, viral packaging RNAs, evolved or SELEX RNAs, synthetic RNAs and structures without classified motifs. Genetic control elements are mRNA regions whose function relates to protein synthesis; the most widely studied RNAs in this category are the HIV Rev response element (RRE) and trans-activating region (TAR) RNAs. Viral packaging RNAs are involved in the formation of functional viruses; the most widely studied of these are RNA elements which bind to bacteriophage coat protein. Synthetic RNAs include those with no natural biological or biochemical function. For each category, a list of subcategories is provided; for example, transfer RNAs are grouped into initiator tRNA, elongator tRNAs, synthetase complexes, complexes with EF-Tu, and individual stems or stem–loops. For each category, the database entries are grouped according to species and particular RNA, such as Saccharomyces cerevisiae tRNA (Phe) under elongator tRNAs, for which seven database entries have been examined.
CLASSIFICATION OF RNA MOTIFS
RNA structural motifs can be partitioned according to the number of strands connecting double helices. The most common motifs are external loops (one helix, capped by one RNA strand) and internal loops (two helices connected by two strands). The internal and external loops in the database entries surveyed are presented in a structural hierarchy. A strict definition has been used to define the loops: external loops are a covalently connected series of residues not Watson–Crick paired to each other, which are closed on one side by a Watson–Crick base pair. We include among internal loops those sometimes described as bulge loops; internal loops consist of one (for bulge loops) or two connected series of residues not Watson–Crick paired and closed on both sides by Watson–Crick base pairs. By this definition, for example, a G-U base pair surrounded on both sides by Watson–Crick pairs is considered an internal loop. This is a base-oriented classification, with particular emphasis on base stacking. We are preparing a detailed discussion of the loop motifs; here we provide only highlights.
Of the structures surveyed, 135 contain internal loops, with a total of 223 internal loops. These have been subclassified into nine classes. The most abundant class of internal loops is non-Watson–Crick paired stacked duplexes (115 of the 223); these are symmetric loops with all bases paired and contained in a single stack. Of these, the most common (52 loops) consist of a single G-U base pair; the longest (7 bp) is the loop E motif from 5S ribosomal RNA.
Our survey also identified 136 structures containing at least one external loop, with a total of 203 external loops. These loops are currently categorized first by size, then by stacking pattern within loops of a given size. For example, the GNRA tetraloops in the standard conformation, of which there are 27 in our sample, are in the tetraloops class, the subclass of tetraloops with four bases in the main stack, and the motif of tetraloops with one base in the 5′ stack and three in the 3′ stack. The most widely conserved external loop in our sample is the tRNA T loop. Of these, 25 are in the same conformation, a U turn with five bases in the main stack of the acceptor stem, and two unpaired bases in the anticodon loop stack. Only one tRNA structure in our sample, PDB ID 1eiy (16), the structure of phenylalanyl-tRNA synthetase complexed with the cognate tRNA (Phe), has a T loop not in this conformation.
The loops in our classification have been carefully filtered for redundancy. Two loops are considered redundant if they have the same structure, and also arise from the same biological source (including species) or have the same sequence (including the same surrounding sequence) in the case of synthetic RNAs. If one or more loops are considered to be redundant with another loop, the loop identifier of the first loop in the redundant group is given beside the loop identifier and sequence of the loops it is redundant with. For example, we have classified six structures containing Escherichia coli tRNA (Gln); the T loops of these structures are all redundant. The entries of the last five structures in the list of T loops all contain the loop identifier, 1euy:b:954–960, of the first entry in the list. This identifier consists of PDB code, chain name and the residue numbers within that chain.
RNA TERTIARY INTERACTIONS
Inspection of the larger known RNA structures has identified several types of inter- and intramolecular interactions that are distinct from either the canonical or non-canonical base pairs found in double helical RNA. These include: helix–helix interactions in the form of coaxial helical stacks and ribose zippers (4); loop–loop interactions such as the D loop:T loop interaction of tRNA and the formation of kissing hairpins between complementary loop sequences (17); ‘A-minor’ interactions (18) that can be between helices, loops or helix–loop; pseudoknots (19); and tetraloop–tetraloop receptor interactions (20). Examples of each of these interactions and structures containing these interactions are provided in the SCOR database.
APPLICATIONS
The SCOR database is intended to have several specific applications. It should be useful in RNA structural and functional analysis, breaking up large structures into functional domains. Based on such analysis, the information in the database should aid in RNA functional prediction, in particular the search for functional RNAs in genomes. Further applications include RNA engineering and design and discovery of RNA protein and small molecule ligands including potential therapeutics. For example, classification of internal loops conforming to that found for the Rev binding element of HIV-1 should allow evaluation of the structural and sequence variation found in this functional motif, provide a consensus target for drug design, and allow analysis of conserved features. Another immediate application should be the design and engineering of chimeric, possibly multi-functional, RNA molecules from component motifs for various uses.
FUTURE DIRECTIONS
SCOR is an evolving resource that will continue to grow as more RNA structures become available and our understanding is enhanced. Classification of the 30S and 50S ribosomal subunit structures is a top priority and is in progress. We will also provide an up-to-date classification of motifs in known RNA structures and a classification of junction loops (RNA strands connecting three or more helical stacks). In order to use the database to find connections between RNA sequence and structure, links to the RNA sequence databases (e.g. tRNA, ribosomal RNA and RNase P RNA) will be provided. This will enable rapid analysis of sequence variation allowed within structurally constrained regions. We plan to make greater use of the directed acyclic graph database topology, which provides a database that is more richly interconnected than a simple hierarchy. Together with analysis of conserved structural motifs, as contained in our database, we hope to be able to predict the presence of motifs from sequence as a major component of RNA three-dimensional structure and function prediction. Other modifications to the database will be incorporated in an evolutionary fashion in conjunction with user needs and growth in the number of RNA structures available. This classification was done manually, by inspecting the RNA structures; we look forward to the development of automated tools to perform this classification in the future.
Acknowledgments
ACKNOWLEDGEMENTS
We particularly thank Ignacio Tinoco for suggesting this project and for valuable advice at various points in carrying it out. We also thank Victor Franco for help setting up the web site and Ramona Pufan for assistance generating structure NDB IDs. The RasMol viewing system is based on code written by Tim Hubbard and Raphaël Leplae; we thank Loredana Lo Conte for assistance in using this code. This research is funded by the Lawrence Berkeley National Laboratory Directed Research and Development Program and by a UC Berkeley COR grant; S.E.B. is supported by NIH grant 1 K22 HG00056.
REFERENCES
- 1.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. Updated article in this issue: Nucleic Acids Res. (2002), 30, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman H.M., Olson,W.K., Beveridge,D.L., Westbrook,J., Gelbin,A., Demeny,T., Hsieh,S.-H., Srinivasan,A.R. and Schneider,B. (1992) The Nucleic Acid Database: a comprehensive relational database of three-dimensional structures of nucleic acids. Biophys. J., 63, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott W.G., Finch,J.T. and Klug,A. (1995) The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell, 81, 991–1002. [DOI] [PubMed] [Google Scholar]
- 4.Cate J.H., Gooding,A.R., Podell,E., Zhou,K., Golden,B.L., Kundrot,C.E., Cech,T.R. and Doudna,J.A. (1996) Crystal structure of a group I ribozyme domain: principles of RNA packing. Science, 273, 1678–1685. [DOI] [PubMed] [Google Scholar]
- 5.Ferre-D’Amare A.R. and Doudna,J.A. (1999) RNA folds: insights from recent crystal structures. Annu. Rev. Biophys. Biomol. Struct., 28, 57–73. [DOI] [PubMed] [Google Scholar]
- 6.Hermann T. and Patel,D.J. (1999) Stitching together RNA tertiary architectures. J. Mol. Biol., 294, 829–849. [DOI] [PubMed] [Google Scholar]
- 7.Batey R.T., Rambo,R.P., and Doudna,J.A. (1999) Tertiary motifs in RNA structure and folding. Angew. Chem. Int. Ed., 38, 2326–2343. [DOI] [PubMed] [Google Scholar]
- 8.Moore P.B. (1999) Structural motifs in RNA. Annu. Rev. Biochem., 68, 287–300. [DOI] [PubMed] [Google Scholar]
- 9.Schluenzen F., Tocilj,A., Zarivach,R., Harms,J., Gluehmann,M., Janell,D., Bashan,A., Bartels,H., Agmon,I., Franceschi,F. and Yonath,A. (2000) Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell, 102, 615–623. [DOI] [PubMed] [Google Scholar]
- 10.Wimberly B.T., Brodersen,D.E., Clemons,W.M.,Jr, Morgan-Warren,R., Carter,A.P., Vonrhein,C., Hartsch,T. and Ramakrishnan,V. (2000) Structure of the 30S ribosomal subunit. Nature, 407, 327–329. [DOI] [PubMed] [Google Scholar]
- 11.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 920–930. [DOI] [PubMed] [Google Scholar]
- 12.Murzin A.G., Brenner,S.E., Hubbard,T.J. and Chothia,C. (1995) SCOP—a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- 13.Ashbuner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T., Harris,M.A., Hill,D.P., Issel-Tarver,L., Kasarskis,A., Lewis,S., Matese,J.C., Richardson,J.E., Ringwald,M., Rubin,G.M. and Sherlock,G. (2000) Gene Ontology: tool for the unification of biology. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rould M.A., Perona,J.J. and Steitz,T.A. (1991) Structural basis of anticodon loop recognition by glutaminyl-tRNA synthetase. Nature 352, 213. [DOI] [PubMed] [Google Scholar]
- 15.Sayle R.A. and Milner-White,E.F. (1995) RasMol: Biomolecular graphics for all. Trends Biochem. Sci., 20, 374–376. [DOI] [PubMed] [Google Scholar]
- 16.Goldgur Y., Mosyak,L., Reshetnikova,L., Ankilova,V. and Safro,M. (1997) The crystal structure of phenylalanyl-tRNA synthetase from Thermus thermophilus complexed with cognate tRNA(Phe). Structure, 5, 59–68. [DOI] [PubMed] [Google Scholar]
- 17.Lee A.J. and Crothers,D.M. (1998) The solution structure of an RNA loop–loop complex: the ColE1 inverted loop sequence. Structure, 6, 993–1005. [DOI] [PubMed] [Google Scholar]
- 18.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Science, 289, 905–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pleij C.W.A. (1994) RNA pseudoknots. Curr. Opin. Struct. Biol., 4, 337–344. [Google Scholar]
- 20.Pley H.W., Flaherty,K.M. and McKay,D.B. (1994) Model for an RNA tertiary interaction from the structure of an intermolecular complex between a GAAA tetraloop and an RNA helix. Nature, 372, 111–113. [DOI] [PubMed] [Google Scholar]