Abstract
The significant structural diversity and potent bioactivity of the fungal indole diterpenes (IDTs) has attracted considerable interest in their biosynthesis. Although substantial skeletal diversity is generated by the action of noncanonical terpene cyclases, comparatively little is known about these enzymes, particularly those involved in the generation of the subgroup containing emindole SA and DA, which show alternate terpenoid skeletons. Here, we describe the IDT biosynthetic machinery generating these unusual IDT architectures from Aspergillus striatus and Aspergillus desertorum. The function of four putative cyclases was interrogated via heterologous expression. Two specific cyclases were identified that catalyze the formation of epimers emindole SA and DA from A. striatus and A. desertorum, respectively. These cyclases are both clustered along with all the elements required for basic IDT biosynthesis yet catalyze an unusual Markovnikov-like cyclization cascade with alternate stereochemical control. Their identification reveals that these alternate architectures are not generated by mechanistically sloppy or promiscuous enzymes, but by cyclases capable of delivering precise regio- and stereospecificities.
The ability of organisms to produce structurally diverse molecules from a small pool of starting metabolites has long inspired study.1,2 For compounds that have the same biosynthetic origin, variation arises from the action of diversity-generating enzymes.3 These enzymes play an important role in secondary metabolism, where structural diversity and associated differential bioactivities may confer survival advantages to an organism in its ecological niche.4
The indole diterpenes (IDTs) are a large class of secondary metabolites that are produced by filamentous fungi.5 IDTs are notable for their potent bioactivities, including fungicidal,5 anticancer,6 insecticidal,5,7 tremorgenic,8 and antiviral9 properties. The potential utility of these compounds and their exquisite structural complexity has led to considerable interest in their biosynthesis.10−13 Knowledge of the biosynthesis of these compounds can enable the production of otherwise inaccessible IDTs and the engineering of novel derivatives with enhanced bioactivities.
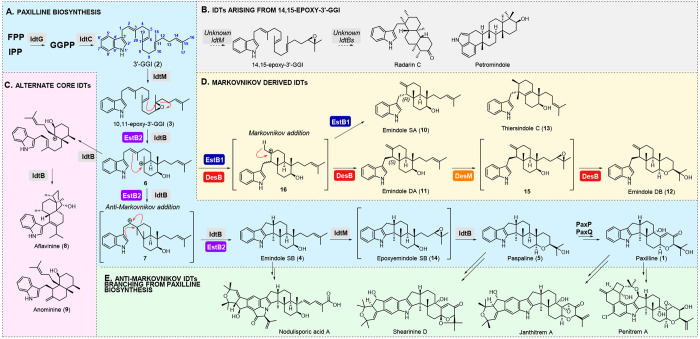
In all IDT biosynthetic pathways, generation of the first stable cyclized IDT is achieved by four enzyme-catalyzed steps which were originally established in the biosynthesis of paxilline (1) (Figure 1A).14,15 The first step is formation of geranylgeranyl pyrophosphate (GGPP). The indole moiety is then introduced by an indole prenyltransferase (IdtC) to afford 3′-geranylgeranylindole (3′-GGI, 2). Next, 3′-GGI (2) is epoxidized by an FAD-dependent monooxygenase (IdtM) and subsequently cyclized by an indole diterpene cyclase (IdtB) to furnish a stable core IDT skeleton.
Figure 1.
Biosynthesis of IDTs. IPP: isopentenyl pyrophosphate, FPP: farnesyl pyrophosphate. (A) Paxilline biosynthesis. (B) Implied biosynthesis of radarins and petromindole. (C) IDTs produced by via alternate arrangement of 6.27 (D) IDTs derived from Markovnikov addition to C-2 alkene of 6. (E) IDT families branching from paxilline.
The structure of this IDT skeleton is determined by the specific function of the IdtM and IdtB enzymes present in the pathway. For paxilline (1) biosynthesis, PaxM (Penicillium paxilli’s IdtM) acts on the C-10 alkene of 3′-GGI (2) to generate 10,11-epoxy-3′-GGI (3), which is cyclized by PaxB to give emindole SB (4). PaxM can also perform a second epoxidation, acting on the C-14 alkene, to facilitate formation of a tetrahydropyran (THP) ring and production of the core IDT paspaline (5) The two epoxidations performed by PaxM are common to all IdtMs characterized to date, apart from NodM, which can only act on the C-10 alkene of 2 and leads to production of nodulisporic acids (Figure 1).16 IdtMs that epoxidize only the C-14 alkene are implied from retrobiosynthetic analyses of the radarins17,18 and petromindole18,19 but have not yet been identified (Figure 1B).
In contrast to the canonical Type I and Type II cyclases that catalyze most terpenoid cyclizations,20 the IdtBs are unusual transmembrane proteins.21,22 These enzymes generate a wide range of architectures that form the basis for structural diversity among IDTs. All characterized IdtBs initiate cyclization by catalyzing attack of C-11 by the C-6 alkene of 3 to form the tertiary carbocation 6. In paxilline biosynthesis, PaxB then facilitates apparent anti-Markovnikov attack of 6 by the C-2 alkene. Although this addition gives secondary carbocation 7, it is expected that this addition is concerted with subsequent migration of C-4 from C-3 to C-2 (to avoid the unstable secondary carbocation)23 and indole addition to generate emindole SB (4).15,16,23 Most characterized IdtBs catalyze this same mechanistic trajectory to give IDT families such as the nodulisporic acids,15,16 penitrems24 lolitrems,25 janthitrems,26 and shearinines (Figure 1).16,26
More recently, Tang et al. characterized three new IdtBs that deliver non-emindole SB skeletons.27 One, which was IDT-cluster-encoded, demonstrated catalytic promiscuity, generating both emindole SB (4) and an emindole PA-precursor through an alternate cyclization cascade via an anti-Markovnikov route from carbocation 6 (Figure S1).27 The other two IDT cyclases were not cluster-encoded and catalyzed alternative rearrangements of carbocation 6 (Figure 1A, 1C).27 Again, these enzymes were remarkable in their mechanistic promiscuity, generating aflavinine (8), anominine (9), and two other congeners.27
IDT architectures that imply a cyclization cascade with Markovnikov addition to the C-2 alkene of 6 have been reported, and include emindole SA (10),28 emindole DA (11),29 emindole DB (12),29 and thiersindole C (13)30 (Figure 1D). However, IdtBs that deliver these IDTs with unusual connectivity have not yet been discovered.
To identify the first examples of indole diterpene cyclases that deliver this unique uncharacterized chemistry we investigated IDT biosynthesis in Aspergillus striatus and Aspergillus desertorum. A. striatus is reported to produce emindole SA (10), paxilline (15), and 1′-O-acetylpaxilline (16),31 whereas A. desertorum produces emindole DA (11), emindole DB (12), and 15.29
To determine the relevant genetic landscapes, whole genome sequencing of A. striatus was carried out for strain NHL 80-NE-22 (ATCC 64988), whereas the genome sequence of A. desertorum CBS 653.73 was accessed from the JGI Genome Portal.32,33 Surprisingly, two putative IDT biosynthetic gene clusters were identified in the genome of A. striatus (Figure 2). The first cluster, EST1, contains only four predicted genes encoding proteins that were homologous to the four core PAX-encoded enzymes required for paspaline (5) biosynthesis. The second cluster, EST2, contains eight open-reading frames (ORFs), all of which were homologous to PAX cluster genes except for estV2 whose protein product displayed 50.2% sequence identity to PtmV, an acetyltransferase involved in penitrem biosynthesis.20 The EST2 cluster also differs from the PAX cluster in the absence of paxD and paxO homologues, and estQ2 is nested between estM2 and estA2. An additional idtB, termed estB3, that is not flanked by other IDT biosynthetic genes, was also identified.
Figure 2.
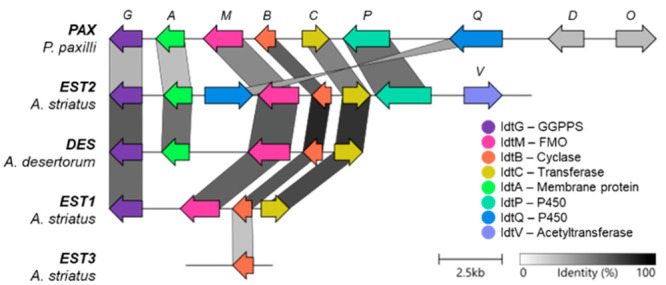
Comparison of IDT clusters identified in A. striatus and A. desertorum with P. paxilli. Note: identity below 30% is not shown. This figure was generated using Clinker and clustermap.js version 0.0.25.7.34 Accession numbers are provided in the Supporting Information. Note paxD and paxO are involved in post-paxilline modifications, which are inefficient in wild-type P. paxilli.35
Interrogation of the genome of A. desertorum revealed a cluster of five ORFs (DES cluster), including four predicted to encode proteins that deliver a cyclized IDT core (desG, desC, desM, and desB). Although A. desertorum reportedly produces paxilline (1) as a minor metabolite,29 homologous ORFs of paxP or paxQ, that are required for the biosynthesis of 1,15,36,29 were not identified.
To determine the function of EST and DES cluster genes a heterologous reconstruction approach was taken, using P. paxilli strains, deleted for specific paxilline biosynthetic genes.35DES or EST cluster genes were assembled into plasmids using the Modular Idempotent DNA Assembly System (MIDAS)37 along with PAX cluster promoter and terminator regions and constructs were introduced into protoplasts of the appropriate P. paxilli strains (Figures S5 and S6). Metabolite profiles were analyzed by liquid chromatography mass spectrometry (LC-MS) and compounds were purified and identified by nuclear magnetic resonance (NMR) spectroscopy.
In every IDT biosynthetic pathway characterized, a prenyltransferase, IdtC, catalyzes the prenylation of the indole moiety with GGPP and represents the commitment of metabolic flux to IDT biosynthesis (Figure 1).35 To identify the genes responsible for this transformation in A. striatus, we expressed estC1 and estC2 separately in a P. paxilli strain deficient in paxC (PN2290), that does not produce paxilline (1). LC-MS analysis of these transformants confirmed the restoration of paxilline biosynthesis demonstrating that both idtCs from A. striatus are orthologues of paxC (Figures 3i–iv, S7, and S8).
Figure 3.
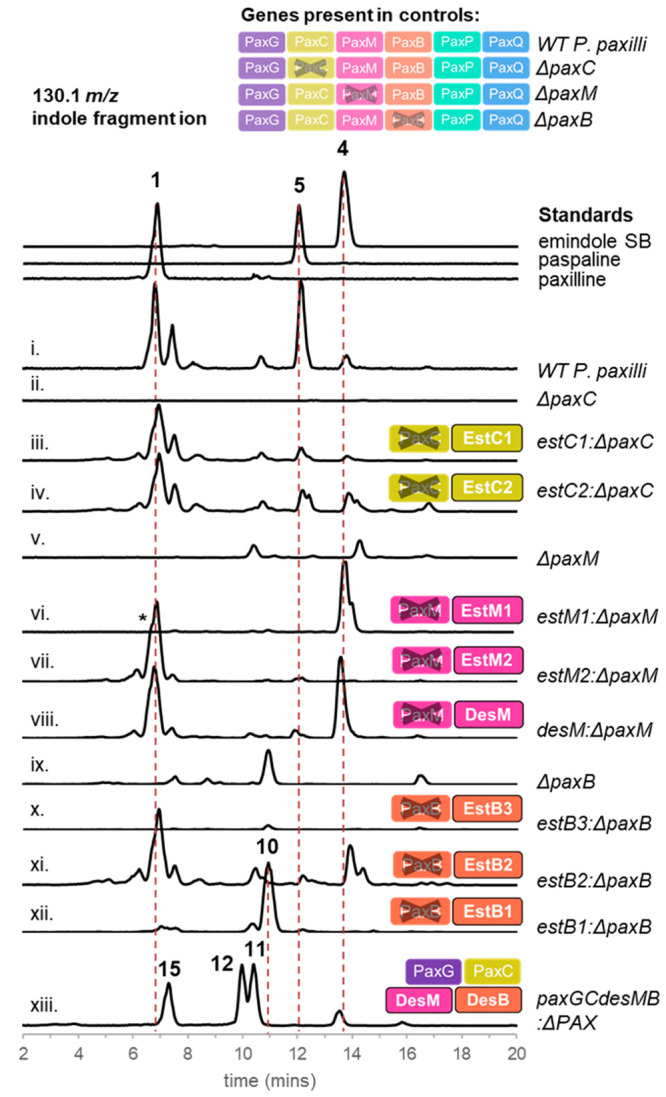
LC-MS analysis of P. paxilli knockout strains complemented by genes from EST1, EST2, or DES using indole fragment ion m/z 130.1 for detection. Peaks were identified by comparison to IDT standards or by NMR (Supporting Information). *Shoulder on paxilline peak is due to the high concentration of this metabolite resulting in poor peak shape.
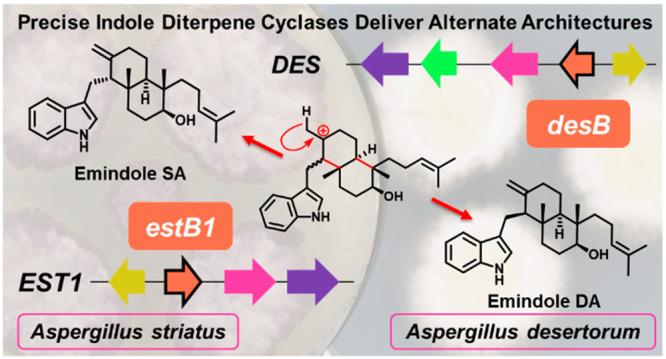
A. striatus produces two distinct IDTs, emindole SB (4) and emindole SA (10);31 therefore, the presence of two separate IDT clusters suggests that each may encode the biosynthetic machinery for the delivery of an alternate terpenoid skeleton.31 The composition of the clusters implies that the EST1 cluster is solely responsible for a simple cyclized IDT product, and that EST2, which includes genes encoding tailoring enzymes, is specifically dedicated to the biosynthesis of paxilline (1) and 1′-O-acetylpaxilline (16). To investigate the biosynthetic output of these two clusters, estM1 and estM2 were separately transformed into a P. paxilli strain lacking paxM (PN2257), that does not produce 1. Intriguingly, paxilline (1) biosynthesis was only restored in transformants harboring estM2, whereas those bearing estM1 produced emindole SB (4) (Figures 3v–vii, S9, and S10). This finding is consistent with EstM1 performing the epoxidation of the C-10 alkene, but not executing the second epoxidation step on the terminal alkene that is required for the generation of the THP ring of paspaline (5) and 1, via epoxyemindole SB (14) (Figure 1). Taken together, these results show that estM2 encodes an epoxidase that is functionally identical to PaxM, whereas EstM1 is functionally identical to NodM, the only previously identified 3′-GGI (2) monoepoxidase.16
Skeletal variation in the IDTs is further dictated by the cyclization cascade controlled by the unusual terpene cyclases. To assign functionality to these IdtBs, we expressed each of the three putative cyclases in a P. paxilli ΔpaxB strain. Strains bearing estB3 did not produce any new IDT compounds compared to the base strain (Figures 3ix,x and S11) although RNA transcripts of estB3 were detected in the mycelia (Figure S16). Conversely, strains harboring estB2 successfully restored paxilline (1) biosynthesis (Figures 3xi and S12), consistent with its genetic context. However, strains bearing estB1 generated a compound that was not present in either P. paxilli wild-type or the ΔpaxB strain (Figures 3xii and S13). This compound was isolated and purified enabling its assignment as emindole SA (10) (Table S11, Figures S17–21). Thus, EstB1 is the first identified IdtB to deliver a Markovnikov-derived IDT.
To investigate the biosynthesis of IDTs in A. desertorum, we employed a similar strategy and introduced the putative idtM (desM) into the P. paxilli strain lacking paxM (PN2257).35 The restoration of paxilline (1) production in desM-expressing transformants confirmed that DesM is functionally equivalent to PaxM (Figures 3viii and S14).
To test the function of DesB, we introduced desB along with other biosynthetic elements, paxG, paxC, and desM into a P. paxilli strain devoid of the entire PAX cluster (CY2).38 LC-MS analysis revealed that two additional compounds were formed ([M + H]+, m/z 406.3 and 422.3) (Figures 3xii,xiii and S15).38 Large scale production and isolation identified these compounds as emindole DA (11) and emindole DB (12) (Tables S12 and S13, Figures S22–31). Thus, DesB catalyzes similar Markovnikov-derived cyclization as EstB1, albeit with different stereospecificity; DesB catalyzes a mechanism where the Re face of the C-2 olefin is attacked by carbocation 6 (Figure 1D). Additionally, the formation of emindole DB by this strain demonstrates the epoxidation of 11 and cyclization of epoxyemindole DA intermediate (15) are delivered by this combination of biosynthetic enzymes, mirroring the equivalent chemistry reported for paspaline (5) biosynthesis.15
This study highlights the power of heterologous gene reconstruction and complementation to functionally characterize biosynthetic gene clusters to reveal new chemistry. DesB and EstB1 are the first identified examples of cyclases capable of catalyzing specific Markovnikov-like chemistry through carbocation (16). While this chemistry was predicted from the known IDT architectures,39 until now it has not been linked to specific sequences or clusters. While previous work had implied some correlations could be made between primary structure and reaction chemistry,27 our sequence and phylogenetic analysis does not reveal clear clustering of EstB1 and DesB, relative to the PaxB-like enzymes (Figure S32). The active site of these unusual transmembrane enzymes is predicted to be in the internal space between the transmembrane helices, where our analysis reveals some sequence differences that may link to mechanistic variation (Figures S33 and S34).
The prior discovery that some unusual IDT architectures were delivered by mechanistically imprecise IDT cyclases raised the possibility that these alternative compounds, and thus an expanded pool of IDTs, are generated by catalytically imprecise enzymes. We demonstrate here that these IDT skeletons are indeed delivered by single precise cyclases, that are encoded along with all the genes required for entry into IDT biosynthesis.
The ability of natural product pathways to produce a vast array of compounds from a small pool of precursors contributes to complete chemical diversity.1,40 IDTs achieve diversity through both alternative tailoring12,13,16,24−26,41 and the production of fundamentally different skeletons.27 Here, we reveal that skeletal change is delivered by specific, clustered cyclases catalyzing alternate cyclization cascades, setting up the remarkable display of compound diversity that IDT arrays demonstrate.
Acknowledgments
This work was financially supported by the New Zealand Ministry of Business, Innovation, and Employment (Grant # RTVU1809) and by the New Zealand Marsden Fund (Grant # VUW1819).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.2c11170.
Molecular biology methods, gene and primer sequences, transcription analysis, transformation methods, analytical methods, and NMR characterization (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Wei X.; Wang W.-G.; Matsuda Y. Branching and converging pathways in fungal natural product biosynthesis. Fungal Biol. Biotechnol. 2022, 9 (1), 6. 10.1186/s40694-022-00135-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigalunas M.; Brakmann S.; Waldmann H. Chemical Evolution of Natural Product Structure. J. Am. Chem. Soc. 2022, 144 (8), 3314–3329. 10.1021/jacs.1c11270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C. T.; Tang Y. Recent Advances in Enzymatic Complexity Generation: Cyclization Reactions. Biochemistry 2018, 57 (22), 3087–3104. 10.1021/acs.biochem.7b01161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller N. P. Translating biosynthetic gene clusters into fungal armor and weaponry. Nat. Chem. Biol. 2015, 11 (9), 671–7. 10.1038/nchembio.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke P. T.; Smith M. M.; Shoop W. L. Nodulisporic acid: its chemistry and biology. Curr. Top. Med. Chem. 2002, 2 (7), 655–74. 10.2174/1568026023393714. [DOI] [PubMed] [Google Scholar]
- Sallam A. A.; Ayoub N. M.; Foudah A. I.; Gissendanner C. R.; Meyer S. A.; El Sayed K. A. Indole diterpene alkaloids as novel inhibitors of the Wnt/beta-catenin pathway in breast cancer cells. Eur. J. Med. Chem. 2013, 70, 594–606. 10.1016/j.ejmech.2013.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondeyka J. G.; Helms G. L.; Hensens O. D.; Goetz M. A.; Zink D. L.; Tsipouras A.; Shoop W. L.; Slayton L.; Dombrowski A. W.; Polishook J. D.; Ostlind D. A.; Tsou N. N.; Ball R. G.; Singh S. B. Nodulisporic Acid A, a Novel and Potent Insecticide from a Nodulisporium Sp. Isolation, Structure Determination, and Chemical Transformations. J. Am. Chem. Soc. 1997, 119 (38), 8809–8816. 10.1021/ja971664k. [DOI] [Google Scholar]
- Kaczorowski G. J.; Knaus H. G.; Leonard R. J.; McManus O. B.; Garcia M. L. High-conductance calcium-activated potassium channels; Structure, pharmacology, and function. J. Bioenerg. Biomembr. 1996, 28 (3), 255–267. 10.1007/BF02110699. [DOI] [PubMed] [Google Scholar]
- Ogata M.; Ueda J.-y.; Hoshi M.; Hashimoto J.; Nakashima T.; Anzai K.; Takagi M.; Shin-ya K. A Novel Indole-diterpenoid, JBIR-03 with Anti-MRSA Activity from Dichotomomyces cejpii var. cejpii NBRC 103559. J. Antibiot. 2007, 60 (10), 645–648. 10.1038/ja.2007.83. [DOI] [PubMed] [Google Scholar]
- Saikia S.; Nicholson M. J.; Young C.; Parker E. J.; Scott B. The genetic basis for indole-diterpene chemical diversity in filamentous fungi. Mycol. Res. 2008, 112 (2), 184–99. 10.1016/j.mycres.2007.06.015. [DOI] [PubMed] [Google Scholar]
- Kozak L.; Szilagyi Z.; Toth L.; Pocsi I.; Molnar I. Tremorgenic and neurotoxic paspaline-derived indole-diterpenes: biosynthetic diversity, threats and applications. Appl. Microbiol. Biotechnol. 2019, 103 (4), 1599–1616. 10.1007/s00253-018-09594-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson A. T.; Cameron R. C.; Stevenson L. J.; Singh A. J.; Lukito Y.; Berry D.; Nicholson M. J.; Parker E. J. Biosynthesis of Nodulisporic Acids: A Multifunctional Monooxygenase Delivers a Complex and Highly Branched Array. Angew. Chem., Int. Ed. 2022, 61 (49), e202213364. 10.1002/anie.202213364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan R. M.; Cameron R. C.; Nicholson M. J.; Parker E. J. Aminoacylation of Indole Diterpenes by Cluster-Specific Monomodular NRPS-like Enzymes. Org. Lett. 2022, 24 (12), 2332–2337. 10.1021/acs.orglett.2c00473. [DOI] [PubMed] [Google Scholar]
- Saikia S.; Parker E. J.; Koulman A.; Scott B. Four gene products are required for the fungal synthesis of the indole-diterpene, paspaline. FEBS Lett. 2006, 580 (6), 1625–1630. 10.1016/j.febslet.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Tagami K.; Liu C.; Minami A.; Noike M.; Isaka T.; Fueki S.; Shichijo Y.; Toshima H.; Gomi K.; Dairi T.; Oikawa H. Reconstitution of biosynthetic machinery for indole-diterpene paxilline in Aspergillus oryzae. J. Am. Chem. Soc. 2013, 135 (4), 1260–3. 10.1021/ja3116636. [DOI] [PubMed] [Google Scholar]
- Van de Bittner K. C.; Nicholson M. J.; Bustamante L. Y.; Kessans S. A.; Ram A.; van Dolleweerd C. J.; Scott B.; Parker E. J. Heterologous Biosynthesis of Nodulisporic Acid F. J. Am. Chem. Soc. 2018, 140 (2), 582–585. 10.1021/jacs.7b10909. [DOI] [PubMed] [Google Scholar]
- Laakso J. A.; Gloer J. B.; Wicklow D. T.; Dowd P. F. Radarins A-D: new antiinsectan and cytotoxic indole diterpenoids from the sclerotia of Aspergillus sulphureus. J. Org. Chem. 1992, 57 (1), 138–141. 10.1021/jo00027a026. [DOI] [Google Scholar]
- Xiong Q.; Zhu X.; Wilson W. K.; Ganesan A.; Matsuda S. P. T. Enzymatic Synthesis of an Indole Diterpene by an Oxidosqualene Cyclase: Mechanistic, Biosynthetic, and Phylogenetic Implications. J. Am. Chem. Soc. 2003, 125 (30), 9002–9003. 10.1021/ja036322v. [DOI] [PubMed] [Google Scholar]
- Ooike M.; Nozawa K.; Udagawa S.-i.; Kawai K.-i. Structures of a New Type of Indoloditerpene, Petromindole, and a New Asterriquinone Derivative, PM-53, from the Ascostromata of Petromyces muricatus. Chem. Pharm. Bull. 1997, 45 (10), 1694–1696. 10.1248/cpb.45.1694. [DOI] [Google Scholar]
- Christianson D. W. Structural and Chemical Biology of Terpenoid Cyclases. Chem. Rev. 2017, 117 (17), 11570–11648. 10.1021/acs.chemrev.7b00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barra L.; Abe I. Chemistry of fungal meroterpenoid cyclases. Nat. Prod. Rep. 2021, 38 (3), 566–585. 10.1039/D0NP00056F. [DOI] [PubMed] [Google Scholar]
- Baunach M.; Franke J.; Hertweck C. Terpenoid biosynthesis off the beaten track: unconventional cyclases and their impact on biomimetic synthesis. Angew. Chem., Int. Ed. 2015, 54 (9), 2604–26. 10.1002/anie.201407883. [DOI] [PubMed] [Google Scholar]
- Tantillo D. J. The carbocation continuum in terpene biosynthesis—where are the secondary cations?. Chem. Soc. Rev. 2010, 39 (8), 2847–2854. 10.1039/b917107j. [DOI] [PubMed] [Google Scholar]
- Liu C.; Tagami K.; Minami A.; Matsumoto T.; Frisvad J. C.; Suzuki H.; Ishikawa J.; Gomi K.; Oikawa H. Reconstitution of biosynthetic machinery for the synthesis of the highly elaborated indole diterpene penitrem. Angew. Chem., Int. Ed. 2015, 54 (19), 5748–52. 10.1002/anie.201501072. [DOI] [PubMed] [Google Scholar]
- Jiang Y.; Ozaki T.; Harada M.; Miyasaka T.; Sato H.; Miyamoto K.; Kanazawa J.; Liu C.; Maruyama J.-i.; Adachi M.; Nakazaki A.; Nishikawa T.; Uchiyama M.; Minami A.; Oikawa H. Biosynthesis of Indole Diterpene Lolitrems: Radical-Induced Cyclization of an Epoxyalcohol Affording a Characteristic Lolitremane Skeleton. Angew. Chem., Int. Ed. 2020, 59 (41), 17996–18002. 10.1002/anie.202007280. [DOI] [PubMed] [Google Scholar]
- Nicholson M. J.; Eaton C. J.; Starkel C.; Tapper B. A.; Cox M. P.; Scott B. Molecular Cloning and Functional Analysis of Gene Clusters for the Biosynthesis of Indole-Diterpenes in Penicillium crustosum and P. janthinellum. Toxins 2015, 7 (8), 2701–22. 10.3390/toxins7082701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M. C.; Lin H. C.; Li D.; Zou Y.; Li J.; Xu W.; Cacho R. A.; Hillenmeyer M. E.; Garg N. K.; Tang Y. Discovery of Unclustered Fungal Indole Diterpene Biosynthetic Pathways through Combinatorial Pathway Reassembly in Engineered Yeast. J. Am. Chem. Soc. 2015, 137 (43), 13724–7. 10.1021/jacs.5b06108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa K.; Yuyama M.; Nakajima S.; Kawai K.-i.; Udagawa S.-i. Studies on fungal products. Part 19. Isolation and structure of a novel indoloditerpene, emindole SA, from Emericella striata. J. Chem. Soc., Perkin Trans. 1 1988, (8), 2155–2160. 10.1039/p19880002155. [DOI] [Google Scholar]
- Nozawa K.; Nakajima S.; Kawai K.-i.; Udagawa S.-i. Isolation and structures of novel indoloditerpenes, emindoles DA and DB, from Emericella desertorum: X-ray molecular structure of emindole DA acetate. J. Chem. Soc., Perkin Trans. 1 1988, (7), 1689–1694. 10.1039/p19880001689. [DOI] [Google Scholar]
- Li C.; Gloer J. B.; Wicklow D. T. Thiersindoles A–C: New Indole Diterpenoids from Penicillium thiersii. J. Nat. Prod. 2003, 66 (9), 1232–1235. 10.1021/np030192m. [DOI] [PubMed] [Google Scholar]
- Nozawa K.; Nakajima S.; Kawai K.-i.; Udagawa S.-i. Isolation and structures of indoloditerpenes, possible biosynthetic intermediates to the tremorgenic mycotoxin, paxilline, from Emericella striata. J. Chem. Soc., Perkin Trans. 1 1988, (9), 2607–2610. 10.1039/p19880002607. [DOI] [Google Scholar]
- Grigoriev I. V.; Nordberg H.; Shabalov I.; Aerts A.; Cantor M.; Goodstein D.; Kuo A.; Minovitsky S.; Nikitin R.; Ohm R. A.; Otillar R.; Poliakov A.; Ratnere I.; Riley R.; Smirnova T.; Rokhsar D.; Dubchak I. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 2012, 40 (D1), D26–32. 10.1093/nar/gkr947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordberg H.; Cantor M.; Dusheyko S.; Hua S.; Poliakov A.; Shabalov I.; Smirnova T.; Grigoriev I. V.; Dubchak I. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 2014, 42 (D1), D26–31. 10.1093/nar/gkt1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist C. L. M.; Chooi Y.-H. clinker & clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics 2021, 37 (16), 2473–2475. 10.1093/bioinformatics/btab007. [DOI] [PubMed] [Google Scholar]
- Scott B.; Young C. A.; Saikia S.; McMillan L. K.; Monahan B. J.; Koulman A.; Astin J.; Eaton C. J.; Bryant A.; Wrenn R. E.; Finch S. C.; Tapper B. A.; Parker E. J.; Jameson G. B. Deletion and gene expression analyses define the paxilline biosynthetic gene cluster in Penicillium paxilli. Toxins 2013, 5 (8), 1422–46. 10.3390/toxins5081422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia S.; Parker E. J.; Koulman A.; Scott B. Defining paxilline biosynthesis in Penicillium paxilli: functional characterization of two cytochrome P450 monooxygenases. J. Biol. Chem. 2007, 282 (23), 16829–37. 10.1074/jbc.M701626200. [DOI] [PubMed] [Google Scholar]
- van Dolleweerd C. J.; Kessans S. A.; Van de Bittner K. C.; Bustamante L. Y.; Bundela R.; Scott B.; Nicholson M. J.; Parker E. J. MIDAS: A Modular DNA Assembly System for Synthetic Biology. ACS Synth. Biol. 2018, 7 (4), 1018–1029. 10.1021/acssynbio.7b00363. [DOI] [PubMed] [Google Scholar]
- Young C.; Itoh Y.; Johnson R.; Garthwaite I.; Miles C. O.; Munday-Finch S. C.; Scott B. Paxilline-negative mutants of Penicillium paxilli generated by heterologous and homologous plasmid integration. Curr. Genet. 1998, 33 (5), 368–377. 10.1007/s002940050349. [DOI] [PubMed] [Google Scholar]
- Parker E. J.; Scott D. B.. Indole-Diterpene Biosynthesis in Ascomycetous Fungi. In Handbook of Industrial Mycology, 1st ed.; CRC Press: 2004; Vol. 22, pp 424–445. [Google Scholar]
- Fewer D. P.; Metsä-Ketelä M. A pharmaceutical model for the molecular evolution of microbial natural products. FEBS J. 2020, 287 (7), 1429–1449. 10.1111/febs.15129. [DOI] [PubMed] [Google Scholar]
- Liu C.; Minami A.; Dairi T.; Gomi K.; Scott B.; Oikawa H. Biosynthesis of Shearinine: Diversification of a Tandem Prenyl Moiety of Fungal Indole Diterpenes. Org. Lett. 2016, 18 (19), 5026–5029. 10.1021/acs.orglett.6b02482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.