Abstract
The Rat Genome Database (RGD, http://rgd.mcw.edu) is an NIH-funded project whose stated mission is ‘to collect, consolidate and integrate data generated from ongoing rat genetic and genomic research efforts and make these data widely available to the scientific community’. In a collaboration between the Bioinformatics Research Center at the Medical College of Wisconsin, the Jackson Laboratory and the National Center for Biotechnology Information, RGD has been created to meet these stated aims. The rat is uniquely suited to its role as a model of human disease and the primary focus of RGD is to aid researchers in their study of the rat and in applying their results to studies in a wider context. In support of this we have integrated a large amount of rat genetic and genomic resources in RGD and these are constantly being expanded through ongoing literature and bulk dataset curation. RGD version 2.0, released in June 2001, includes curated data on rat genes, quantitative trait loci (QTL), microsatellite markers and rat strains used in genetic and genomic research. VCMap, a dynamic sequence-based homology tool was introduced, and allows researchers of rat, mouse and human to view mapped genes and sequences and their locations in the other two organisms, an essential tool for comparative genomics. In addition, RGD provides tools for gene prediction, radiation hybrid mapping, polymorphic marker selection and more. Future developments will include the introduction of disease-based curation expanding the curated information to cover popular disease systems studied in the rat. This will be integrated with the emerging rat genomic sequence and annotation pipelines to provide a high-quality disease-centric resource, applicable to human and mouse via comparative tools such as VCMap. RGD has a defined community outreach focus with a Visiting Scientist program and the Rat Community Forum, a web-based forum for rat researchers and others interested in using the rat as an experimental model. Thus, RGD is not only a valuable resource for those working with the rat but also for researchers in other model organisms wishing to harness the existing genetic and physiological data available in the rat to complement their own work.
INTRODUCTION
The rat has been the subject of scientific research for over 150 years and in that time it has developed as a highly popular organism for physiological, pharmacological and genetic research in a wide variety of areas fundamental to human health. It is also extensively used in the study of the genetics of hypertension, autoimmune disease, diabetes, obesity, cancer and many other diseases that severely impact our general population (1–4). Recognizing the importance of the rat as a model organism, the NIH funded the Rat Genome Database (RGD, http://rgd.mcw.edu) to ‘collect, consolidate and integrate data generated from ongoing rat genetic and genomic research efforts and make these data widely available to the scientific community’ (http://grants.nih.gov/grants/guide/rfa-files/RFA-HL-99-013.html). RGD is based at the Medical College of Wisconsin (MCW) in Milwaukee and is a direct collaboration between the MCW, the Mouse Genome Database (MGD) (5) and the National Center for Biotechnology Information (NCBI). RGD also coordinates all nomenclature for genes, strains and QTLs with Ratmap (http://ratmap.gen.gu.se), a European effort to manage related rat data and the international Rat Genome and Nomenclature Committee (http://rgnc.gen.gu.se).
RGD is a community resource and is publicly accessible on the World Wide Web at http://rgd.mcw.edu. The fundamental goal of RGD is to facilitate the discovery of the genetic basis of disease in the rat. A major requirement of this is to make the database and the genetic resources contained therein, and hence the genetic and genomic experiments they support, accessible to the community of all rat researchers. This article discusses the data, tools and approaches that RGD provides in order to make the database useful to the genomic expert and novice alike, plus the RGD outreach programs designed to increase community involvement in this field.
EXPERIMENTAL APPROACHES: AN INTRODUCTION
As a model, the rat is well suited to the study of complex genetic diseases, those diseases caused by the combined effects of multiple individual genes, none of which is responsible for the disease phenotype alone. Narrowing down the regions containing these genes entails comparing and contrasting rat strains presenting the disease phenotype with those resistant to the disease using a combination of phenotyping, genotyping and statistical approaches. The regions identified by these approaches are termed quantitative trait loci (QTL). The QTL define broad regions of the genome, which are then subjected to a detailed examination to identify candidate genes likely to play a role in the disease phenotype. Subsequent experimental verification of this involvement is the final step in the gene identification process. The following sections describe RGD resources applicable to each of these steps, (see 1 for a detailed discussion of these approaches and their use in the study of hypertension in the rat.
Rat strains, phenotypes, genotypes and maps
Selection of the appropriate rat strains is of prime importance and RGD contains records of over 250 rat strains many of which include detailed information on physiology, genetics and disease susceptibility (6). These records are searchable by keyword and as with all data in RGD, links to original references are provided. Also included are existing QTL that have been reported in the literature, allowing the researcher to see which rat strains have been used to map existing traits, the locations of these QTL and potential candidate genes. Following identification of the strains to be used, the next requirement is the selection of polymorphic markers that can be used to distinguish between the genomes of the two strains using a simple PCR-based assay. RGD contains information on almost 10 000 simple sequence length polymorphisms (SSLP), also known as microsatellite markers, with map locations, primer and clone sequences where available. A related resource is the allele size data for approximately 4500 SSLPs genotyped in 48 commonly used rat strains as part of the US–German rat genome project in the late 1990s (7). RGD currently has three major maps providing locations of many SSLPs, two of which were created from genetic mapping crosses, the third being a radiation hybrid map which not only maps 4615 SSLPs but also has 5682 ESTs placed upon it (8).
Genes, homologs and nomenclature
Rat gene records form the final major area of RGD curation. As with the other curated information, gene data is gathered through a combination of automatic processing of large datasets released by other databases and manual curation of journal articles by our scientific curation team. Gene records include symbol, name, any retired symbols and names, mapping data where available and a comprehensive set of external database links. These include links to Ratmap (9), NCBI’s LocusLink (10) and UniGene resources and PubMed for related references. Ensuring correct symbol and name nomenclature for curated genes is a major concern and all new genes are checked against existing rat, human and mouse guidelines by experienced nomenclature curators. Researchers deciding on nomenclature for new genes or who have questions about existing nomenclature are encouraged to submit their proposals and questions to the RGD nomenclature team via our web site. Curated ortholog assignment between rat, mouse and human genes is also included where available and follows the comprehensive set of criteria developed by the MGD (http://www.informatics.jax.org/userdocs/homology_criteria.shtml).
Data availability
All data in RGD is available on a public FTP site (http://rgd.mcw.edu/pub) as tab-delimited text files updated on the same schedule as the database itself.
ONLINE TOOLS
In addition to creating a repository of rat genetic and genomic data as described above, our philosophy has been to also create informatic tools that allow researchers to further explore and manipulate the data. To this end RGD also provides a number of popular tools that build upon the resources within the database and support comparative mapping, radiation hybrid and genetic mapping and gene prediction. Whilst these tools are rat-centric and are housed on the RGD, the comparative mapping tool VCMap and the gene prediction tool MetaGene are of use to researchers working in many other systems, particularly human and mouse.
Virtual Comparative Map (VCMap)
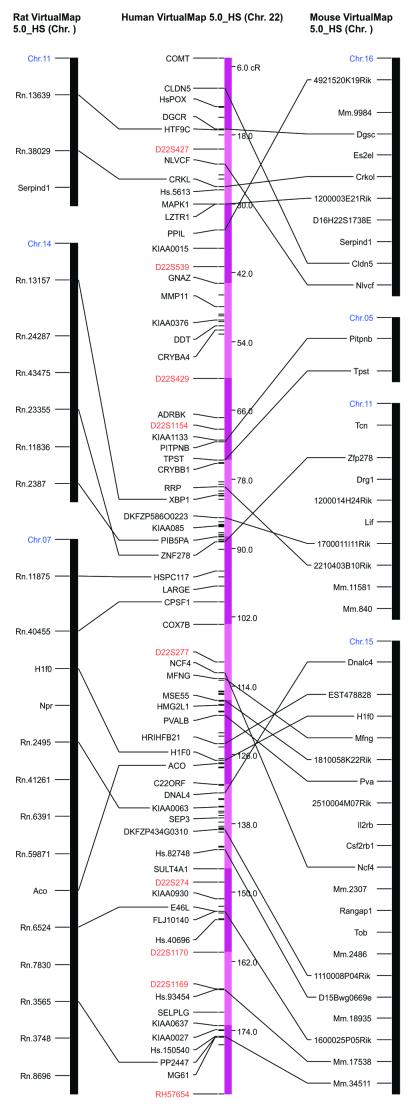
VCMap (http://rgd.mcw.edu/VCMAP) is a sequence-based algorithm that determines likely regions of synteny between the rat, mouse and human genomes (11). By comparing all human, mouse and rat ESTs and cDNAs using the Blast algorithm (12), homologous ESTs between the three organisms are determined. These individual results are then regrouped according to their UniGene cluster and in the instances where an unambiguous relationship between two genomes can be created, a syntenic anchor is formed. Further annotation with known radiation hybrid map locations allows conserved syntenic regions to be determined and it is from this that comparative maps are created (Fig. 1). An extension of this concept is the prediction of map locations for markers that are unmapped in one organism but for which a map location is known for the predicted homolog in another organism (virtual mapping). VCMap provides 64 comparative maps of the syntenic relationships between rat, mouse and human and is fully searchable by GenBank accession number, UniGene ID and gene symbol.
Figure 1.
Comparative maps of rat, human and mouse.
MetaGene
MetaGene (http://rgd.mcw.edu/METAGENE) is a unique gene prediction tool that acknowledges the well known fact that there is no one single perfect gene prediction algorithm. It analyses the submitted sequence by using a neural network to combine the gene prediction results of up to nine popular algorithms such as Genscan (13) and Grail (14) to provide a consensus MetaGene prediction.
RH Mapserver
RH Mapserver (http://rgd.mcw.edu/RHMAPSERVER) (15) is a radiation hybrid mapping tool that allows the user to submit radiation hybrid vectors created in the lab using the T55v3 rat radiation hybrid mapping panel. Instead of having to install and learn complex radiation hybrid mapping software, RH Mapserver allows the researcher to simply submit scored PCR results and in return they will receive graphic maps and placement statistics describing the map location of each marker relative to the published MCW radiation hybrid map (8).
Genome Scanner
Genome Scanner (http://rgd.mcw.edu/GENOMESCANNER) combines microsatellite allele size data available for over 4500 markers in 48 rat strains with the available genetic and radiation hybrid maps. By selecting two stains to be used in a genetic cross, genome scanner can be used to select polymorphic markers between the two selected strains. These markers can then be used in genotyping experiments such as whole genome scans or when increasing marker density in specific chromosomal regions.
OUTREACH AND EDUCATION
RGD aims to make rat genetic and genomic information available to the widest audience possible and has several programs to encourage community involvement to achieve these ends. RGD hosts the Rat Community Forum, an online bulletin board system for researchers to ask and answer questions relating to rat research. The RGD Visiting Scientist program is available to researchers who wish to take their involvement somewhat further and is designed to allow researchers to visit RGD and spend time with the curation and bioinformatics staff working on projects of mutual benefit to the researcher and the database. This has been a very successful program providing robust interaction between the rat research community and the members of RGD.
CURRENT PROJECTS
The focus of RGD is to provide a disease-centric perspective of the rat genome for the community, which is generally organized scientifically by disease speciality. To this end, we are prioritizing our curation efforts based on diseases identified as having the greatest impact on humans. A complex semi-automated methodology developed in the past year has been used to identify references and data objects such as genes, sequences, QTLs, strains and SSLPs related to a few diseases. This library of genetic and genomic information is reviewed by RGD’s curation staff and disease experts from the community to derive a complete picture of current rat research related to this disease. Data objects identified in this process are being loaded into RGD on a priority basis. In addition, sequence, QTL and map data identified as related to a specific disease is being used to mine rat genomic sequence for processing in a genome assembly and annotation pipeline developed at RGD in order to provide access to genome annotation of the disease regions of highest interest to the community as early as possible. This approach gives immediate focus to the analysis and annotation of sequence data currently underway by the RGSP (http://rgd.mcw.edu/sequences/rgp_info.shtml). This methodology will be applied to six to eight diseases per year to provide specific curated disease data and relationships valuable to the rat community. The VCMap tool is also being used as part of the total disease data analysis pipeline to provide users with a comparative view of the rat disease data with known human and mouse data.
Two other types of data annotation will be available in early 2002. Because expression profiling (microarray) experiments are becoming increasingly prevalent as a tool for the identification of differentially expressed genes in disease states, we have annotated all rat cDNA libraries and this information will be available in the RGD in January 2002. RGD will be annotating genes using the Gene Ontology (16), a controlled vocabulary in use in many organism databases to allow cross-organism searches for genes with shared cellular location, biological processes and molecular function.
IMPLEMENTATION
RGD is developed and maintained on the Sun/Solaris platform using Oracle 8i as the object relational database management system. All CGI scripts are written in Perl and use the Perl DBI for data retrieval. Schema documentation is available on request.
CONTACTING THE RGD
Rat Genome Database, Bioinformatics Research Center, 8701 Watertown Plank Road, Milwaukee, WI, 53226, USA. Please use our contact form available at http://rgd.mcw.edu/contact. Tel: +1 414 456 8871; Fax: +1 414 456 6595; Email: help@rgd.mcw.edu.
SUPPLEMENTARY MATERIAL
A table of relevant URL links is available as Supplementary Material at NAR Online.
Acknowledgments
ACKNOWLEDGEMENT
RGD is funded by grant HL64541 from the National Heart, Lung and Blood Institute on behalf of the NIH.
REFERENCES
- 1.Rapp J.P. (2000) Genetic analysis of inherited hypertension in the rat. Physiol. Rev., 80, 135–172. [DOI] [PubMed] [Google Scholar]
- 2.Shepel L.A., Lan,H., Haag,J.D., Brasic,G.M., Gheen,M.E., Simon,J.S., Hoff,P., Newton,M.A. and Gould,M.N. (1998) Genetic identification of multiple loci that control breast cancer susceptibility in the rat. Genetics, 149, 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Remmers E.F., Longman,R.E., Du,Y., O‘Hare,A., Cannon,G.W., Griffiths,M.M. and Wilder,R.L. (1996) A genome scan localizes five non-MHC loci controlling collagen-induced arthritis in rats. Nature Genet., 14, 82–85. [DOI] [PubMed] [Google Scholar]
- 4.Jacob H.J., Pettersson,A., Wilson,D., Mao,Y., Lernmark,A. and Lander,E.S. (1992) Genetic dissection of autoimmune type I diabetes in the BB rat. Nature Genet., 2, 56–60. [DOI] [PubMed] [Google Scholar]
- 5.Blake J.A., Eppig,J.T., Richardson,J.E., Bult,C.J. and Kadin,J.A. (2001) The Mouse Genome Database (MGD): integration nexus for the laboratory mouse. Nucleic Acids Res., 29, 91–94. Updated article in this issue: Nucleic Acids Res. (2002), 30, 113–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenhouse D.D., Festing,M.F.W., Hasan,S. and Cohen,A.L. (1990) Inbred strains of rats and mutants. In Hedrich,H.J. (ed.), Genetic Monitoring of Inbred Strains of Rats. Gustav Fischer Verlag, Stuttgart, pp. 410–480.
- 7.Jacob H.J., Lindpaintner,K., Lincoln,S.E., Kusumi,K., Bunker,R.K., Mao,Y.P., Ganten,D., Dzau,V.J. and Lander,E.S. (1991) Genetic mapping of a gene causing hypertension in the stroke-prone spontaneously hypertensive rat. Cell, 67, 213–224. [DOI] [PubMed] [Google Scholar]
- 8.Steen R.G., Kwitek-Black,A.E., Glenn,C., Gullings-Handley,J., Van Etten,W., Atkinson,O.S., Appel,D., Twigger,S., Muir,M., Mull,T. et al. (1999) A high-density integrated genetic linkage and radiation hybrid map of the laboratory rat. Genome Res., 9, AP1–AP8. [PubMed] [Google Scholar]
- 9.Stahl F. (1999) RATMAP Report—resources on the net. Rat Genome, 5, 103–108. [Google Scholar]
- 10.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwitek-Black A.E., Tonellato,P.J., Chen,D., Gullings-Handley,J., Cheng,Y.S., Twigger,S., Scheetz,T.E., Casavant,T.L., Stoll,M., Nobrega,M.A. et al. (2001) Automated construction of high-density comparative maps between rat, human and mouse. Genome Res., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burge C. and Karlin,S. (1997) Prediction of complete gene structures in human genomic DNA. J. Mol. Biol., 268, 78–94. [DOI] [PubMed] [Google Scholar]
- 14.Xu Y., Mural,R., Shah,M. and Uberbacher,E. (1994) Recognizing exons in genomic sequence using GRAIL II. Genet. Eng., 16, 241–253. [PubMed] [Google Scholar]
- 15.Lu J., Kwitek-Black,A.E., Jacob,H.J. and Tonellato,P.J. (1999) A web-based radiation hybrid map server. Rat Genome, 5, 97–102. [Google Scholar]
- 16.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T. et al. (2000) Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data in RGD is available on a public FTP site (http://rgd.mcw.edu/pub) as tab-delimited text files updated on the same schedule as the database itself.