Abstract
The ever increasing rate at which whole genome sequences are becoming accessible to the scientific community has created an urgent need for tools enabling comparison of chromosomes of different species. We have applied biometric methods to available chromosome sequences and posted the results on our Comparative Genometrics (CG) web site. By genometrics, a term coined by Elston and Wilson [Genet. Epidemiol. (1990), 7, 17–19], we understand a biometric analysis of chromosomes. During the initial phase, our web site displays, for all completely sequenced prokaryotic genomes, three genometric analyses: the DNA walk [Lobry (1999) Microbiology Today, 26, 164–165] and two complementary representations, i.e. the cumulative GC- and TA-skew analyses, capable of identifying, at the level of whole genomes, features inherent to chromosome organization and functioning. It appears that the latter features are taxon-specific. Although primarily focused on prokaryotic chromosomes, the CG web site contains genometric information on paradigm plasmids, phages, viruses and eukaryotic organelles. Relevant data and methods can be readily used by the scientific community for further analyses as well as for tutorial purposes. Our data posted at the CG web site are freely available on the World Wide Web at http://www.unil.ch/comparativegenometrics.
INTRODUCTION
By the end of August 2001, complete and nearly complete sequences of, respectively, 55 prokaryotic and five eukaryotic genomes, were available at the genome database of the National Center for Biotechnology Information (NCBI) (1), while the recent development of genometrics, i.e. biometric analyses of chromosomes (2), allows the study of the so-called architectural patterns along chromosomes. However, work in the emerging field of biometrics is seriously hampered by the absence of centralization of the relevant information. The aim of our Comparative Genometrics web site (CG) is to provide standardized descriptions of chromosomes at the genome level.
Inherent to the double helix structure, nucleotides of one strand are complementary to those of the opposite strand according to the pairing rules G-C and T-A, implying that, for any double-stranded chromosome, the total number of Gs is equal to that of Cs like the total number of Ts is equal to that of As (3,4). Surprisingly, even at the level of single-stranded DNA, as first observed for the Bacillus subtilis chromosome (5), the latter rule is almost rigorously respected. However, within each strand for all organisms for which the information is currently known, there are significant local deviations from the average number of nucleotides belonging to one or the other pairing couple. Accurate information on local nucleotide distribution has been used for the identification of, for instance, (i) pathogenicity islands in Yersinia pestis (6), (ii) integrated foreign DNA like prophages such as SPβ in B.subtilis (see below; Fig. 1), or (iii) internal chromosomal recombination events, as observed in Pseudomonas aeruginosa (7). Most importantly, nucleotide distributions along the chromosomes, expressed as DNA walk graphs, have allowed the origin of DNA replication of some of the examined organisms to be identified.
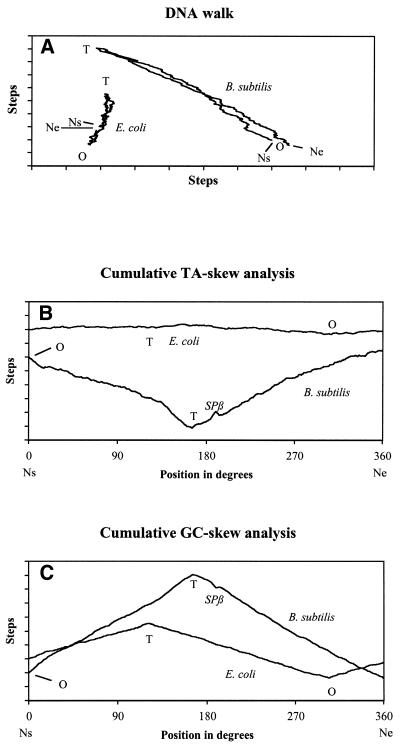
Figure 1.
Genometric characterizations of B.subtilis and E.coli. Bacillus subtilis and E.coli chromosomes comprise 4 214 814 and 4 639 221 bp, respectively. For a given chromosome, Ns and Ne refer to the nucleotides at the start and the end of the sequence, respectively. O and T correspond to the experimentally determined origin and terminus of chromosome replication (15,17). SPβ is an integrated prophage of B.subtilis of 134 416 nt (16). One division on the step scale corresponds to 10 000 steps (16). For both chromosomes, positions are indicated in degrees. (A) DNA walk: processing of a given genome, nucleotide by nucleotide, generates the displayed paths which measure the internal deviation of pairing nucleotides. Reading of one nucleotide, i.e. G, T, C and A, corresponds to one step towards north, east, south and west, respectively. Escherichia coli and B.subtilis are examples of organisms presenting a path with a positive and a negative slope, respectively. (B) Cumulative TA-skew graph is obtained by replacing G by T and C by A in the GC-skew description. In E.coli, the origin and the terminus of DNA replication correspond to the minimum and the maximum of the curve, respectively, while in B.subtilis, the reverse is true. (C) Cumulative GC-skew graph: the algorithm is similar to a DNA walk. A step eastwards is assigned to each nucleotide, while for a G or a C, a concomitant step to the north or the south, respectively, is performed. Whenever available, experimental evidence reveal that, for both bacteria, the origin and the terminus of chromosome replication correspond to the minimum and the maximum of the cumulative GC-skew graphs, respectively.
At present, determination of the origin of DNA replication is one of the central problems amenable to genometric analyses. In 1981, Smithies et al. (8) deduced from the nucleotide sequence of the SV40 virus that genes situated to the left or to the right of the origin of DNA replication are transcribed towards the left or to the right, respectively. However, at the left or right of the origin of DNA replication the frequencies of Cs or Gs, respectively, were slightly more abundant. Lobry (9) was the first to apply local deviation analyses to whole bacterial genomes. He devised an algorithmic tool measuring local deviations of one nucleic base in the GC or the TA couple which enabled him to identify a single origin of DNA replication on circular chromosomes of several prokaryotic organisms (9). Although his method allowed the identification of the then unknown origin of replication of Mycoplasma genitalium (10), it failed for nearly half of the so far completely sequenced prokaryotic genomes. Lobry’s original algorithmic approach was subsequently improved by the use of a sliding window (11), or by analyses of the distribution of one or several chosen oligonucleotides (12).
Our CG web site described here is initially focused on prokaryotic chromosomes and on paradigm plasmids, phages, viruses and eukaryotic organelles. Over the past year, part of our DNA walk data, now posted on CG, was already available at a previous web site: http://www.unil.ch/igbm/genomics/genometrics.html.
SOURCES OF GENOMIC DATA AND METHODS
Complete sequences of all examined organisms have been obtained from NCBI. Analyses consist of different comparisons of nucleotide distributions and their alignment along genomes. The DNA walk, a term defined and coined by Lobry (13,14), was performed on all sequenced chromosomes. It provides a quantification of internal deviations of pairs of nucleotides chosen according to pairing rules: G versus C, and T versus A, respectively. Two complementary methods (14), the cumulative GC- and the TA-skew analyses, were also used. We describe on the CG web site the algorithmic method which offers a way of drawing any of the graphs with spreadsheet software.
Actually, for the DNA walk, we use a shareware developed by the Genometrician’s Company S.A., and freely available at www.genometrician.com. This shareware shortens the time-consuming task of drawing DNA walks, and publishes the results directly as web pages. However, for the cumulative GC- and TA-skews, we use a new software developed by the Genometrician’s Company S.A. None of the latter computer tools is available on our web site.
POSTED GENOMETRIC RESULTS
Up to now, partial genometric databases only have been available. However, a complete database encompassing Lobry’s biometric analyses is presently not available. This prompted us to create a new web site making available (i) textfiles containing the nucleotide distributions at a local level, and (ii) the genometric information based on these local distributions obtained by DNA walks (Fig. 1A) as well as complementary analyses, i.e. the cumulative TA- and GC-skew plots (Fig. 1B and C).
Figure 1 reveals striking differences between Escherichia coli and B.subtilis, in particular at the level of the slopes, the amplitude of the curves, and the noise. Interestingly, inspection of our web site display reveals that these aspects are taxon-specific. A further feature clearly discernable on the graphs is the location of SPβ, due to the fact that most of the genes of this prophage are characterized by a nucleotide usage distinct from that of its bacterial host (15,16).
DEVELOPMENT AREAS
As mentioned above, the DNA walk analysis was able to uncover a single origin of DNA replication for only about a half of the sequenced prokaryotic genomes. The successfully analyzed chromosomes appeared divided into two arms called replichores (17) or chirochores (9). Improved algorithms have revealed that this pattern is widespread among prokaryotes. Indeed, it was shown that about half of completely sequenced archaebacterial chromosomes are similarly organized (12). However, several organisms, the complete genomes of which have been sequenced, remain refractory to the presently available methods of analysis. Therefore, we are attempting to improve genometric algorithms with the aim of finding out if some of the circular prokaryotic chromosomes may have more than one origin of replication.
Additional data will be posted to our database in the near future. In particular, (i) analyses obtained with the same strategy of over 1000 small genomes, i.e. plasmid, phage, virus and organelle DNA molecules, and (ii) analyses performed on either intergenic or coding sequences of all available genomes.
TUTORIAL ASPECTS
In addition to DNA walk graphs, textfiles describing the local nucleotide content used to draw the graphs are posted on our web site. They should enable not only scientists but also students, able to use a spreadsheet software, to perform other genometric analyses like, for instance, those presented by Freeman et al. (18): http://bmerc-www.bu.edu/information/all_genomes.html.
USER SUPPORT
We encourage the scientific community to contribute genomic data sets to our database. While respecting confidentiality, analyses may still be performed on tables summarizing the averaged local nucleotide content. Please contact wwwcogen@unil.ch to get instructions on how to proceed.
CITATION OF CG
We trust that users of the CG database will refer to this contribution in their publications. The following reference format is suggested: Comparative Genometrics Database (CG), Genometrics Group, IGBM, Lausanne University, Switzerland. World Wide Web, http://www.unil.ch/comparativegenometrics. The date (month/year) of data retrieval should be specified.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful to O. Braissant, C. Mauël, K. Minnig and R. E. Studer, for helpful discussions, and to F. Adamer, A. Bachelard, N. Bodenhausen, M. Haenni, D. Hofmann, M. Huguenin, N. Iglesias, D. Michod, C. Tauxe, A. Troxler and L. Waselle for critical comments relative to analysis displays. The Genometrician’s Company is thanked for generously providing their software free of charge.
REFERENCES
- 1.Wheeler D.L., Church,D.M., Lash,A.E., Leipe,D.D., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Tatusova,T.A., Wagner,L. et al. (2001) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 29, 11–16. Updated article in this issue: Nucleic Acids Res. (2002), 30, 142–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elston R.C. and Wilson,A.F. (1990) Genetic linkage and complex disease: a comment. Genet. Epidemiol., 7, 17–19. [Google Scholar]
- 3.Chargaff E. (1950) Chemical specificity of nucleic acids and mechanism of their enzymatic degradation. Experientia, 6, 201–240. [DOI] [PubMed] [Google Scholar]
- 4.Watson J.D. and Crick,F.H.C. (1953) Molecular structure of nucleic acids. Nature, 171, 737–738. [DOI] [PubMed] [Google Scholar]
- 5.Rudner R., Karkas,J.D. and Chargaff,E. (1968) Separation of B. subtilis DNA into complementary strands. III. Direct Analysis. Proc. Natl Acad. Sci. USA, 60, 921–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parkhill J., Wren,B.W., Thomson,N.R., Titball,R.W., Holden,M.T.G., Prentice,M.B., Sebaihia,M., James,K.D., Churcher,C., Mungal,K.M. et al. (2001) Genome sequence of Yersinia pestis, the causative agent of plague. Nature, 413, 523–527. [DOI] [PubMed] [Google Scholar]
- 7.Stover C.K., Pham,X.Q., Erwin,A.L., Mizoguchi,S.D., Warrener,P., Hickey,M.J., Brinkman,F.S., Hufnagle,W.O., Kowalik,D.J., Lagrou,M. et al. (2000) Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature, 406, 959–964. [DOI] [PubMed] [Google Scholar]
- 8.Smithies O., Engels,W.R., Devereux,J.R., Slightom,J.L. and Shen,S. (1981) Base substitutions, length differences and DNA strand asymmetries in the human Gγ and Aγ fetal globin gene region. Cell, 26, 345–353. [DOI] [PubMed] [Google Scholar]
- 9.Lobry J.R. (1996) Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol., 13, 660–665. [DOI] [PubMed] [Google Scholar]
- 10.Lobry J.R. (1996) Origin of replication of Mycoplasma genitalium. Science, 272, 745–746. [DOI] [PubMed] [Google Scholar]
- 11.Grigoriev A. (1998) Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res., 26, 2286–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myllykallio H., Lopez,P., Lopez-Garcia,P., Heilig,R., Saurin,W., Zivanovic,Y., Philippe,H. and Forterre,P. (2000) Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science, 288, 2212–2215. [DOI] [PubMed] [Google Scholar]
- 13.Lobry J.R. (1996) A simple vectorial representation of DNA sequences for the detection of replication origins in bacteria. Biochimie, 78, 323–326. [DOI] [PubMed] [Google Scholar]
- 14.Lobry J.R. (1999) Genomic landscapes. Microbiology Today, 26, 164–165. [Google Scholar]
- 15.Kunst F., Ogasawara,N., Moszer,I., Albertini,A.M., Alloni,G., Azevedo,V., Bertero,M.G., Bessières,P., Bolotin,A., Borchert,S. et al. (1997) The complete genome sequence of the Gram positive bacterium Bacillus subtilis. Nature, 390, 249–256. [DOI] [PubMed] [Google Scholar]
- 16.Lazarevic V., Düsterhöft,A., Soldo,B., Hilbert,H., Mauël,C. and Karamata,D. (1999) Nucleotide sequence of the Bacillus subtilis temperate bacteriophage SPβc2. Microbiology, 145, 1055–1067. [DOI] [PubMed] [Google Scholar]
- 17.Blattner F.R., Plunkett,G.,III, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 18.Freeman J.M., Plasterer,T.N., Smith,T.F. and Mohr,S.C. (1998) Patterns of genome organization in bacteria. Science, 279, 1825. [Google Scholar]