Abstract
FANTOM DB, the database of Functional Annotation of RIKEN Mouse cDNA Clones, is designed to store sequence information of RIKEN full-length enriched mouse cDNA clones, graphical views of sequence analysis results, curated functional annotation information and additional descriptions, including Gene Ontology terms. RIKEN’s Mouse Gene Encyclopedia Project aims to collect full-length enriched cDNA clones from various mouse tissues, determine the full-length nucleotide sequences, infer their chromosomal locations by computer and characterize gene expression patterns. FANTOM DB has been developed to facilitate this work and to facilitate functional genomic studies such as positional candidate cloning, cDNA microarrays and protein interaction analyses. FANTOM DB contains 21 076 full-length cDNA sequences with rich functional annotations and is publicly available. FANTOM DB thus provides curated functional annotation to RIKEN full-length enriched mouse clones, and has links to other public resources. FANTOM DB can be accessed at http://fantom.gsc.riken.go.jp/db/.
INTRODUCTION
The RIKEN Mouse Gene Encyclopedia Project, a systematic approach to determining the full coding potential of the mouse genome, involves collecting and sequencing full-length complementary DNAs (cDNAs), physical mapping of the corresponding genes to the mouse genome, determining gene expression patterns in all mouse tissues and determining interactions among all transcripts. FANTOM DB has been developed to facilitate work on the project as well as functional genomic studies such as positional candidate cloning, cDNA microarrays and protein interaction analyses. FANTOM DB provides users with well-curated functional annotation of the RIKEN full-length cDNA clones together with a systematic search tool and BLAST search. The annotation process and the vocabulary used were determined at the international meeting of Functional Annotation of RIKEN full-length Mouse cDNAs (FANTOM meeting) (1). The data are now stored as a database, accessible worldwide. These data are available both in a web page generated on-the-fly and in concatenated text in MaXML (Mouse Annotation XML) format.
IMPLEMENTATION
Curators annotated the clones with the help of the FANTOM+ system, which allowed them to view pre-computed sequence similarity and motif search results, to launch additional searches and to transfer the annotation from any of these to the FANTOM DB. FANTOM+ system has the following features:
1. A simple interface with HTML and JavaScript.
2. A graphical user interface.
3. A controlled system of annotation records.
4. A parser for XML output (MaXML).
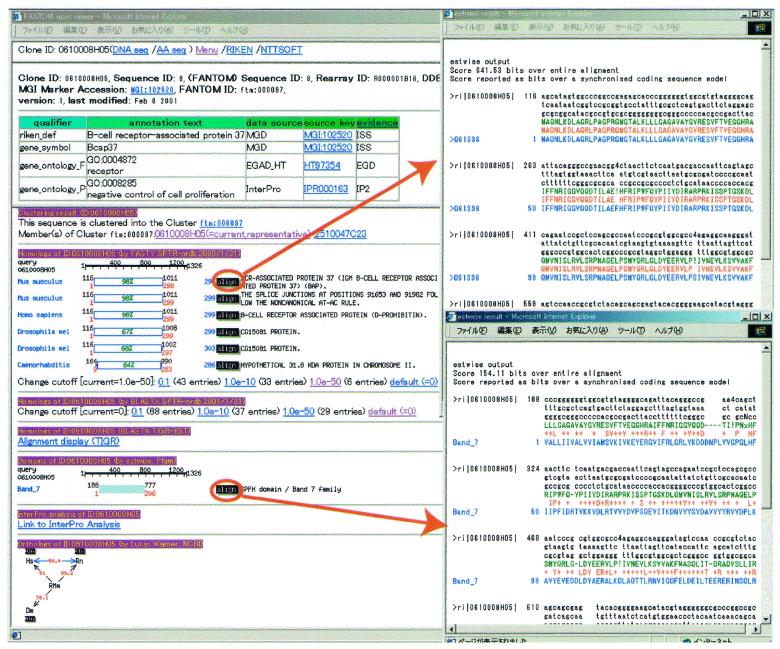
The top page of the FANTOM DB (http://fantom.gsc.riken.go.jp/db/) has a menu (Search, MaXML and BLAST search) and a link to the FANTOM main page. The left side of Figure 1 shows a search result page containing three frames. The top frame shows a summary of the current object (RIKEN clone, database record, protein motif, etc.) (summary field). The middle frame shows the results of curated functional annotation of the current cDNA clone (annotation field). The curation has been performed using the FANTOM+ system which provides an annotation editor. The lower frame displays pre-computed sequence similarity and motif search results (reference field). The results of clustering, FASTA search against SPTR-nrdb (SWISS-PROT and TrEMBL non-redundant database, ftp://ftp.ebi.ac.uk/pub/databases/st_tr_nrdb/), BLAST search against SPTR-nrdb and TIGR-EST (http://www.tigr.org/tdb/tgi.shtml), Wise2 search against Pfam and InterPro search, etc., are displayed. The orthologue search using HomoloGene database is also shown at the bottom. The alignment of each sequence can be browsed by clicking the ‘align’ link button (right upper and lower window). MaXML provides all the functional annotation information described in the annotation field in XML format. The BLAST search allows the users to search against all the sequences included in the FANTOM DB.
Figure 1.
A search result page containing three frames is shown. The top frame is the summary field, with links to DNA and amino acid sequences. The middle frame is the annotation field, where functional annotation of a cDNA is described. The lower frame is the reference field, which shows sequence analysis results and source information. The right windows show alignments with SPTR-nrdb (upper) and Pfam (lower). The alignment windows appear when the ‘align’ buttons in the reference frame are selected. Links in the reference and annotation frames take users to MGD, InterPro, LocusLink, GenPept, EGAD_HT, TIGR-MGI (TIGR-Mouse Gene Index), DDBJ, GenBank, EMBL, NCBI-nr, PIR (Protein Information Resource), UniGene and UTRdb (Untranslated Region database, http://bighost.area.ba.cnr.it/BIG/UTRHome/). The reference field also has links to Mapping (http://www.gsc.riken.go.jp/e/FANTOM/map/) and Superfamily (http://stash.mrc-lmb.cam.ac.uk/SUPERFAMILY/).
Sequence data
Full-length mouse cDNA sequences were produced as a Phase II activity of the RIKEN Mouse Encyclopedia Project (2).
Functional annotation
In the FANTOM DB, each RIKEN clone is assigned a RIKEN definition (riken_def) to indicate the most likely gene function or status of the sequence on the basis of similarity to known genes. A supplementary RIKEN definition line (riken_def_suppl) is available for additional pertinent annotation. Biological function (Biological Process, Molecular Function and Cellular Component) is described by assigning Gene Ontology (GO) terms to the clones (3). The IDs of these GO terms can be referred to as the attributes of database, whose qualifiers are gene_ontology_P (Biological Process), gene_ontology_F (Molecular Function) and gene_ontology_P (Cellular Process). The database contains curators’ statements in note and comment fields.
Search
Annotation data for mouse clones can be seen via the Search menu in FANTOM+. Currently, users can search clones by ID search, keyword search, Pfam domain search and TIGR Cellular Role search.
ID search. Each entry has many kinds of IDs, including Clone ID, Sequence ID and FANTOM ID. Users can search for entries by ID.
Keyword search. Users can search for entries by functional annotation keywords with target qualifiers. Both GO terms and IDs can be used as keywords.
Pfam domain. Users can search for entries with results of estwisedb [a program in the Wise2 package (http://www.sanger.ac.uk/Software/Wise2/)] against the Pfam database (4). If they know the Pfam name to retrieve, they can enter it in the search form. If they do not know it, they can see a list of all Pfam names matched to sequences in the FANTOM database.
Cellular Role search. Users can search for entries with the results of TIGR Cellular Role prediction by homology searches against the EGAD HT database (Expressed Gene Anatomy Database, http://www.tigr.org/tdb/egad/egad.shtml).
Data availability
All sequences and functional annotations are available from our web and FTP sites. For further information please write to fantom-help@gsc.rike.go.jp.
FUTURE DIRECTIONS
We are continuously updating functional annotations in the FANTOM DB with new results of sequence analyses. We are organizing a FANTOM2 meeting to annotate an additional sequence set. After this meeting, additional sequences and functional annotations will be released. We have been developing an automatic functional annotation system (FIND: the Functional Inference Descriptor) to determine RIKEN definitions. FIND will be used at the FANTOM2 meeting and the results will be compared to those obtained by human curators.
CITING FANTOM
The following citation format is suggested when referring to the RIKEN FANTOM set annotation: FANTOM, Genome Exploration Research Group, Genomic Sciences Center, Yokohama Institute, RIKEN, Yokohama, Kanagawa, Japan (http://fantom.gsc.riken.go.jp/) and: The RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium (2001) Functional annotation of a full-length mouse cDNA collection. Nature, 409, 685–690.
Acknowledgments
ACKNOWLEDGEMENTS
We acknowledge all the members of the FANTOM consortium for the use of FANTOM annotation. We also acknowledge Dr M. Brownstein for comments and editing the manuscript. This study was supported by a Research Grant for the RIKEN Genome Exploration Research Project from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government and by ACT-JST (Research and Development for Applying Advanced Computational Science and Technology) of the Japan Science and Technology Corporation (JST).
REFERENCES
- 1.Quackenbush J. (2000) Viva la revolution! A report from the FANTOM meeting. Nature Genet., 26, 255–256. [DOI] [PubMed] [Google Scholar]
- 2.The RIKEN Genome Exploration Research Group Phase II Team and The FANTOM Consortium (2001) Functional annotation of a full-length mouse cDNA collection. Nature, 409, 685–690. [DOI] [PubMed] [Google Scholar]
- 3.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T. et al. (2000) Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Howe,K.L. and Sonnhammer,E.L. (2000) The Pfam protein families database. Nucleic Acids Res., 28, 263–266. Updated article in this issue: Nucleic Acids Res. (2002), 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequences and functional annotations are available from our web and FTP sites. For further information please write to fantom-help@gsc.rike.go.jp.