Abstract
The Pharmacogenetics Knowledge Base (PharmGKB; http://www.pharmgkb.org/) contains genomic, phenotype and clinical information collected from ongoing pharmacogenetic studies. Tools to browse, query, download, submit, edit and process the information are available to registered research network members. A subset of the tools is publicly available. PharmGKB currently contains over 150 genes under study, 14 Coriell populations and a large ontology of pharmacogenetics concepts. The pharmacogenetic concepts and the experimental data are interconnected by a set of relations to form a knowledge base of information for pharmacogenetic researchers. The information in PharmGKB, and its associated tools for processing that information, are tailored for leading-edge pharmacogenetics research. The PharmGKB project was initiated in April 2000 and the first version of the knowledge base went online in February 2001.
INTRODUCTION
Variability in response to pharmaceutical drugs poses a significant problem for physicians, patients and pharmaceutical companies (1). The variation has several known causes including environmental factors, ongoing medical conditions and inherited genetic differences. The field of pharmacogenetics (in this paper, the terms ‘pharmacogenetics’ and ‘pharmacogenomics’ are used interchangeably) studies response variability related to inherited genetic differences. The National Institutes of Health (NIH) is sponsoring the Pharmacogenetics Research Network (http://www.nigms.nih.gov/pharmacogenetics/) to accumulate, store and process pharmacogenetic-related results. The Pharmacogenetics Knowledge Base (PharmGKB; http://www.pharmgkb.org/) (2) provides a central repository for data collected by laboratories in the research network and provides tools for submitting, editing viewing and processing that information. In the future, we anticipate having to restrict access to some data (e.g. clinical records), but at the time of writing this paper all of the submitted data is available to the public.
PharmGKB is organized as a knowledge base (3) with an ontology (4) that contains a network of genetic, clinical and cellular phenotype knowledge, interconnected by relations and organized by levels of abstraction. Experimental data and results are added to the knowledge base as instances of terms in the ontology (5). This will allow more complex data processing and inferencing than is allowed by traditional databases.
KNOWLEDGE BASE CONTENT
Much of the current research in the Pharmacogenetics Research Network focuses on identifying single nucleotide polymorphisms (SNPs) in people. PharmGKB contains SNP information and provides several tools described below to both submit and access information in the dbSNP database (6). PharmGKB also accepts other variants such as insertions, deletions and repeats. The knowledge base content pertaining to genetics includes genes, proteins, reference sequences, regions of interest, haplotypes, coordinate systems and populations of individuals.
PharmGKB also models information about cellular phenotypes, including pharmacokinetics and enzyme kinetics information. PharmGKB will also include clinical content, such as descriptions of drugs, information about clinical studies, drug regimes, observations of drug kinetics and various clinical parameters specific to particular studies. The most commonly reported data at this time is clinical data relating to drug metabolism.
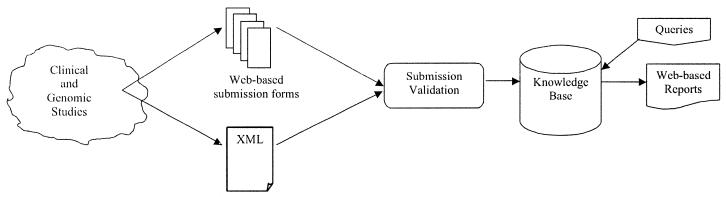
Currently, researchers can submit 38 different types of data containing genomic information, drugs, diseases, populations and so on. Submissions are made individually through web forms or in uploaded files containing PharmGKB-defined XML elements. Once a submission has been validated and accepted, it is merged into the existing body of PharmGKB knowledge where it is made accessible. This process is shown in Figure 1.
Figure 1.
The flow of data into and out of PharmGKB.
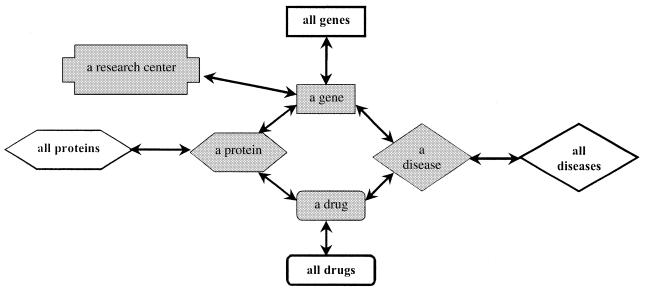
Each data submission contains data elements that are merged into the knowledge base in various ways. Some elements are grouped to form new knowledge base objects. Other elements are added to existing objects. Related objects are linked together to form a network of information. For example, genes are related to the proteins they express, to the research centers studying them, and to the drugs that interact with them. In addition, drugs are related to various information, including the diseases they treat. Data retrieval programs find related information by traversing the network of objects starting from a known object. One advantage of this representation is that the same information can be accessed from different starting objects, allowing queries to be formulated from different viewpoints. There are different types of relationships among the objects, including ‘expresses’, as in ‘a gene expresses a protein’, and ‘studied by’, as in ‘a gene is studied by a research center’. There are currently over 600 different relationships used in PharmGKB. A part of the network of PharmGKB information is illustrated in Figure 2.
Figure 2.
Some of the relationships among data objects in PharmGKB.
Research centers can request that specific categories of knowledge, such as cardiovascular drugs or receptor proteins, be modeled so that data in, or relating to, that category can be submitted to PharmGKB. This ongoing process requires considerable effort by both the PharmGKB staff and the requesting investigator in order for the modeling to be done correctly. Once the modeling is complete, the research center often must reformat their collected data to match the PharmGKB model before submitting it. PharmGKB has extensively modeled several categories of basic sequence-related genomic information, enzyme kinetics, pharmacokinetics and some miscellaneous information including literature citations and external databases.
ONGOING STUDIES
Researchers in the Pharmacogenetics Research Network are studying a variety of genotype–phenotype relationships including genotypic effects on the metabolism, pharmacokinetics, and clinical efficacy of drugs as well as on the function of metabolizing enzymes and drug targets in cell systems. Drugs currently under study include tamoxifen and other anti-cancer agents, asthma medications, antiarrythmics, antidepressants, statins and ACE inhibitors. PharmGKB contains both abstract and detailed information in all of these areas, and the relationships between the ontological terms form a rich web of information. Details of ongoing research are available at http://www.pharmgkb.org/information.html.
APPLICATIONS FOR PHARMACOGENETIC RESEARCHERS
PharmGKB provides a number of useful tools, including KBQuery, which allows the viewer to formulate complex queries on the knowledge base and displays information in a tabular form, which can be downloaded. KBEditor allows selected users to edit the knowledge base. The dbSNP submission program automatically submits any new PharmGKB SNPs to the dbSNP database, while a dbSNP surveillance program generates weekly reports to research members of new submissions to dbSNP involving their genes of interest. Similarly, the PubMed surveillance program generates weekly reports to research network members of new publications related to their areas of interest. For researchers interested in comparative genomics, PharmGKB contains an interface to VISTA (7) that allows genes in PharmGKB to be compared to the same genes in a mouse.
INTERFACING WITH RELATED DATABASES
The information in PharmGKB is related to data in other online databases such as dbSNP, LocusLink and OMIM. The automatic submission and surveillance tools allow PharmGKB to both contribute to external databases and to correlate its information with theirs.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Shuo Liu, Yueyi Liu, Charity Y. Lu and Marshall R. Mayberry III for their assistance in developing the PharmGKB software. PharmGKB is financially supported by grants from the National Institute of General Medical Sciences (NIGMS), Human Genome Research Institute (NHGRI) and National Library of Medicine (NLM) within the National Institutes of Health (NIH) and the Pharmacogenetics Research Network and Stanford University’s Children’s Health Initiative (Russ Altman, PI). This work is supported by the NIH/NIGMS Pharmacogenetics Research Network and Database (U01GM61374; Russ Altman, PI).
REFERENCES
- 1.Evans W.E. and Relling,M.V. (1999) Pharmacogenomics: translating functional genomics into rational therapeutics. Science, 286, 487–491. [DOI] [PubMed] [Google Scholar]
- 2.Klein T.E., Chang,J.T., Cho,M.K., Easton,K.L., Fergerson,R., Hewett,M., Lin,Z., Liu,Y., Liu,S., Oliver,D.E. et al. (2001) Integrating genotype and phenotype information: an overview of the PharmGKB project. Pharmacogenom. J., 1, 167–170. [DOI] [PubMed] [Google Scholar]
- 3.Karp P.D., Ouzounis,C. and Paley,S.M. (1996) HinCyc: a knowledge base of the complete genome and metabolic pathways of H. influenzae. In Proceedings of the Fourth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA, pp. 116–124. [PubMed]
- 4.Hafner C.D. and Fridman,N. (1996) Ontological foundations for biology knowledge models. In Proceedings of the Fourth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA, pp. 78–87. [PubMed]
- 5.Rubin D.L., Hewett,M., Oliver,D.E., Klein,T.E. and Altman,R.B. (2002) Automating data acquisition into ontologies from pharmacogenetics relational data sources using declarative object definitions and XML. In Altman,R.B., Dunker,A.K., Hunter,L., Lauderdale,K. and Klein,T.E. (eds), Proceedings of the Seventh Pacific Symposium on Biocomputing. World Scientific Publishing Co. Pte. Ltd, Singapore, vol. 7, 88–99. [PubMed]
- 6.Sherry S.T., Ward,M.H., Kholodov,M., Baker,J., Phah,L., Smigielski,E.M. and Sirotkin,K. (2001) The NCBI database of genetic variation. Nucleic Acids Res., 29, 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayor C., Brudno,M., Schwartz,J.R., Poliakov,A., Rubin,E.M., Frazer,K.A., Pachter,L.S. and Dubchak,I. (2000) VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics, 16, 1046–1047. [DOI] [PubMed] [Google Scholar]