Abstract
The DNA Data Bank of Japan (DDBJ, http://www.ddbj.nig.ac.jp) has made an effort to collect as much data as possible mainly from Japanese researchers. The increase rates of the data we collected, annotated and released to the public in the past year are 43% for the number of entries and 52% for the number of bases. The increase rates are accelerated even after the human genome was sequenced, because sequencing technology has been remarkably advanced and simplified, and research in life science has been shifted from the gene scale to the genome scale. In addition, we have developed the Genome Information Broker (GIB, http://gib.genes.nig.ac.jp) that now includes more than 50 complete microbial genome and Arabidopsis genome data. We have also developed a database of the human genome, the Human Genomics Studio (HGS, http://studio.nig.ac.jp). HGS provides one with a set of sequences being as continuous as possible in any one of the 24 chromosomes. Both GIB and HGS have been updated incorporating newly available data and retrieval tools.
INTRODUCTION
Since the whole genome of Haemophilus influenzae was sequenced (1), the object of DNA sequencing has been turned from gene to genome. The success in sequencing of this microbial genome has proven that the shotgun method (2) is efficient enough for sequencing the whole genome not only of prokaryote but also more complex eukaryote species (3,4). It can be said that the scope of research in life science has then been shifted from the gene scale to the genome scale. This shift has long been awaited, because of the assertion that the genes in an individual organism do not function independently but do interdependently in complex functional networks. Therefore, even if one studies the function of a particular gene, one may have to extend one’s study to other related genes in the same genome.
A series of the whole-genome sequencing endeavours have resulted in sequencing through the whole human genome. The genome data have then immediately been published both in the journal (5) and the International Nucleotide Sequence Databases (INSD) that are composed of DDBJ, the EMBL Bank and GenBank. It is noteworthy that the immediate publication of such enormous-scale data was possible perhaps only through a well-established collaboration among the INSD members. INSD have also provided retrieval and analysis tools by which one can make the data useful for one’s study. In fact, a large number of researchers have enjoyed the simultaneous publications of the human genome data in print and online.
As a member of INSD, DDBJ has made an effort to collect the original DNA sequence data, and releasing them to the public after annotation. The majority of the data have been submitted directly from Japanese researchers. We have exchanged the data thus released with the EMBL Bank and GenBank on a daily basis. This practice allows the three data banks to serve users worldwide with the same quality and quantity of data. We have also developed the Genome Information Broker (GIB) and the Human Genomics Studio (HGS). Both databases are available now at DDBJ.
DATA COLLECTION AT DDBJ
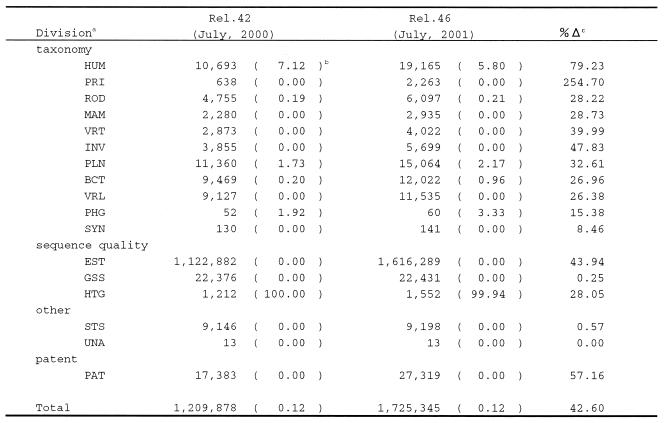
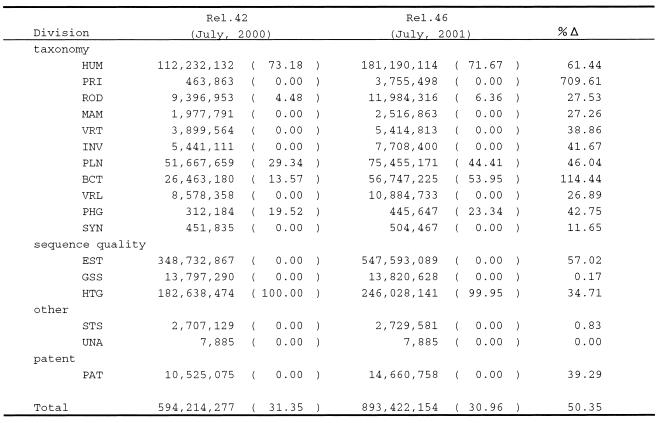
Tables 1 and 2 show the amounts of data released to the public from the DDBJ submissions at two time points, July 2000 and July 2001, respectively. On the whole, the rate of increase in the past year for the number of entries is 43.0% and for the number of bases is 51.6% indicating that the number of bases in an entry has been increased. This implies that the contribution from various genome groups in Japan has been increased in the past year. Usually, a genome team tends to submit an entry including a larger number of bases than other entries. This tendency is clarified more if we focus on the items (taxonomic divisions) in the tables.
Table 1. The number of entries made public from the DDBJ submissions.
aEach division is as follows. (taxonomy) HUM: human, PRI: primate, ROD: rodent, MAM: mammal, VRT: vertebrate, INV: invertebrate, PLN: plant and fungi, BCT: bacteria, VRL: virus, PHG: bacteriophage, SYN: synthesized sequence; (sequence quality) EST: expressed sequence tag, GSS: genome survey sequence, HTG: high-throughput genomic sequence; (other) STS: sequence tagged site, UNA: unannotated sequence; (patent) PAT: patent data.
bThe number in parentheses is the percentage of genome data to the total of each division.
c%Δ is the rate of increase in the year.
Table 2. The number of bases (bp) made public from the DDBJ submissions.
In the past year the Japanese human genome teams made a significant contribution to the international human genome sequencing consortium. We at DDBJ established a good collaboration with them to make it possible to collect and immediately release their large-scale data to the public. The data include a part of the entire HLA region in chromosome 6 (6), parts of chromosomes 22 (7) and 21 (8). In Tables 1 and 2, the HUM division shows the number of entries and the number of bases, respectively, of human sequence data. The number in the parentheses is the ratio of the data submitted from the Japanese human genome teams in percentage. The HUM division in Table 2 indicates that >70% of the bases have been submitted from the genome teams. In contrast, the corresponding division in Table 1 shows that <10% of the entries have been submitted from the genome teams. These two contrasting figures confirm the above-mentioned tendency. The rates of increase for this division both in the number of entries and of bases exceed the average rates over the all divisions. It is also noted that the majority of the HTG division is human genome sequence data in the pre-assembled status. As is clear in Table 1, all data in this division have been contributed from the genome teams.
Similar observations to the HUM division can be made for the ROD, PLN and BCT divisions in Tables 1 and 2. Among them, it is particularly noteworthy that the rice gemone team of the National Institute of Agrobiological Resources and the Arabidopsis team of the Kazusa DNA Institute (9) have made significant contributions to the PLN division. Also noteworthy is that several genomes of bacterial species, Buchunera sp. (10), Bacillus halodurans (11), Chlamydia pneumoniae (12), Mesorhizobium loti (13), Staphylococcus aureus (14) and others, have been sequenced and submitted in the past year. Consequently, the BCT division has been on a remarkable increase both in the number of bases and the ratio of genome sequence data as shown in Table 2.
Most of the Japanese genome teams now use our mass submission tool, MST (15), for submitting their data to DDBJ. In this case we can easily sort out the genome data from the others in a division by searching for particular accession numbers given to a genome team. There are, however, cases in which sorting out cannot be carried out easily. The Silver Genome Project of our institute for sequencing ape genome (http://sayer.lab.nig.ac.jp/~silver/) is one of them. That is why the PRI division in Tables 1 and 2 shows no ratio of genome data in spite of the fact that the project has made an outstanding contribution to the PRI division.
SPECIALIZED DATABASES DEVELOPED AT DDBJ
Our prime mission is of course to collect the original DNA data, and release them to the public after annotation. In addition to the mission we have extended our annotation capacity to collaborating with the Riken mouse project (16) and to developing and updating several specialised databases on the basis of our DNA database. One of these databases is the Genome Information Broker of the complete genome sequence data (GIB, http://gib.genes.nig.ac.jp) (17). Since its original version, GIB has been updated incorporating newly published complete genome sequence data and new retrieval tools. GIB at present includes more than 50 complete bacterial genome sequences and the complete yeast and Arabidopsis genome sequences. One can carry out homology and keyword searches for one particular species or all across the species in GIB. Therefore, for instance, GIB may enable one to simultaneously investigate the evolution of a set of functionally related genes both in prokaryote and eukaryote species. It may also help one with studying the relationships between the location of related genes and their functional aspects.
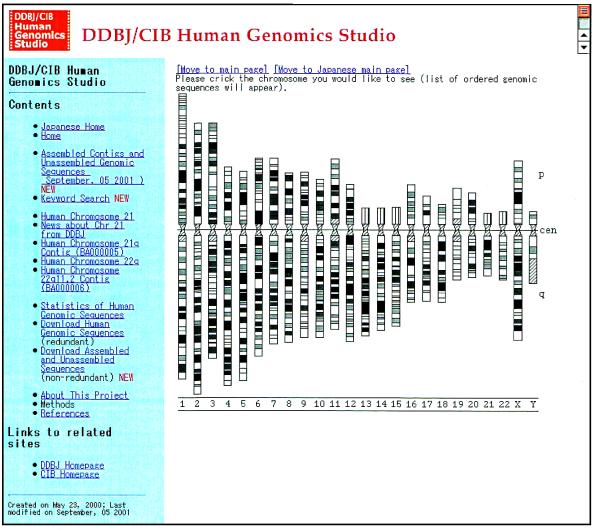
Another specialised database is the Human Genomics Studio (HGS, http://studio.nig.ac.jp). The home page of HGS is shown in Figure 1. We started developing HGS just before the whole human genome sequence data were published. There are mainly four aims of developing HGS: (i) to reconstruct the whole human chromosomes collecting and assembling all the available sequence data, (ii) to map all the available genes on the chromosomes, (iii) to compile a human gene catalog containing information about the function, relationships among genes, size, polymorphism and so forth, and (iv) to provide anyone with those data being as complete and updated as possible. We have provided part of the first aim through the above web site and are working on the rest.
Figure 1.
The homepage of the Human Genomics Studio.
CONCLUDING REMARKS
Genome sequencing has raised one problem despite having made a profound contribution to life science. It is noted first that a submitted genome sequence often includes possible genes, which were inferred by computer tools. These genes may be called in silico genes. The problem is that an in silico gene is determined by the algorithm and arbitrarily given parameters of the computer tool not by experiment.
Moreover, a different tool produces different genes for the same sequence. This creates the undesirable chain reaction that one user writes a paper mistakenly regarding in silico genes as real ones and then another follows citing this spurious paper without carefully examining it. There is, however, an international movement which would issue the recommendation that INSD should encourage a data submitter to distinguish in silico genes from experimentally obtained ones. If this recommendation is implemented, DDBJ will act on it.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the rest of the DDBJ members for making it possible to run well this international DNA data bank. We are also grateful to the Ministry of Education, Science, Culture, Sports and Technology for their enduring financial support.
REFERENCES
- 1.Fleischmann R.D., Adams,M.D., White,O., Clayton,R.A., Kirkness,E.F., Kerlavage,A.R., Bult,C.J., Tomb,J.F., Dougherty,B.A., Merrick,J.M. et al. (1995) Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science, 269, 496–512. [DOI] [PubMed] [Google Scholar]
- 2.Anderson S. (1981) Shotgun DNA sequencing using cloned DNase I-generated fragments. Nucleic Acids Res., 9, 3015–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pennisi E. (1998) Worming secrets from the C. elegans genome. Science, 282, 1972–1974. [DOI] [PubMed] [Google Scholar]
- 4.Adams M.D., Celniker,S.E., Holt,R.A., Evans,C.A., Gocayne,J.D., Amanatides,P.G., Scherer,S.E., Li,P.W., Hoskins,R.A., Galle,R.F. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 5.International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 6.MHC Sequencing Consortium (1999) Complete sequence and gene map of a human major histocompatibility complex. Nature, 401, 921–923. [DOI] [PubMed] [Google Scholar]
- 7.Dunham I., Shimizu,N., Roe,B.A., Chissoe,S., Hunt,A.R., Collins,J.E., Bruskiewich,R., Beare,D.M., Clamp,M., Smink,L.J. et al. (1999) The DNA sequence of human chromosome 22. Nature, 402, 489–495. [DOI] [PubMed] [Google Scholar]
- 8.Hattori M., Fujiyama,A., Taylor,T.D., Watanabe,H., Yada,T., Park,H.S., Toyoda,A., Ishii,K., Totoki,Y., Choi,D.K. et al. (2000) The DNA sequence of human chromosome 21. Nature, 405, 311–319. [DOI] [PubMed] [Google Scholar]
- 9.Tabata S., Kaneko,T., Nakamura,Y., Kotani,H., Kato,T., Asamizu,E., Miyajima,N., Sasamoto,S., Kimura,T., Hosouchi,T. et al. (2000) Sequence and analysis of chromosome 5 of the plant Arabidopsis thaliana. Nature, 408, 823–826. [DOI] [PubMed] [Google Scholar]
- 10.Shigenobu S., Watanabe,H., Hattori,M., Sakaki,Y. and Ishikawa,H. (2000) Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature, 407, 81–86. [DOI] [PubMed] [Google Scholar]
- 11.Nakasone K., Masui,N., Takaki,Y., Sasaki,R., Maeno,G., Sakiyama,T., Hirama,C., Fuji,F. and Takami,H. (2000) Characterization and comparative study of the rrn operons of alkaliphilic Bacillus halodurans C-125. Extremophiles, 4, 209–214. [DOI] [PubMed] [Google Scholar]
- 12.Shirai M., Hirakawa,H., Kimoto,M., Tabuchi,M., Kishi,F., Ouchi,K., Shiba,T., Ishii,K., Hattori,M., Kuhara,S. et al. (2000) Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res., 28, 2311–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaneko T., Nakamura,Y., Sato,S., Asamizu,E., Kato,T., Sasamoto,S., Watanabe,A., Idesawa,K., Ishikawa,A., Kawashima,K. et al. (2000) Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res., 7, 331–338. [DOI] [PubMed] [Google Scholar]
- 14.Kuroda M., Ohta,T., Uchiyama,I., Baba,T., Yuzawa,H., Kobayashi,I., Cui,L., Oguchi,A., Aoki,K., Nagai,Y. et al. (2001) Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet, 357, 1225–1240. [DOI] [PubMed] [Google Scholar]
- 15.Sugawara H., Miyazaki,S., Gojobori,T. and Tateno,Y. (1999) DNA Data Bank of Japan dealing with large-scale data submission. Nucleic Acids Res., 27, 25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawai J., Shinagawa,A., Shibata,K., Yoshino,M., Itoh,M., Ishii,Y., Arakawa,T., Hara,A., Fukunishi,Y., Konno,H. et al. (2001) Functional annotation of a full-length mouse cDNA collection. Nature, 409, 685–690. [DOI] [PubMed] [Google Scholar]
- 17.Tateno Y., Fukami-Kobayashi,K., Miyazaki,S., Sugawara,H. and Gojobori,T. (1998) DNA Data Bank of Japan at work on genome sequence data. Nucleic Acids Res., 26, 16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]