Abstract
The PRINTS database houses a collection of protein fingerprints. These may be used to make family and tentative functional assignments for uncharacterised sequences. The September 2001 release (version 32.0) includes 1600 fingerprints, encoding ∼10 000 motifs, covering a range of globular and membrane proteins, modular polypeptides and so on. In addition to its continued steady growth, we report here its use as a source of annotation in the InterPro resource, and the use of its relational cousin, PRINTS-S, to model relationships between families, including those beyond the reach of conventional sequence analysis approaches. The database is accessible for BLAST, fingerprint and text searches at http://www.bioinf.man.ac.uk/dbbrowser/PRINTS/.
INTRODUCTION
Fingerprints are groups of motifs observed in sequence alignments; taken together, the motifs characterise the aligned family and hence provide a specific diagnostic signature. Fingerprints thus derive much of their potency from the biological context afforded by matching multiple motifs; this makes them at once more flexible and more powerful than single-motif approaches. The technique further departs from other pattern-matching methods by readily allowing the creation of discriminators at super-family-, family- and sub-family-specific levels. Such a hierarchical approach has been used, for example, to resolve G protein-coupled receptor (GPCR) super-families into their constituent families and receptor sub-types (1), and to classify a variety of channel proteins, transporters and enzymes.
To date, 1600 fingerprints have been developed, manually annotated and deposited in the PRINTS database (2). Overall, the database is still relatively small, largely because the detailed annotation of entries is extremely time-consuming. However, the extent of manually-crafted annotations sets the database apart from the growing number of automatically derived ‘family’ resources, for which there is no biological documentation and no result validation, and in which family groupings may change between database releases.
PRINTS was originally built as a single ASCII (text) file. However, with the continued growth of the database, maintenance was becoming inefficient and error-prone. We therefore migrated the resource to the PostgreSQL relational database management system (DBMS) in order to both address the maintenance issues and allow more complex queries of the underlying data (3). Here we describe recent progress and new developments with the database, and highlight the power of relational PRINTS (PRINTS-S) in exploring evolutionary relationships that reach beyond the limits of sequence similarity search tools.
SOURCE DATABASE AND SEARCH TOOLS
PRINTS is released in major and minor versions: minor releases reflect updates, bringing the contents in line with the current version of the source database [a SWISS-PROT/TrEMBL composite (4)]; major releases denote the addition of new material to the resource. Major releases are made quarterly, each release including 50 new annotated families. Eight major releases have been made since the last report.
The two main tools available for searching PRINTS are: (i) a BLAST (5) server, which allows similarity searches against ‘sequences’ matched in the current version of the database (6); and (ii) the FingerPRINTScan suite (7), which allows sequence searches against ‘fingerprints’ contained in the current release. FingerPRINTScan, now used within the EDITtoTrEMBL (8) package as part of the EBI’s automatic protocol to annotate TrEMBL, is a powerful diagnostic tool, affording greater specificity than the BLAST implementation (6).
A particular strength of PRINTS is that its motifs are stored in the form of un-gapped, local sequence alignments. This allows different implementations to be established with alternative scoring methods. Thus, a Blocks-format version of the resource that exploits Blocks scoring methods is available at the Fred Hutchinson Cancer Research Center (9). In addition, the EMOTIF database at Stanford overlays a permissive regular expression approach over PRINTS’ multiply-aligned motifs, offering different levels of stringency from which to infer the significance of matches (10).
RELATIONAL PRINTS
One of the most important developments of the database has been its migration to a relational DBMS. This ‘streamlined’ version of the database, PRINTS-S (3), reduces redundancy, maintains consistency and facilitates routine maintenance. It also permits more complex queries, and allows us to support both new display and flat-file formats. PRINTS-S is accessible for interactive use via the Web. The interface allows both strict keyword searching and more powerful queries using a combination of regular expressions and logical operators.
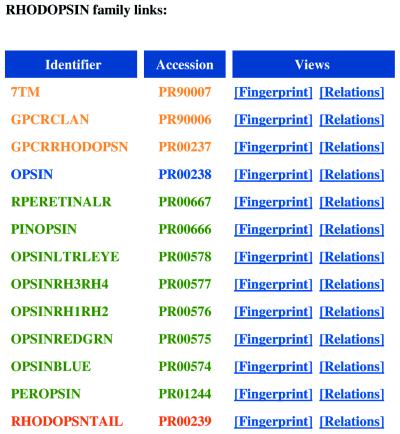
A valuable attribute of PRINTS-S is the ability to model relationships explicitly by defining parent–child and sibling relations within, and implied by, the PRINTS family hierarchy (Fig. 1). This means, for example, that members of a clan (a group of families for which there are indications of an evolutionary relationship, but between which there is no statistically significant similarity in sequence) can nevertheless be linked. Thus, it is possible to transcend relationships evident at the sequence level and gain structural insights from a realm beyond the theoretical limits of conventional sequence analysis tools [this is the so-called ‘Midnight Zone’, the region of identity where sequence comparisons fail completely to detect structural similarities (11)].
Figure 1.
The rhodopsin family hierarchy depicted by PRINTS-S. The different levels of the hierarchy are colour-coded as follows: red, children; green, siblings; purple, parents; brown, grandparents (or great-grandparents).
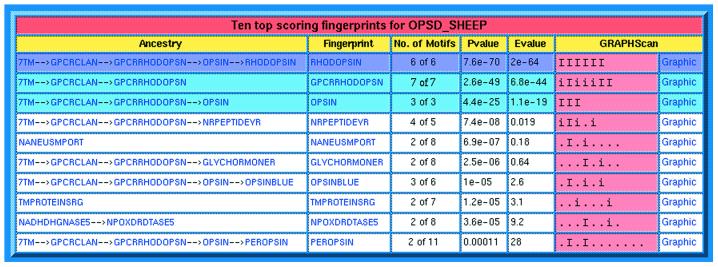
As an illustration, consider the relationships encoded in the database for the rhodopsin-like GPCRs shown in Figure 1. The FingerPRINTScan suite has been modified to exploit these relationships in such a way that when we search the database with a query sequence, all child/sibling/parent/grandparent relations between matched fingerprints are revealed. For example, take the result of searching with the sequence of ovine rhodopsin, shown in Figure 2. For each matched fingerprint (RHODOPSIN, GPCRRHODOPSN and OPSIN are the only matches with significant E-values highlighted in the table), the relationships between them are traced back through the family hierarchy to the most remote putative ancestor. Thus, we see that RHODOPSIN is a child of the OPSIN family, which is a child of GPCRRHODOPSN (the rhodopsin-like GPCR super-family), whose parent is the GPCR clan (which includes the secretin-like receptors, metabotropic receptors, etc.), which is derived from a putative ‘7TM’ architectural ancestor. Such an ‘ancestral perspective’ is only possible because PRINTS-S models the biological associations between families within an internal relational structure, allowing a hierarchical representation of connections between database entries, including those outside the realm of sequence similarity searches.
Figure 2.
Result of searching PRINTS-S with the sequence of ovine rhodopsin using FingerPRINTScan. The table shows the top 10 matches (significant matches are highlighted, the best is coloured purple), and traces the relationships between each matched fingerprint from its position in the familial hierarchy back to its most distant ancestor. Here, each rhodopsin-like GPCR match can be traced back through its parent super-family, through the ancestral GPCR clan, ultimately to a presumed ‘7TM’ architectural predecessor.
RELATED DATABASE DEVELOPMENTS
Another landmark in the evolution of PRINTS has been the international collaborative project to integrate the database with PROSITE (12), profiles (12), Pfam (13) and ProDom (14) within a unified protein family annotation resource. This initiative, known as InterPro (15), which primarily exploits the detailed family annotations provided by PROSITE and PRINTS, aims both to reduce duplication of effort in the laborious, bottle-necking process of annotation, and to facilitate communication between disparate resources. InterPro provides a convenient one-stop shop for the analysis of newly determined sequences and has already found application in the analysis of the Drosophila and human genomes.
A more recent development is a pilot project to provide an automatic supplement to PRINTS, termed prePRINTS. This exploits an automatic pipeline for sequence alignment, motif detection, iterative database searching and annotation. Interactive versions of parts of the pipeline are also being developed to: (i) allow users to create their own fingerprints for use in conjunction with FingerPRINTScan; and (ii) generate annotation for groups of user-specified sequences. This is PRECIS [Protein Reports Engineered from Concise Information in SWISS-PROT (16); http://bioinf.man.ac.uk/cgi-bin/dbbrowser/precis/precis.cgi]. prePRINTS and its associated tools will ultimately help to increase the family coverage of PRINTS and so improve its effectiveness as a sequence analysis tool.
AVAILABILITY
For local installation, PRINTS flat-files may be retrieved from the anonymous FTP servers at Manchester (ftp://ftp.bioinf.man.ac.uk/pub/prints), HGMP-RC (ftp://ftp.hgmp.mrc.ac.uk/pub/database/prints), EBI (ftp://ftp.ebi.ac.uk/pub/databases), EMBL (ftp://ftp.embl-heidelberg.de) and NCBI (ftp://ncbi.nlm.nih.gov).
CONCLUSIONS
PRINTS is an evolving resource and the new developments help to increase its utility as a tool for protein sequence analysis and genome annotation. In addition, PRINTS-S sheds light on evolutionary relationships between families that were formerly hidden in PRINTS. Together, PRINTS and PRINTS-S are thus complementary tools that facilitate genome annotation, and add greater depth to sequence analyses by offering new ancestral perspectives on protein family relationships.
Acknowledgments
ACKNOWLEDGEMENTS
PRINTS is built and maintained at the University of Manchester with support from the Royal Society (T.K.A. is a Royal Society University Research Fellow). We are grateful for individual support from the MRC (A.G.), the BBSRC (J.E.M. and A.L.M.), the EPSRC (N.M.), Cambridge Drug Discovery (G.M.) and Roche Discovery Welwyn (P.S.).
REFERENCES
- 1.Attwood T.K. (2001) A compendium of specific motifs for diagnosing GPCR subtypes. Trends Pharmacol. Sci., 22, 162–165. [DOI] [PubMed] [Google Scholar]
- 2.Attwood T.K., Flower,D.R., Lewis,A.P., Mabey,J.E., Morgan,S.R., Scordis,P., Selley,J.N. and Wright,W. (1999) PRINTS prepares for the new millennium. Nucleic Acids Res., 27, 220–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Attwood T.K., Croning,M.D.R., Flower,D.R., Lewis,A.P., Mabey,J.E., Scordis,P., Selley,J. and Wright,W. (2000) PRINTS-S: the database formerly known as PRINTS. Nucleic Acids Res., 28, 225–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 6.Wright W., Scordis,P. and Attwood,T.K. (1999) BLAST PRINTS – an alternative perspective on sequence similarity. Bioinformatics, 15, 523–524. [DOI] [PubMed] [Google Scholar]
- 7.Scordis P., Flower,D.R. and Attwood,T.K. (1999) FingerPRINTScan: intelligent searching of the PRINTS motif database. Bioinformatics, 15, 799–806. [DOI] [PubMed] [Google Scholar]
- 8.Moeller S., Leser,U., Fleischmann,W. and Apweiler,R. (1999) EDITtoTrEMBL: a distributed approach to high-quality automated protein sequence annotation. Bioinformatics, 15, 219–227. [DOI] [PubMed] [Google Scholar]
- 9.Henikoff J. Greene,E.A., Pietrokovski,S. and Henikoff,S. (2000) Increased coverage of protein families with the Blocks Database servers. Nucleic Acids Res., 28, 228–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang J.Y. and Brutlag,D.L. (2001) The EMOTIF database. Nucleic Acids Res., 29, 202–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rost B. (1998) Marrying structure and genomics. Structure, 6, 259–263. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann K., Bucher,P., Falquet,L. and Bairoch,A. (1999) The PROSITE database, its status in 1999. Nucleic Acids Res., 27, 215–219. Updated article in this issue: Nucleic Acids Res. (2002), 30, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Howe,K.L. and Sonnhammer,E.L.L. (2000) The Pfam Protein Families Database. Nucleic Acids Res., 28, 263–266. Updated article in this issue: Nucleic Acids Res. (2002), 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corpet F., Servant,F., Gouzy,J. and Kahn,D. (2000) ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res., 28, 267–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ApweilerR., Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,L, Corpet,F., Croning,M.D.R. et al. (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res., 29, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reich J.R., Mitchell,A., Goble,C.A. and Attwood,T.K. (2001) PRECIS: Protein Reports Engineered from Concise Information in SWISS-PROT. IEEE Intelligent Systems, in press. [DOI] [PubMed] [Google Scholar]