Abstract
A large collection of T-DNA insertion transformants of Arabidopsis thaliana has been generated at the Institute of Agronomic Research, Versailles, France. The molecular characterisation of the insertion sites is currently performed by sequencing genomic regions flanking the inserted T-DNA (FST). The almost complete sequence of the nuclear genome of A.thaliana provides the framework for organising FSTs in a genome oriented database, FLAGdb/FST (http://genoplante-info.infobiogen.fr). The main scope of FLAGdb/FST is to help biologists to find the FSTs that interrupt the genes in which they are interested. FSTs are anchored to the genome sequences of A.thaliana and positions of both predicted genes and FSTs are shown graphically on sequences. Requests to locate the genomic position of a query sequence are made using BLAST programs. The response delivered by FLAGdb/FST is a graphical representation of the putative FSTs and of predicted genes in a 20 kb region.
INTRODUCTION
Gene disruption is a powerful tool to discover and analyse gene functions, particularly in organisms for which the complete genome sequence is available. However, in plants, homologous recombination is an inefficient process, preventing large-scale targeted gene disruption (1). Therefore, collections of T-DNA (2–7) or transposon insertion (8–13) mutants are useful alternatives as a first step in functional genomics. A large collection of 60 000 T-DNA insertion transformants of Arabidopsis thaliana plants, ecotype Wassilevskija, has been generated, at the Institut National de la Recherche Agronomique (INRA), Versailles, by vacuum filtration of an Agrobacterium tumefaciens strain containing the pGKB5 vector (14). T-DNA lines contain an average of 1.5 insertions per line (4). Apparently, T-DNA integrates randomly in the Arabidopsis genome (15) and therefore most genes can be disrupted using this method. T-DNA insertion into a given gene may be searched for in the collection by PCR screening (16,17) starting with DNA extracted from organised pools of seedlings and ending with individual plants. This successful strategy (18,19) is now complemented by a project undertaking an exhaustive characterisation of the Versailles transformant collection. The molecular characterisation of T-DNA insertion sites (FSTs) is currently underway by PCR-walking and sequencing (20,21) and the genome sequence of this organism (22) provides the framework for organising the FSTs. Arabidopsis thaliana is one of the model genomes for plants, and the Arabidopsis Genome Initiative (AGI; 22) published the almost complete sequence of the 125 Mb of its five chromosomes. The nuclear genome of A.thaliana contains approximately 25 500 predicted genes, which have been annotated by the AGI (22). The FST project intends to produce 30 000 FSTs before next year, which will provide a significant number of disrupted genes and regulatory regions.
The main goal of FLAGdb/FST is to help biologists find the FSTs that interrupt genes in which they are interested.
FLAGdb/FST CONTENT
FLAGdb/FST contains approximately 6000 FSTs as of September 2001. Each is long enough to be safely mapped. The average size of the FSTs is 250 bp. Approximately 300 novel FSTs are being produced per week. They are generated automatically from raw sequencing data files and then curated by human experts. Approximately 85% of the obtained sequences are considered to be useful FSTs and recorded in FLAGdb/FST. Discarded sequences are either of poor quality or do not contain the expected residual T-DNA border and may therefore exist from non-specific PCR amplifications. The T-DNA insertion lines of the Versailles collection carry an average of 1.5 insertions (4). In a line with more than one insertion, only the T-DNA insertion giving the smallest PCR product is sequenced and therefore only one FST is obtained per T-DNA line (21). Only in a few cases has an independent laboratory informed us of their inability to recover the T-DNA border tagged by an FST in our laboratory. FLAGdb/FST uses the up-to-date nuclear genome sequences of A.thaliana and their gene predictions, retrieved frequently from TAIR (23). A future version will soon use the pseudo chromosome sequences generated by the AGI.
DATABASE IMPLEMENTATION
FLAGdb/FST has been developed in the Relational DataBase Management System ORACLE v8.i. In order to manage sequence information of the A.thaliana genome, a conceptual model has been adapted from a model previously constructed for a genome-oriented database dedicated to micro-organisms, MICADO (24). The interface of FLAGdb/FST has been set up on a World Wide Web server (http://genoplante-info.infobiogen.fr) allowing Internet access with a Web client.
SEARCHING FLAGdb/FST
In FLAGdb/FST the FSTs are anchored to the genome sequences of A.thaliana and positions of both predicted genes and FSTs are shown graphically on sequences. Requests are made using BLAST programs (25) to locate the genomic position of a query sequence. Included BLAST programs are the standard nucleotide–nucleotide BLASTn, the standard protein–protein BLASTp and the nucleotide query against translated DB tBLASTx. A 1 kb query is most often sufficient to unequivocally locate the right locus, and queries with ESTs or cDNAs are generally successful. The following summarizes the response delivered by FLAGdb/FST.
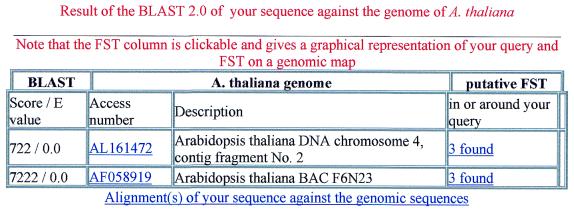
1. A list (Fig. 1) points to loci with identity or high similarity to the query sequence.
Figure 1.
Locating a query sequence within the genome sequences. Users search the database with a query sequence and a BLAST program that locates its genomic position. In this example, the query sequence is cognate to the same genome sequence present in two different accessions. Activating the link in the FST column gives access to the graphical display of the putative FSTs and to the predicted genes in a 20 kb region around the BLAST hit as shown in Figure 2. The name given to the query is automatically loaded and appears in the title. Other links lead to the BLAST output and the GenBank file corresponding to the accession number.
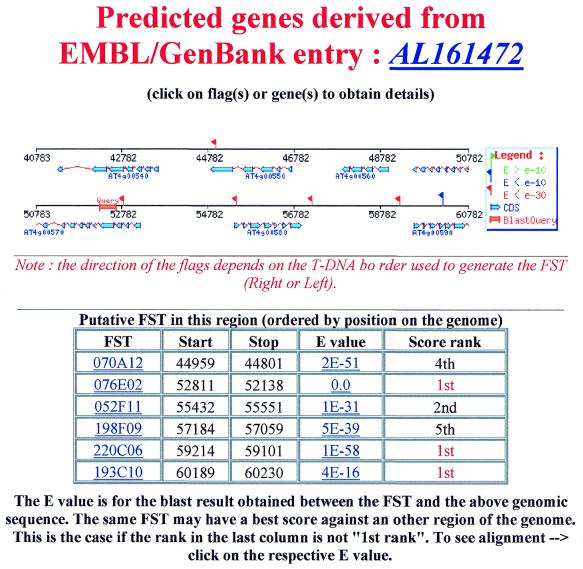
2. Users select a region in the list of loci to access a graphical representation of the putative FSTs and of predicted genes in 20 kb surounding the locus of the query sequence (Fig. 2). A colour code for BLAST e-values gives a rough estimation of the quality of alignments. In order to help further experimental validation, the location of the FST with respect to the right or the left border of the T-DNA is indicated by the direction of the flags.
Figure 2.
Graphical display of the FSTs and predicted genes in a 20 kb genomic region around the query sequence location. In the score rank column, ‘1st’ indicates that the query sequence is cognate to the referred location; lower ranks point to similar regions. When the query sequence is perfectly repeated in the genome, such conflicts cannot be sorted by the database. FSTs that are in highly repeated regions are filtered out. Activating the link in the FST column gives access to the FST page containing the FST sequence, its features and the accession number of the corresponding T-DNA line.
3. Flags are links to the FST files. Besides the sequence, the FST file contains the length of the sequence, its base composition, the side of the T-DNA from which the FST has been sequenced (right or left border) and whether or not the adaptator, ligated to the genomic DNA for PCR walking, has been found in the sequence. Furthermore, the FST file contains links to the BLAST outputs for alignments of the query sequence against all the A.thaliana DNA, EST and protein sequences.
4. The graphical display of the intron–exon structure of genes is a link to a file giving the precise limits of the predicted exons, allowing a rapid and exact location of the T-DNA insertion.
5. A description of the procedure for obtaining seeds from the Versailles collection is available by a click on the ‘Ask for line’ button.
Notable advantages of data representation in FLAGdb/FST include the following.
1. Mapping of FSTs is completely independent of the gene predictions.
2. T-DNA insertions in gene promoters and intergenic regions that may have an effect on gene expression are also indicated.
3. Results are given not only for the genome region cognate to the query sequence but also for related loci. Therefore, FLAGdb is efficient not only for genes within families of duplicated genes, frequently occurring in the genome of A.thaliana, but also when searches are performed with query sequences from species other than A.thaliana.
FUTURE DEVELOPMENTS
A Java version of the interface is in preparation, together with a visualisation of FST locations within the chromosomes. The database will be implemented progressively to give access to observations of phenotypes of the transformants.
CITING FLAGdb/FST
Please refer to this article when citing FLAGdb/FST.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the staff of Génoplante-Info and Infobiogen for their help and cooperation in maintaining FLAGdb/FST on their site. This work was supported by the French Génoplante Program.
REFERENCES
- 1.Kempin S., Liljegren,S.J., Block,L.M., Rounsley,S.D. and Yanofsky,M.F. (1997) Targeted disruption in Arabidopsis. Nature, 389, 802–803. [DOI] [PubMed] [Google Scholar]
- 2.Feldmann K.A. (1991) T-DNA insertion mutagenesis in Arabidopsis: mutational spectrum. Plant J., 1, 19–22. [Google Scholar]
- 3.Koncz C., Nemeth,K., Redei,G.P. and Schell,J. (1992) T-DNA insertional mutagenesis in Arabidopsis. Plant Mol. Biol., 20, 963–976. [DOI] [PubMed] [Google Scholar]
- 4.Bechtold N., Ellis,J. and Pelletier,G. (1993) In planta Agrobacterium mediated gene transfer by filtration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. (Paris), 316, 1194–1199. [Google Scholar]
- 5.Krysan P.J., Young,J.C. and Sussman,M.R. (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell, 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeon J.-S., Lee,S., Jung,K.H., Jun,S.H., Jeong,D.H., Lee,J., Kim,C., Jang,S., Yang,K., Nam,J. et al. (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J., 22, 561–570. [DOI] [PubMed] [Google Scholar]
- 7.Zhao Z.Y., Cai,T., Miller,M., Wang,N., Pang,H., Rudert,M., Schroeder,S., Hondred,D., Seltzer,J. and Pierce,D. (2000) Agrobacterium-mediated sorghum transformation. Plant Mol. Biol., 44, 789–798. [DOI] [PubMed] [Google Scholar]
- 8.Dubois P., Cutler,S. and Belzile,F.J. (1998) Regional insertional mutagenesis on chromosome III of Arabidopsis thaliana using the maize Ac element. Plant J., 13, 141–151. [DOI] [PubMed] [Google Scholar]
- 9.Martienssen R.A. (1998) Functional genomics: probing plant gene function and expression with transposons. Proc. Natl Acad. Sci. USA, 95, 2021–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wisman E., Hartmann,U., Sagasser,M., Baumann,E., Palme,K., Hahlbrock,K., Saedler,H. and Weisshaar,B. (1998) Knock-out mutants from an En-1 mutagenized Arabidopsis thaliana population generate phenylpropanoid biosynthesis phenotypes. Proc. Natl Acad. Sci. USA, 95, 12432–12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parinov S., Sevugan,M., Ye,D., Yang,W.-C., Kumaran,M. and Sundaresan,V. (1999) Analysis of flanking sequences from Dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell, 11, 2263–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Speulman E., Metz,P.L.J., van Arkel,G., Lintel Hekkert,B., Stiekema,W.J. and Pereira,A. (1999) A two-component Enhancer–Inhibitor transposon mutagenesis system for functional analysis of the Arabidopsis genome. Plant Cell, 11, 1853–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tissier A.F., Marillonnet,S., Klimyuk,V., Patel,K., Torres,M.A., Murphy,G. and Jones,J.D.G. (1999) Multiple independent defective Suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell, 11, 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouchez D., Camilleri,C. and Caboche,M. (1993) A binary vector based on Basta resistance in planta transformation of Arabidopsis thaliana. C. R. Acad. Sci. Ser. III Sci. Vie, 316, 1188–1193. [Google Scholar]
- 15.Barakat A., Gallois,P., Raynal,M., Mestre-Ortega,D., Sallaud,C., Guiderdoni,E., Delseny,M. and Bernardi,G. (2000) The distribution of T-DNA in the genomes of transgenic Arabidopsis and rice. FEBS Lett., 471, 161–164. [DOI] [PubMed] [Google Scholar]
- 16.Bouchez D. and Hofte,H. (1998) Functional genomics in plants. Plant Physiol., 118, 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winkler R.G. and Feldmann,K.A. (1998) PCR-based identification of T-DNA insertion mutants. Methods Mol. Biol., 82, 129–136. [DOI] [PubMed] [Google Scholar]
- 18.Winkler R.G., Frank,M.R., Galbraith,D.W., Feyereisen,R. and Feldmann,K.A. (1998) Systematic reverse genetics of transfer-DNA-tagged lines of Arabidopsis. Plant Physiol., 118, 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meissner R.C., Jin,H., Cominelli,E., Denekamp,M., Fuertes,A., Greco,R., Kranz,H.D., Penfield,S., Petroni,K., Urzainqui,A. et al. (1999) Function search in a large transcription factor gene family in Arabidopsis: assessing the potential of reverse genetics to identify insertional mutations in R2R3 MYB genes. Plant Cell, 11, 1827–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Devic M., Albert,S., Delseny,M. and Roscoe,T. (1997) Efficient PCR walking on plant genomic DNA. Plant Physiol. Biochem., 35, 331–339. [Google Scholar]
- 21.Balzergue S., Dubreucq,B., Chauvin,S., Le-Clainche,I., Le Boulaire,F., deRose,R., Samson,F., Biaudet,V., Lecharny,A., Cruaud,C. et al. (2001) Improved PCR-walking for large-scale isolation of plant T-DNA borders. Biotechniques, 30, 496–504. [DOI] [PubMed] [Google Scholar]
- 22.The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana.Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 23.Huala E., Dickerman,A.W., Garcia-Hernandez,M., Weems,D., Reiser,L., LaFond,F., Hanley,D., Kiphart,D., Zhuang,M., Huang,W. et al. (2001) The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res., 29, 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biaudet V., Samson,F. and Bessières,Ph. (1997) Micado—a network-oriented database for microbial genomes. Comp. Appl. Biosci., 13, 431–438. [DOI] [PubMed] [Google Scholar]
- 25.Altschul S.F., Madden,T.I., Schaffer,A.A., Zhang,J., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]