Abstract
euGenes is a genome information system and database that provides a common summary of eukaryote genes and genomes, at http://iubio.bio.indiana.edu/eugenes/. Seven popular genomes are included: human, mouse, fruitfly, Caenorhabditis elegans worm, Saccharomyces yeast, Arabidopsis mustard weed and zebrafish, with more planned. This information, automatically extracted and updated from several source databases, offers features not readily available through other genome databases to bioscientists looking for gene relationships across organisms. The database describes 150 000 known, predicted and orphan genes, using consistent gene names along with their homologies and associations with a standard vocabulary of molecular functions, cell locations and biological processes. Usable whole-genome maps including features, chromosome locations and molecular data integration are available, as are options to retrieve sequences from these genomes. Search and retrieval methods for these data are easy to use and efficient, allowing one to ask combined questions of sequence features, protein functions and other gene attributes, and fetch results in reports, computable tabular outputs or bulk database forms. These summarized data are useful for integration in other projects, such as gene expression databases. euGenes provides an extensible, flexible genome information system for many organisms.
INTRODUCTION
The euGenes genome information service and database are useful to bioscientists and students looking for gene and genome relationships across several eukaryote species. This is a summary of genome information of human, fruitfly, Caenorhabditis elegans worm, mouse, Saccharomyces yeast, Arabidopsis mustard weed, zebrafish, with rice genome data in the planning stages. Primary information on all known and inferred protein coding genes of these genomes is included, with whole chromosome DNA and feature annotations. There are flexible, usable, whole-genome map displays with feature annotations, chromosome locations and molecular data integration. Consistent gene symbols, identifiers and synonyms are used for the known, predicted and orphan coding genes from these organisms. Gene homologies are calculated using BLAST among these genomes. Standard vocabulary of molecular function, biological process and cell location is integrated into data searching and reporting.
The informatics underpinnings of this information service provide users with quick, efficient search, retrieval and display methods that work for any web browser. Current information is automatically drawn from several public sources. Integration of diverse data into a common format, suitable for use with other projects such as gene expression research, makes this database useful to more than web browsers. Extension of FlyBase (1) genome database technology to several genomes provides the mechanics for this. Genome data can be viewed graphically, by chromosome location and through searches of other gene information. Genome sequence can be extracted with any set of feature annotations, in a variety of formats. Gene region maps included in gene reports show the structure of the gene and neighboring sequence region. Many users find that the database compares favorably to other genome information systems in terms of content, comprehensiveness and usability.
As with any derived data set, the euGenes summarization doesn’t improve source data sets. It seeks to organize and integrate somewhat diverse sources into a common structure, with a common interface to search and retrieve, or to extract for further use in projects needing eukaryote genome data. euGenes started as a pilot project in 1999, arising from recommendations of the NIH Model Organism Database Workshop (http://iubio.bio.indiana.edu/eugenes/docs/). In July 2000, this project became public, and usage has grown rapidly, as bioscientists in commercial biotechnology, academic and government institutions learn of its utility. The service is built from many of the same component parts used in the successful FlyBase genome information service for Drosophila, with a goal of extending these methods to any group of organisms with genome information.
Gene searching and reporting
The summary information available, of the order of 150 000 genes of human and model eukaryote organisms, provides essential data and links to fuller information from source databases. euGenes is a good entry point for one who doesn’t need all details of an organism, or who has expertise with one organism and wants to find gene data on related organisms. It is designed for ease of use and understanding. Each organism has a section in euGenes, listing source data and the derived and computed summaries, genome features and homologies. Computations and tables of comparison across all species are also provided.
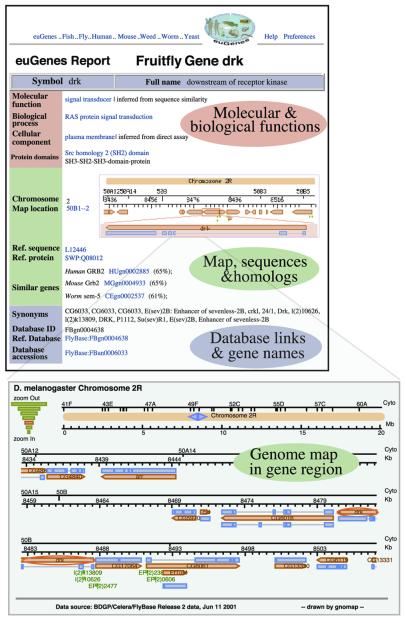
Using the simple web forms, one can find, for instance, hundreds of worm genes involved in signal transduction that also show homology to fruitfly genes, or just the few which show homology to fruitfly, human, mouse and yeast genes. In those gene reports, the function, cell location and process of the genes are listed, both experimentally determined or estimated from computation. If known, the location on chromosomes is provided as well as a genome map of the region, and access to the DNA sequence from this gene and region. Homologous genes computed from the euGenes reference protein set are also listed, giving access to related information from other species. Known gene synonyms, database identifiers and other summary information are provided. Figure 1 shows these main contents of a summary gene report, for the fruitfly gene drk, along with the genome map of this gene’s region on chromosome arm 2L.
Figure 1.
euGenes gene report and genome map. Summary gene information for the fruitfly gene drk, with function, chromosome location and gene region map, homologous genes and related database links.
The results of this and other searches are available not just as hypertext documents, but in spreadsheet table formats for your data manipulations, and database formats including XML. Whether a handful or thousands of genes match your question, the information retrieval system allows rapid navigation through all results, and an option to refine your query by adding more restrictions, if the initial question yields more results than desired.
Source genome data
Genome data is continuously extracted from current public sources, including Drosophila melanogaster from FlyBase (1), C.elegans from WormBase (2), Homo sapiens from LocusLink and RefSeq (3) and UCSC Golden Path (4), mouse from Mouse Genome Database (MGD; 5), Saccharomyces yeast from Saccharomyces Genome Database (SGD; 6), zebrafish from Zebrafish Information Network (ZFIN; 7), Arabidopsis thaliana from the TIGR ATH database (8; http://www.tigr.org/tdb/e2k1/ath1/) and The Arabidopsis Information Resource (TAIR; 9). Gene classifications by biological process, molecular function and cell location are provided from the Gene Ontology consortium (10), and sequence data sources include GenBank, SWISS-PROT, TrEMBL and PIR databanks. The source data are continually improving and changing; annotation of these genomes will continue for years. Over the time span of this project, several sources have been added or removed, as data changed or became out-of-date. Improvements and additions from sources are added as they become available. The results reported here are from a July 2001 snapshot of the database.
Table 1 lists overall genome attribute counts gathered from these sources. The genes reported include experimental, predicted as well as orphan genes. Especially for fruitfly reported genes, many from older literature will never be located on genome sequence. The percent of these genes located on genome sequence is zero for mouse and zebrafish as their whole genome sequence is not yet available. Of those genes with a reference protein for homology calculations, the percent showing homology to one or more other genes is indicated. For mouse and zebrafish especially, this percent does not include genes from a full genome set, leading to a higher apparent overall homology. The percent of reported genes with a gene ontology (GO) classification is given in this table. For those with whole genome sequence, the total kilobase count of the genomes and number of features annotated on these genomes and extracted for use in euGenes is listed.
Table 1. Genome attribute counts in euGenes, July 2001.
| Genes reported | Located (%) | Homology (%) | GO data (%) | Genome size (kb) | Genome features | |
|---|---|---|---|---|---|---|
| Fruitfly | 23 649 | 56 | 44 | 31 | 116 094 | 41 570 |
| Human | 37 049 | 66 | 76 | 0 | 3 310 005 | 1 575 667 |
| Mouse | 28 210 | 88 | 20 | |||
| Weed | 26 819 | 100 | 18 | 14 | 116 702 | 54 053 |
| Worm | 21 881 | 100 | 27 | 27 | 100 090 | 207 478 |
| Yeast | 7226 | 90 | 30 | 88 | 12 155 | 13 594 |
| Zebrafish | 1221 | 87 | 0 |
Data management
euGenes uses automated data collection, rather than curation by people, not quite like Googlebot or Alta-Vista Web robots, but using bioinformatics savvy to choose among the best genome data sets. This is done like several common genome and sequence databases that extract data from other sources. As data is collected through the Internet from sources, it requires checking and validation of gene symbols, identifiers and other values. Data indexing is done to permit searching by fields and words. Identifiers are assigned, where possible using ones compatible with source data, and data values are crosschecked for consistency. The product is a ‘read-only’ database, stored primarily in key-value text object records, though future versions may move to relational database software. Though this final product amounts to ~5 GB of storage, more than half of which is human DNA sequence, >80 GB of source data is needed to create and maintain it. This database is maintained with a collection of data management tools, including standard public software components and some designed for use with euGenes and FlyBase, which are ‘glued’ together with Perl programs. The term genome information system is used to describe this, though database also applies. As an information system, its focus is on efficient data search and retrieval, where methods of data management are selected from those best suited to various kinds of data. This project aspires to eventually provide a similar product to AceDB (http://www.acedb.org/): a database in common terms, as well as a set of software that can be applied to other genome data sets.
Maps and sequences
For those organisms with whole genome sequence (Table 1), the genome sequence and features, or annotations, are accessible as display web maps with hyperlinks to detailed reports, and as annotated sequence in a variety of standard biosequence formats. Figure 1 shows the map of a gene region, which links to a larger genome map. The maps can be traversed by location, can be set to show large or small regions of chromosomes, and the set of features to display is under one’s control. Features include genes, mRNA, tRNA and other RNAs, cDNA, CDS, repeat regions, gaps, insertions and other annotations as provided from source databanks.
Gene associations
GO classifications are managed with a hierarchical, object-oriented gene and vocabulary database built for use with this project and with FlyBase. This classification database provides the ability for one to find not just those genes that have been assigned a particular function, but also all genes that have functions in sub-categories of an overall classification—such as all enzymes, or all cell cycle regulators. For instance, 27 weed genes are classified as cell cycle regulators, but 26 of these are specifically cyclin genes. This function of euGenes uniquely allows one to combine searching for GO classifications with other gene attributes.
DATA ACCESS
Human interface
Web functions for search, retrieval, map display and summaries of genome data are found at http://iubio.bio.indiana.edu/eugenes/. Combined queries with gene ontology, homologies, features and other attributes are feasible. Public usage has risen from <1% of the FlyBase system in late 2000 to 5% in early 2001. Many people use this system on a daily basis. These include bioscientists from all parts of the globe, in academia, government, and biotechnology and pharmaceutical industries. The genome maps are particularly popular, presumably due to their ease of use and functionality.
Reference database links are included for source data. Cross-links to euGenes from WormBase (2) and GeneCards (11) are currently available. Web logs show frequent and returning use by those referred from these services, an indication that this enhances WormBase and GeneCards services. Single-organism databases can offer benefits to their clients with the addition of cross-links to this multi-organism summary information.
Automation and distribution
One goal is to offer easier mechanisms to extract subsets of information for use in other services and databases. Query and retrieval by automated means can be done now, to extract subsets of these basic gene data for use in other databases, spreadsheets and other data analyses. Internet file transfer of the full database is available at ftp://iubio.bio.indiana.edu/eugenes/. The Sequence Retrieval System (SRS; 12) search engine is used in this system, and this multi-organism genome data set can be incorporated within other SRS-federated data systems. Plans are underway to distribute euGenes to other sites around the world. The software to produce and maintain euGenes data is available free for academic use, though copyrighted. Commercial interests need to license its use. Much of the software is written in Perl, with use of bioinformatics tools such as SRS, BLAST and Unix tools including MySQL and Berkeley DB. The Java-based programs (gnomap and Readseq; D. Gilbert, unpublished data) that produce genome maps and extract annotated sequence from genomes are component tools which can be used in other contexts.
RELATED PROJECTS
euGenes is not a unique source for multi-eukaryote genome summary information. This is an active area of development among public and industrial bioinformatics groups. LocusLink (3) at NCBI, integrated with RefSeq, Entrez and other NCBI managed data, is the closest to euGenes in the summary of gene data it provides for five organisms. These two services provide the research community with useful choices in access and organization of the same important data. The Ensembl project (http://www.ensembl.org) has potential and goals that are similar, though it is focused more on annotation of the human and mouse genome at the sequence level. euGenes has more emphasis on supra-sequence and multiple organism data integration, rather than detailed evidence of genome annotation. TIGR (http://www.tigr.org/tdb/) provides several eukaryotic and microbial genome data services, such as Gene Indices that provide multi-genome analyses at the sequence level. A publicly distributed genome annotation system (DAS; http://biodas.org) is under development among project members from WormBase (2), Ensembl, UCSC Human Genome Project (4), TIGR and FlyBase (1). DAS will offer reference genome annotation, with Internet client and server software, to provide bioscientists ready access to current genome maps, sequence and annotations. Proprietary genome systems from Celera, DoubleTwist, Proteome and others offer similar and distinct views of summary genome information. These choices have values and drawbacks compared to public information systems, but fill an important role for understanding genomes. The Proteome company selection of yeast, worm and human databases are similar in offerings to euGenes; they have been loosely co-developed following similar goals for genome and proteome information services.
It is likely that many of these systems will converge to provide the biosciences community with a very good range of options for reading, extracting and computing on these important sets of genome data. Improvements in these systems are rapidly appearing, the product of cooperation, friendly competition and uniquely developed options among these groups. These choices provide the bioscience community with increasingly accurate and up-to-date genome data in many useful ways.
FUTURE DIRECTIONS
The euGenes project is a work in progress, as are the genome data that it draws on. General improvements in display, sequence retrieval, integration of map searching with other gene attribute searches will be forthcoming. Enhancements will be made for multi-organism data extraction, to aid researchers who are actively analyzing and mining such data. The selection of eukaryote genomes included in euGenes has been practically oriented to address immediate needs for summarized, current data of several widely studied eukaryote genomes. The information system described here can be adapted to any organism set with similar data.
Comparative or synteny maps will be added, showing genome regions among organisms with conserved sequence regions. Along with improvements in extracting sequence data from these genomes, these synteny data will help scientists study evolution of genomes. A sequence similarity (BLAST) search of genomes contained in euGenes is planned. A summary of research publications on genes, and metabolic path information are other planned additions.
Much of the future of growth of genome databases and information systems is to extend past single organism contents to provide methods, software and data management that works for any range of organisms. Much of the software and data management methods used for a single genome system can be applied with some added effort to any set of organisms. This was found during the building of euGenes from software components designed for FlyBase by the author. Currently, the software engineering of public interfaces in euGenes and FlyBase are co-developed; a tool or improvement is integrated into both systems.
Acknowledgments
ACKNOWLEDGEMENTS
William Gelbart has been instrumental in providing an initial outline for a model eukaryotic organism service and collaborating on its prototype service. Thanks are due to the many bioinformaticians and bioscientists involved in providing public genome information and in offering advice in development of this project, including, but not limited to, E. Doerry, J. Sprague, L. Stein, S. Martinelli, D. Maglott, M. Ashburner, M. Rebhan, M. Safran and B. Haas. This project is supported by Indiana University, the NSF (DBI-0090782 and DBI-9982851) and the NIH funded FlyBase project.
REFERENCES
- 1.The FlyBase Consortium (1999) The FlyBase database of the Drosophila Genome Projects and community literature. Nucleic Acids Res., 27, 85–88. Updated article in this issue: Nucleic Acids Res. (2002), 30, 106–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stein L., Sternberg,S., Durbin,R., Thierry-Mieg,J. and Spieth,J. (2001) WormBase: network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res., 29, 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 5.Blake J.A., Eppig,J.T., Richardson,J.E., Bult,C.J. and Kadin,J.A. (2001) The Mouse Genome Database (MGD): integration nexus for the laboratory mouse. Nucleic Acids Res., 29, 91–94. Updated article in this issue: Nucleic Acids Res. (2002), 30, 113–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherry J.M., Adler,C., Ball,C., Chervitz,S.A., Dwight,S.S., Hester,E.T., Jia,Y., Juvik,G., Roe,T., Schroeder,M., Weng,S. and Botstein, D (1998) SGD: Saccharomyces Genome Database. Nucleic Acids Res., 26, 73–79. Updated in this issue: Nucleic Acids Res. (2002), 30, 69–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sprague J., Doerry,E., Douglas,S. and Westerfield,M. (2001) The Zebrafish Information Network (ZFIN): a resource for genetic, genomic and developmental research. Nucleic Acids Res., 29, 87–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bevan M., Mayer,K., White,O., Eisen,J.A., Preuss,D., Bureau,T., Salzberg,S.L. and Mewes,H. (2001) Sequence and analysis of the Arabidopsis genome. Curr. Opin. Plant Biol., 4, 105–110. [DOI] [PubMed] [Google Scholar]
- 9.Huala E., Dickerman,A.W., Garcia-Hernandez,M., Weems,D., Reiser,L., LaFond,F., Hanley,D., Kiphart,D., Zhuang,M., Huang,W., Mueller,L.A. et al. (2001) The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res., 29, 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Gene Ontology Consortium (2000) Gene Ontology: tool for the unification of biology. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rebhan M., Chalifa-Caspi,V., Prilusky,J. and Lancet,D. (1998) GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics, 14, 656–664. [DOI] [PubMed] [Google Scholar]
- 12.Etzold T. and Argos,P. (1993) SRS – an indexing and retrieval tool for flat file data libraries. Comp. Appl. Biosci., 9, 49–58. [DOI] [PubMed] [Google Scholar]