Abstract
STUDY QUESTION
Are single nucleotide variants (SNVs) in Aurora kinases B and C (AURKB, AURKC) associated with risk of aneuploid conception?
SUMMARY ANSWER
Two SNVs were found in patients with extreme aneuploid concepti rates with respect to their age; one variant, AURKC p.I79V, is benign, while another, AURKB p.L39P, is a potential gain-of-function mutant with increased efficiency in promoting chromosome alignment.
WHAT IS KNOWN ALREADY
Maternal age does not always predict aneuploidy risk, and rare gene variants can be drivers of disease. The AURKB and AURKC regulate chromosome segregation, and are associated with reproductive impairments in mouse and human.
STUDY DESIGN, SIZE, DURATION
An extreme phenotype sample selection scheme was performed for variant discovery. Ninety-six DNA samples were from young patients with higher than average embryonic aneuploidy rates and an additional 96 DNA samples were from older patients with lower than average aneuploidy rates.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Using the192 DNA samples, the coding regions of AURKB and AURKC were sequenced using next generation sequencing. To assess biological significance, we expressed complementary RNA encoding the human variants in mouse oocytes. Assays such as determining subcellular localization and assessing catalytic activity were performed to determine alterations in protein function during meiosis.
MAIN RESULTS AND THE ROLE OF CHANCE
Ten SNVs were identified using three independent variant-calling methods. Two of the SNVs (AURKB p.L39P and AURKC p.I79V) were non-synonymous and identified by at least two variant-identification methods. The variant encoding AURKC p.I79V, identified in a young woman with a higher than average rate of aneuploid embryos, showed wild-type localization pattern and catalytic activity. On the other hand, the variant encoding AURKB p.L39P, identified in an older woman with lower than average rates of aneuploid embryos, increased the protein’s ability to regulate alignment of chromosomes at the metaphase plate. These experiments were repeated three independent times using 2–3 mice for each trial.
LARGE SCALE DATA
N/A.
LIMITATIONS, REASONS FOR CAUTION
Biological significance of the human variants was assessed in an in vitro mouse oocyte model where the variants are over-expressed. Therefore, the human protein may not function identically to the mouse homolog, or the same in mouse oocytes as in human oocytes. Furthermore, supraphysiological expression levels may not accurately reflect endogenous activity. Moreover, the evaluated variants were identified in one patient each, and no trial linking the SNV to pregnancy outcomes was conducted. Finally, the patient aneuploidy rates were established by performing comprehensive chromosome screening in blastocysts, and because of the link between female gamete aneuploidy giving rise to aneuploid embryos, we evaluate the role of the variants in Meiosis I. However, it is possible that the chromosome segregation mistake arose during Meiosis II or in mitosis in the preimplantation embryo. Their implications in human female meiosis and aneuploidy risk remain to be determined.
WIDER IMPLICATIONS OF THE FINDINGS
The data provide evidence that gene variants exist in reproductively younger or advanced aged women that are predictive of the risk of producing aneuploid concepti in humans. Furthermore, a single amino acid in the N-terminus of AURKB is a gain-of-function mutant that could be protective of euploidy.
STUDY FUNDING/COMPETING INTERESTS
This work was supported by a Research Grant from the American Society of Reproductive Medicine and support from the Charles and Johanna Busch Memorial Fund at Rutgers, the State University of NJ to K.S. and the Foundation for Embryonic Competence, Inc to N.T. The authors declare no conflicts of interest.
Keywords: Aurora kinase, meiosis, aneuploidy, oocyte, infertility, single nucleotide variants.
Introduction
Infertility affects around 15% of humans of reproductive age (Thoma et al., 2013). The leading genetic abnormality causing this disease is embryonic aneuploidy, in which the developing offspring has an abnormal chromosome compliment (Hassold and Hunt, 2001). Most of these chromosomal imbalances are maternally derived and result from mis-segregation of chromosomes in the first meiotic division (Hassold et al., 1987, 1991; May et al., 1990; Takaesu et al., 1990; Nagaoka et al., 2012; Fragouli et al., 2013). Meiosis is the cellular process during which gametes undergo two rounds of chromosome segregation without an intervening round of DNA replication to become haploid. While it is well characterized that these chromosomal mis-segregations frequently occur during the first meiotic division, the source of these errors is still largely unknown.
The chance of generating aneuploid embryos increases with maternal age (Lejeune et al., 1959; Hassold et al., 2007; Jones, 2008; Penrose, 2009). By performing comprehensive chromosome screening (CCS) on trophectoderm biopsies of in vitro-fertilized embryos, we observed that some women have rates of aneuploid concepti that deviate from the average for their given age (Franasiak et al., 2014). Therefore, age alone is not always predictive of the embryonic aneuploidy risk and, these data suggest that other factors, such as genetic predisposition or environmental exposures, account for the altered aneuploidy levels. Because rare genetic variants can be drivers of disease (Pritchard, 2001; Bodmer and Bonilla, 2008; Stankiewicz and Lupski, 2010), we hypothesized that genetic variants contribute to these altered risks in some patients. To identify these rare variants, we used an extreme phenotyping strategy that increases the likelihood of gene variant discovery (Cohen et al., 2004; Ahituv et al., 2007; Bruse et al., 2016). This strategy utilizes sequences from young patients who had higher than average levels of aneuploid concepti to identify causative variants, and sequences from older patients who had lower than average levels of aneuploid concepti to identify protective variants.
We focused on the Aurora kinases (AURK), a conserved family of serine/threonine protein kinases critical for regulating chromosome segregation in both mitosis and meiosis as a candidate approach to test this genetic variant hypothesis (Shuda et al., 2009; Solc et al., 2012; Nikonova et al., 2013; Balboula and Schindler, 2014; Quartuccio and Schindler, 2015; van der Horst et al., 2015). In mammalian meiosis, Aurora kinase B (AURKB) and Aurora kinase C (AURKC) have essential roles in regulating chromosome alignment, sister chromatid cohesion, and the spindle assembly checkpoint; functions critical to the generation of euploid gametes (Tang et al., 2006; Swain et al., 2008; Shuda et al., 2009; Vogt et al., 2009; Sharif et al., 2010; Schindler et al., 2012; Yoshida et al., 2015). Several AURKB and AURKC variants in humans correlate with reduced fertility (Lopez-Carrasco et al., 2013) or male sterility (Dieterich et al., 2007a; Ben Khelifa et al., 2011, 2012; Fellmeth et al., 2016). Therefore, we hypothesized that mutations in either of these genes could affect the accuracy of chromosome segregation during gamete development and consequently serve as predictors of embryonic aneuploidy rates. In this study, we used next generation sequencing to identify variants in AURKB or AURKC that could predict the meiotic-origin embryonic aneuploidy risk for patients undergoing infertility treatment. We then used mouse oocytes as a model to assess the biological significance of the variants during mammalian female meiosis.
Materials and Methods
Ethical approval
Analysis of DNA samples from Reproductive Medicine Associates of New Jersey (RMANJ) DNA Bank was approved by the Institutional Review Board #RMA1-09–165. All animals were maintained following the Rutgers Institutional Animal Use and Care Committee (#11–032) and the National Institutes of Health guidelines.
Sample selection
One hundred and ninety-two female patients of European descent who underwent CCS as part of their IVF procedure at RMANJ were selected for the study. These samples were selected under the extreme phenotype sample selection scheme. The first group included 96 patients older than 35 years with a reported embryo aneuploidy rate lower than 50%; the second group included 96 patients younger than 35 years with an embryo aneuploidy rate higher than 50%. Women with fewer than four blastocysts after IVF and embryo culture were excluded from the sampling to rule out patients that were poor hormone responders or had diminished ovarian reserve. The average anti-Müllerian hormone level for the young cohort was 3.56 ± 3.37 and the level for the old cohort was 2.61 ± 2.95. There was no difference in BMI between cohort (young = 23.07 ± 3.96 versus old = 24.95 ± 5.59). The aneuploidy rate was transformed by a binomial proportional method yielding adjusted Agresti–Coull aneuploidy ratios (Agresti and Coull, 1998). Thresholds were established by a joint analysis to rank the patients by age and Agresti–Coull aneuploidy rate (ACR). The young cohort was identified from the top 96 of the youngest 30% of patients ranked by decreasing ACR (high to low). The old cohort was identified from the top 96 of the oldest 30% of patients ranked by increasing ACR (low to high).
Primer design and sequencing
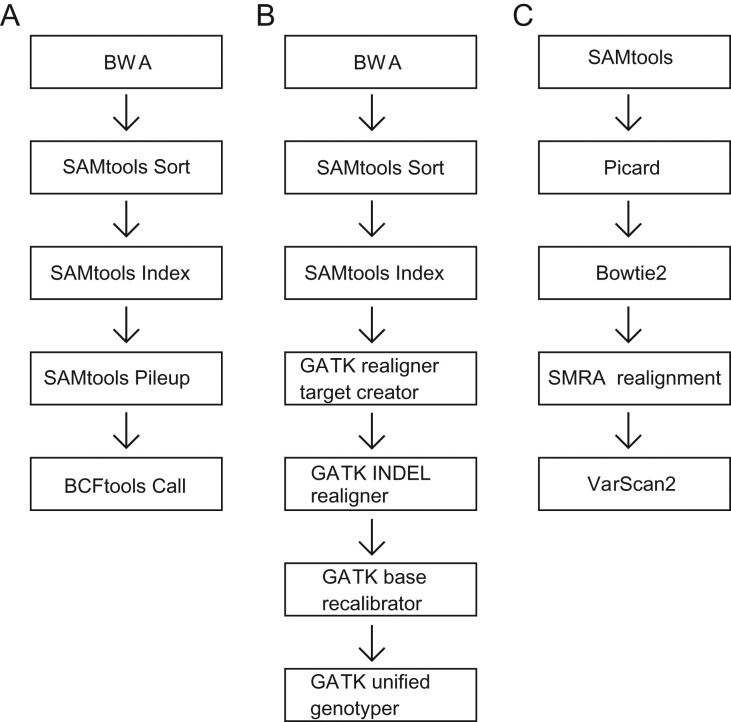
Primers were designed using the Life Technologies Ion Torrent AmpliSeq tool on the human reference genome version hg19 for AURKB (chr17:8 103 049–8 114 944) and AURKC (chr19:57 737 377–57 747 916). After excluding repetitive and low GC content regions, a total of 71 amplicons were designed to sequence the two genes. The primer sequences and targeted regions of the 71 amplicons are shown in Supplementary data, Table SI. Libraries were prepared using the AmpliSeq protocol as recommended by the supplier (Thermofisher Scientific, CA, USA). The molar concentration of each amplicon was obtained using a Bioanalyzer on the Agilent High Sensitivity DNA microfluidic chip (Agilent Technologies, Santa Clara, CA, USA), and the samples were then normalized to 20 pM. The Ion OneTouch 200 Template Kit was used for template preparation and the Ion Sequencing Kit v2.0 was used for the Ion 318 Chip based sequencing as recommended (Thermofisher Scientific, USA). For barcoding purposes, Ion Xpress Barcodes were used. Sequence data were produced by the Ion Torrent Suite Software version 3.6.2 and exported for in-house analysis using three methods (Fig. 1).
Figure 1.
Schematic of the three variant-calling methods: (A) SAMtools pileup; (B) GATK; (C) VarScan 2.
Variant calling and annotation
Because Ion Torrent data are particularly sensitive to insertion/deletion (INDEL) errors (Bragg et al., 2013), we focused on single nucleotide variants (SNVs) within the coding region. Additional details of the three variant discovery methods used to identify variants (Fig. 1) can be found in supplemental methods.
Variant validation
Amplicons of interest were pre-amplified by PCR. Primers for sequencing were designed using Primer Express software (Thermofisher Scientific, USA). The following primers were used: For AURKB, forward 5′-GGATGACCTTCTCATCCCTAAAC-3′ and reverse 5′-AATACTGCTTGCAGCACATTC-3′; for AURKC, forward 5′-CACAGTCGATGACTTTGAAATCG-3′ and reverse 5′-GGTACAAGTGCAGGGTTTACT-3′. Sanger sequencing was performed in an ABI 3730X analyzer using a BigDye Direct Sanger Sequencing Kit (Thermofisher Scientific, USA) and following manufacturer's instructions.
Variant screening
DNA samples were collected from 2030 patients who underwent preimplantation genetic screening at RMANJ. DNA extraction from either blood or follicular fluid samples was performed with QIAamp DNA Blood Maxi Kit (Qiagen, Germany) following manufacturer's instructions. DNA samples were stored at −80°C.
A TaqMan custom single nucleotide polymorphism (SNP) genotyping assay (Thermofisher Scientific, USA) was used to screen for each of the variants in 2030 patient samples following manufacturer's instructions. Primers and probes for each assay were as follows: AURKB L39P assay, forward (5′-GGACATTGGAGCGGCTCAT-3′), reverse (5′-CTCCGGAAAGAGCCTGTCA-3′), probe for wild-type allele (5′-CATCTGCACTTGTCCTC-3′), probe for variant allele (5′-ATCTGCACCTGTCCTC-3′); AURKC I79V assay, forward (5′-CATTGTGGCCCTGAAGGTTCT-3′), reverse (5′-GCTGGTGCTCCAGTCCTT-3′), probe for wild-type allele (5′-CAAGTCGCAGATAGAGAA-3′), probe for variant allele (5′-AAGTCGCAGGTAGAGAA-3′). The PCR step was performed in 384-well reaction plates in final 5 μl reaction volume. A pre- and post-PCR plate read was performed in a 7900HT Real time PCR system for generating allelic discrimination plots.
Mouse oocyte collection, microinjection and culture
Six to nine-week old CF1 mice (Harlan Laboratories, #NSA-CF1, IN, USA) and Aurkc−/− mice with a 129/SVpas/C57Bl/6 background (Kimmins et al., 2007; Schindler et al., 2012) were stimulated 48 h before oocyte collection with pregnant mare's serum gonadotropin (5 IU) (Calbiochem #367 222, Darmstadt, Germany). Prophase I-arrested oocytes were collected as previously described (Anger et al., 2005). In vitro culture and maturation was performed as described previously (Schindler et al., 2012) in a humidified incubator at 37°C and 5% CO2 in air. Bicarbonate-free minimal essential medium (with 25 mM Hepes, pH 7.3 and 3 mg/ml polyvinylpyrollidone) supplemented with 2.5 μM milrinone (Sigma–Aldrich, #M4659, St. Louis, MO, USA) was used for collection and microinjection of oocytes so as to prevent meiotic resumption (Tsafriri et al., 1996). Prophase I-arrested oocytes were microinjected as previously described (Anger et al., 2005) with the synthesized complementary RNAs (cRNAs) (500 ng/μl in Fig. 3; 10 ng/μl in Figs. 4 and 5) and incubated overnight to allow sufficient expression. Oocytes were matured for 7 h to Met I in Chatot, Ziomek and Bavister (CZB) medium (Chatot et al., 1989) without milrinone.
Figure 3.
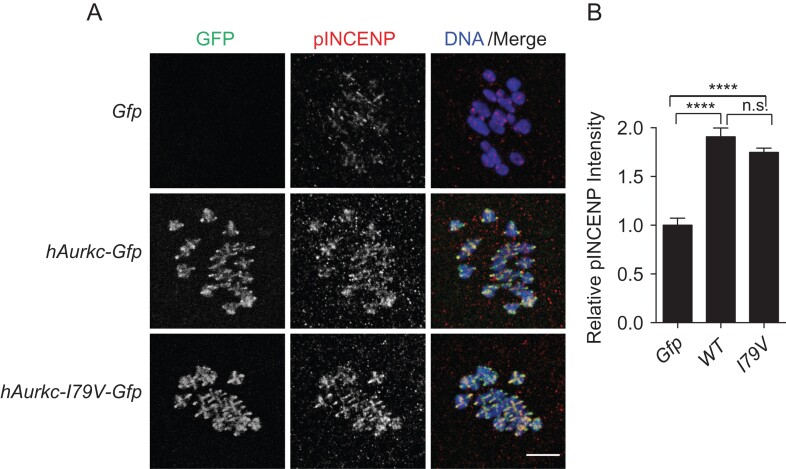
AURKC-I79V is indistinguishable from wild-type. (A) Prophase I-arrested oocytes from Aurkc−/− mice were microinjected with the indicated cRNAs in each row. After expression, the oocytes were matured in vitro to Met I (7 h) prior to fixation and immunocytochemistry to detect human AURKC (GFP; green in merge), phosphorylated INCENP (pINCENP; red in merge) and DNA (DAPI, blue in merge). (B) Quantification of the pixel intensity of pINCENP in (A) normalized to the Gfp-injected control. ****P < 0.0001 by one-way ANOVA. n.s. = not significant. The error bars are standard error. Scale bar represents = 10 μm.
Figure 4.
AURKB-L39P localization is normal. (A and B) Prophase I-arrested oocytes from CF1 mice were microinjected with the indicated cRNAs. After expression, the oocytes were matured in vitro to Met I (7 h) prior to fixation and immunocytochemistry to detect AURKB (GFP; green in merge) and kinetochores (CREST antiserum, red in merge). DNA detected by DAPI staining (blue in merge). Scale bars represent = 10 μm.
Figure 5.
AURKB-L39P corrects misaligned chromosomes more efficiently that WT AURKB. (A–C) Prophase I-arrested oocytes from CF1 mice were microinjected with the indicated cRNAs. After maturation and recovery from nocodazole as described in Materials and Methods section, oocytes were fixed and processed for immunocytochemistry to detect AURKB (GFP; green in merge) and kinetochores (CREST antiserum, red in merge). DNA detected by DAPI staining (blue in merge). (B) Chromosome misalignment was determined by confocal microscopy as described in Materials and Methods section. (C) Gfp expression levels of each oocyte examined. Red color indicates oocytes scored with misaligned chromosomes. Scale bar represents = 10 μm. *P < 0.05 by Student's t-test. The error bars are standard error.
To assess ability to correct kinetochore-microtubule attachments, prophase I-arrested oocytes were microinjected with the synthesized cRNAs (10 ng/μl) and incubated overnight to allow sufficient expression. Oocytes were then washed free of milrinone and matured for 5 h in CZB medium. To generate unstable kinetochore-microtubule (K-MT) attachments oocytes were transferred to CZB medium supplemented with 5 μM nocodazole (Calbiochem; 487928) for 1 h. Oocytes were then washed free of nocodazole and continued maturation in CZB medium supplemented with 5 μM MG132 (Calbiochem; 474791), to inhibit anaphase onset, to recover for 2 h prior to fixation.
cRNA preparation
mAurkb-Gfp cDNA was generated as previously described (Shuda et al., 2009). hAURKB cDNA was purchased from Sino Biological Inc (#HG11415-M, China) and hAURKC_v1 from Genecopeia (#EXQ0034-M02-B, USA). The human genes were subsequently PCR-amplified and ligated into pIVT-EGFP vectors (Igarashi et al., 2007; Fellmeth et al., 2015) Mutagenesis reactions were performed with QuikChange multi-site directed mutagenesis kit (Agilent Technologies, #210 514, USA). Mutagenic primers for the variants were hAURKB-L39P (5′-GCCTGTCACCCCATCTGCACCTGTCCTCATGAGCCGC-3′); hAURKC-I79V (5′-GGCCCTGAAGGTTCTCTTCAAGTCGCAGGTAGAGAAGGAAGGACTGG-3′). Plasmids were isolated (Qiagen, Hilden, Germany), from which a fraction was sequenced to confirm the presence of desired mutation. Linearization was performed with NdeI (New England BioLabs, Ipswich, MA, USA) and the product was purified using Qiagen PCR purification kit. In vitro transcription was performed with the linearized product using the mMessage mMachine kit (Thermofisher Scientific, #AM1344M, USA) followed by cRNA purification using a RNA-Easy purification kit (Qiagen, #74 104, Germany).
Immunocytochemistry and imaging
After meiotic maturation, oocytes were fixed in PBS with 2% w/v paraformaldehyde for 20 min for CREST (Anti-centromeric antigen) (Antibodies Incorporated, Davis, CA, USA #15–234; 1:30), pINCENP (a gift from Dr Lampson at the University of Pennsylvania, 1:1000) and acetylated tubulin (Sigma–Aldrich, Davis, CA, USA, #T7451; 1:1000) immunostaining, or with 3.7% w/v paraformaldehyde for 1 h for γ-tubulin detection (Sigma–Aldrich, USA, #T6557; 1:100). Oocytes were subsequently washed in blocking buffer (PBS + 0.3% (w/v) bovine serum albumin + 0.01% (v/v) Tween-20) and permeabilized as described previously (Fellmeth et al., 2015). The aforementioned antibodies and dilutions were used for primary antibody immunostaining (1 h). Next, oocytes were washed in blocking solution followed by immunostaining for 1 h with the secondary antibodies anti-human (Thermofisher Scientific,#A21091, USA), anti-rabbit (Thermofisher Scientific, #A10042, USA), anti-mouse (Thermofisher Scientific, #A10037 and #A21050, USA) diluted 1:200 in blocking solution. Following a final wash in blocking solution, oocytes were mounted on microscope slides in VectaShield (Vector Laboratories, #H-1000, Burlingame, CA, USA) containing DAPI (Thermofisher Scientific, #D1306; 1:170, USA). Fluorescence was detected using Zeiss 510 Meta laser-scanning confocal microscope under a 40× objective. When comparison of intensity measurements was required, the laser power was kept constant between groups. Images were processed and analyzed using Image J (NIH, Bethesda, MD, USA). Expression levels were normalized by measuring the pixel intensity of AURKB-L39P-Gfp and normalizing to the average intensity in wild-type (WT) AURKB-Gfp-injected controls. Quantification of chromosome misalignment was performed as described previously (Lane et al., 2012).
Statistical analysis
Statistical analysis was carried out using Prism GraphPad Software (La Jolla, CA, USA). One-way ANOVA with Tukey post-tests or Student's t-test, as indicated in the legend, was used to calculate the differences between groups. Differences with P < 0.05 were considered significant.
Results
Identification of AURKB and AURKC variants
To understand the genetic contribution to the risk of producing aneuploid embryos, we identified 192 women who were at the extremes for producing aneuploid concepti for their age. Because AURKB and AURKC are known regulators of chromosome segregation and because variants have previously been shown to affect human fertility (Dieterich et al., 2007b; Lopez-Carrasco et al., 2013), we sequenced the coding regions of these two genes using Ion Torrent sequencing technology.
The three variant-identification methods, GATK, SAMtools and VarScan 2, identified nine, four and three high-quality coding SNVs, respectively (Table I). Together, the three methods identified 10 unique SNVs, three of which were not previously reported in dbSNP (Table I).
Table I.
SNVs identified by the three variant-calling pipelines GATK, SAMtools and VarScan 2.
| Type of SNV | Chromosome | Position | dbSNP138 | Y/H | O/L | GATK | SAMtools | VarScan 2 |
|---|---|---|---|---|---|---|---|---|
| Synonymous | chr17 | 8 108 190 | Yes | 1 | 0 | No | No | Yes |
| Non-synonymous | chr17 | 8 108 331 | Yes | 165 | 149 | Yes | No | No |
| Synonymous | chr17 | 8 108 339 | Yes | 128 | 110 | Yes | Yes | Yes |
| Synonymous | chr17 | 8 108 552 | No | 77 | 87 | Yes | No | No |
| Synonymous | chr17 | 8 108 690 | No | 94 | 83 | Yes | No | No |
| Synonymous | chr17 | 8 109 901 | Yes | 0 | 1 | Yes | Yes | No |
| Synonymous | chr17 | 8 110 622 | Yes | 1 | 0 | Yes | No | No |
| Non-synonymous | chr17 | 8 111 091* | No | 0 | 1 | Yes | Yes | No |
| Non-synonymous | chr19 | 57 743 531* | Yes | 1 | 0 | Yes | Yes | Yes |
| Synonymous | chr19 | 57 744 009 | Yes | 1 | 0 | Yes | No | No |
Chromosome 17 contains AURKB and Chromosome 19 contains AURKC.
Y/H, Number of alternative alleles in the young patient; high aneuploidy rate. O/L, Number of alternative alleles in the old patient, low aneuploidy rate; SNV, single nucleotide variant.
*Indicates the SNVs further studied in this report.
Next, we inspected the two non-synonymous SNVs that were identified in at least two methods (Table I). Both of these SNVs were A-to-G mutations (Table II). One was in AURKB (chr17:8 111 091; AURKB c.116T > C; note that AURKB is encoded on the minus strand), and the other was in AURKC (chr19:57 743 531; AURKC c.235A > G). According to the Exome Aggregation Consortium (ExAC) database that contains SNV information for 60 000 individuals (Lek et al., 2015), the AURKB mutation is a rare variant and it is identified as heterozygous in four individuals of Non-Finnish European origin with an allele frequency of 6.02 × 10−5 in that population (3.32 × 10−5 in the total study population). This mutation causes a leucine-to-proline change at amino acid position 39. The AURKC mutation is more common than the AURKB mutation, present 60 times in the ExAC African population (allele frequency 5.81× 10−3) and it is not present in other populations (4.96 × 10−4 in the total study population). The AURKC mutation is also reported in the NHLBI Exome Sequencing Project (ESP), the 1000 Genomes Project and dbSNP build 138 (rs61736320) (Sherry et al., 2001; Abecasis et al., 2010; Norton et al., 2012). This mutation causes an isoleucine-to-valine change at amino acid position 79. Functional prediction from SIFT (Ng and Henikoff, 2001) software program that predicts changes in protein function due to missense mutations, predicts that the AURKB mutation is tolerated, and that the AURKC mutation is damaging. Polyphen2 (Adzhubei et al., 2010), an alternative prediction program, however, predicts that both of the mutations are benign (Table II).
Table II.
Annotation of selected variants.
| Gene | Chromosome | Position | Ref. | Alt | CytoBand | Esp6500si | 1000G | SNP138 | ExAC | SIFT | Polyphen2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AURKB | chr17 | 8 111 091 | A | G | 17p13.1 | 3.322−5 | T | B | |||
| AURKC | chr19 | 57 743 531 | A | G | 19q13.43 | 0.002153 | 0.0037 | rs61736320 | 4.959−4 | D | B |
ExAC, Exome Aggregation Consortium; SNP, single nucleotide polymorphism.
Ref. indicates the nucleotide in the reference genome (hg19) and Alt denotes the nucleotide identified in the sequencing. Esp6500si (Norton et al., 2012), 1000G (Abecasis et al., 2010), SNP138 (Sherry et al., 2001) and ExAC (Lek et al., 2015) are databases describing variants identified in human populations. Known allele frequencies for variants in Esp6500si, 1000G, and ExAC and known dbSNP identifier in dbSNP138 are shown. SIFT (Ng and Henikoff, 2001) and Polyphen2 (Adzhubei et al., 2010) predict whether the nucleotide change is tolerated and benign (T and B) or detrimental (D) to protein function.
Confirmation of variants and determination of allele frequencies
To validate the two non-synonymous SNV candidates (Fig. 2A), we used Sanger sequencing and a TaqMan genotyping assay. First, we confirmed the presence of the two SNVs using Sanger sequencing (Fig. 2B). Both patients were heterozygous for the SNV. The TaqMan allelic discrimination plot (Fig. 2C) demonstrated that the affected patients harbor the variants (blue dots for the patient with AURKB A-G and red dots for the patient with AURKC A-G). Next, we genotyped 1838 additional patient samples for these variants using the same TaqMan assay (Fig. 2C). One additional heterozygous carrier of the AURKC variant was found, resulting in an allele frequency of 4.93 × 10−4 for this variant in this study. No other carriers were found for the AURKB variant (allele frequency of 2.46−4 in this study). Therefore, these variants are rare in general populations in the ExAC database (Table II) and in the IVF clinic setting used in this study.
Figure 2.
Detection and confirmation of AURKB and AURKC variants. Images of the raw data obtained from Ion Torrent sequencing (A), Sanger sequencing (B) and TaqMan genotyping (C). Left column: AURKB A-G (chr17:8 111 091) Right column: AURKC A-G (chr19:57 743 531). In panel A, horizontal gray bars in the IGV represent a subset of the read depth, 9/297 for AURKB and 9/95 for AURKC. For the TaqMan assay allelic discrimination plots in panel C, blue dots represent four replicates for the AURKB variant carrier, red dots for the AURKC carrier and black dots negative controls with no template. X and Y-axes are the reporter dye fluorescent intensities for each probe targeting the wild-type allele (A) and the variant (G). IGV, Integrative Genomics viewer; WT, wild-type.
AURKC-I79V is a benign variant
In human oocytes, AURKC localizes at centromeres and along chromosome arms (Avo Santos et al., 2011), similar to mouse AURKC (Tang et al., 2006; Shuda et al., 2009; Sharif et al., 2010). We previously demonstrated that exogenous expression of human AURKC localizes to the same regions of metaphase I (Met I) chromosomes in mouse oocytes (Fellmeth et al., 2015). Furthermore, expression of human AURKC can complement the phenotypic defects of oocytes from Aurkc−/− mice. This model can be used to assess the functional consequences of variants of human AURKC variants (Fellmeth et al., 2016).
To determine if the isoleucine-to-valine variant alters AURKC activity, we expressed AURKC-I79V-GFP in Aurkc−/− oocytes and assessed phosphorylation of an AURKB/C substrate, INCENP by immunocytochemistry. As controls, oocytes were injected with human AURKC-Gfp or Gfp alone. In the Gfp-alone group, GFP signal was dispersed evenly throughout the oocyte. Compared to the WT control, the AURKC-I79V variant had an identical localization pattern at Met I: to centromeres and along the chromosome axes (Fig. 3A). Furthermore, the immunoreactivity of the phospho-specific INCENP antibody was not statistically different between WT and variant-injected oocytes (P = 0.12) (Fig. 3A and B). In both of these groups phosphorylated INCENP was significantly increased compared to the Gfp-injected controls (P < 0.0001) due to exogenous expression of AURKC. Taken together, these data suggest that the valine substitution does not alter AURKC localized activity, and is likely a benign mutation.
AURKB-L39P may be a gain-of-function allele
In human mitotic cells, AURKB localizes to centromeres where it regulates chromosome alignment and kinetochore-microtubule attachments (Kallio et al., 2002; Cimini et al., 2006; Carmena et al., 2009; van der Horst et al., 2015). In mouse oocytes at Met I, exogenous mouse AURKB-Gfp (mAURKB-Gfp) also localizes to centromeres (Shuda et al., 2009; Sharif et al., 2010). To determine if human AURKB (hAURKB) behaves similar to mAURKB in mouse oocytes, we first assessed hAURKB localization. Wild-type mouse oocytes injected with either mAurkb-Gfp or hAURKB-Gfp cRNAs were subsequently matured in vitro to Met I prior to fixation and immunocytochemistry. As previously reported, mAURKB-Gfp localizes to centromeres (Fig. 4A) (Shuda et al., 2009; Vogt et al., 2009, 2010; Sharif et al., 2010). Similarly, hAURKB-Gfp localized to centromeres, but also localized along the arms of the chromosomes, more similar to human and mouse AURKC localization (compare Figs. 3A to 4A).
In conducting these analyses, we also observed a dose-dependent change in human AURKB localization (Supplementary data, Fig. S1). When expressed at a higher level (50 ng/μl), hAURKB-Gfp localization was now observed at spindle poles in addition to the chromosome and centromere localization. At the highest expression level tested (100 ng/μl), the chromosome localization was absent, while the centromeric and spindle pole localization was retained. Therefore, the abundance of human AURKB expressed in mouse oocytes changes the subcellular localization from chromosomes to spindle poles.
We next compared the localization of hAURKB-Gfp to hAURKB-L39P-Gfp in wild-type mouse oocytes using a low concentration of cRNA (10ng/μl) that would be as comparable to endogenous levels as possible. Localization of the human AURKB variant was identical to that of wild-type AURKB because both localized to MI bivalent chromosomes. Therefore, mutation of leucine 39 to proline does not alter subcellular localization of AURKB at this low concentration.
In mitotic cells, AURKB promotes proper chromosome segregation in part by regulating chromosome alignment at the metaphase plate (Ditchfield et al., 2003) The human AURKB-L39P variant was identified in an older woman with lower than average rate of aneuploid concepti, so we hypothesized that it is a gain-of-function allele. To test this hypothesis, we first needed to develop a functional assay for human AURKB activity in mouse oocytes. To evaluate the catalytic activity of human AURKB, we designed an assay to assess the ability of the kinase to support chromosome alignment at the Met I plate. To this end, oocytes expressing WT or mutant human AURKB were matured to proMet I followed by treatment with nocodazole to depolymerize the meiotic spindle to induce chromosome misalignment. Next, we assessed the ability to recover by fixing oocytes 2 h after washing out the drug and performed confocal microscopy to analyze chromosome alignment. At this time point, ~80% of oocytes expressing WT hAURKB still had at least one chromosome not yet aligned in the Met I plate (Fig. 5A and B). The kinetics in the ability to correct misaligned chromosomes within 2 h significantly improved when oocytes expressed hAURKB-L39P (only ~60% still misaligned). This improvement was independent on kinase expression level (Fig. 5C). These data support the hypothesis that AURKB p.L39P has enhanced ability to promote chromosome alignment.
Discussion
Because maternal age does not always predict the likelihood of generating aneuploid embryos (Franasiak et al., 2014), we sought to identify and characterize genetic variants associated with a patient's embryonic aneuploidy rate. To test the hypothesis that there is a genetic link to altered aneuploidy levels, here, we describe results from a pilot study focused on Aurora kinase B and C gene variants, which encode two proteins known to regulate chromosome segregation.
Mutations in AURKC cause detrimental consequences in male meiosis, and they were the first known genetic cause of macrozoospermia and tetraploidy in sperm (Dieterich et al., 2007a, 2009; Ben Khelifa et al., 2011, 2012). Furthermore, these mutations were characterized in a mammalian meiosis model where their expression caused a failure to complete Meiosis I, thus producing tetraploid gametes (Fellmeth et al., 2016). Nonetheless, two women who were homozygous carriers for the variant AURKC c.144delC were fertile (having two and six children) (Harbuz et al., 2009), suggesting a sexually dimorphic requirement for AURKC in meiosis. Similarly, our findings indicate that the variant that encodes AURKC p.I79V (AURKC c.235A > G) does not have a deleterious impact on oocyte meiosis (Fig. 3). We note that the aneuploidy was detected in blastocyst embryos, and, therefore, there is the formal possibility that the chromosome segregation mistake occurred during embryonic mitosis, which was not evaluated here. The patient identified harboring this variant was 32 years old, but had higher than average embryonic aneuploidy rates. On the other hand, a subsequent patient identified in the larger, population-based analysis was a 35 year-old patient with two aneuploid embryos out of 8 (25%). This patient is at the other extreme of the aneuploidy phenotype: reproductively advanced age but with low rates of embryonic aneuploidy. Taken together, the data suggest that AURKC c.235A > G is unlikely to be predictive of embryonic aneuploidy risk in women or in preimplantation embryogenesis.
Five exonic variants of AURKB were previously detected in individuals with reproductive impairments, two of which were considered common polymorphisms given their reported high frequencies (Lopez-Carrasco et al., 2013). In addition, variants in this gene have been detected in patients with breast cancer, although they did not consistently correlate with the disease (Tchatchou et al., 2007; Kabisch et al., 2015). The AURKB variant reported here was previously found in four individuals out of ~60 000 (Lek et al., 2015). The single amino acid change of leucine 39 to a proline resulted in a gain in an increased ability for AURKB to promote chromosome alignment when oocytes were challenged with conditions that promote gross chromosome misalignment (Fig. 5).
The variant encoding AURKB-L39P was identified in a 39-year-old patient with a lower than average embryonic aneuploidy rate of 33%. Consequently, our interpretation is that this variant may be protective of embryonic euploidy. One function of AURKB is to promote chromosome alignment by sensing improper K-MT attachments, and cause their destabilization (Lampson et al., 2004). Expression of AURKB-L39P allowed for faster chromosome alignment when mouse oocytes were challenged with nocodazole. Therefore, it is possible that this variant has a higher affinity for the substrates that trigger microtubule depolymerization or that it is more responsive to fixing unattached kinetochores than is WT AURKB. Future studies examining the molecular mechanisms governing this increase in kinetics for the AURKB-L39P variant, will help further establish the extent to which this specific variant is protective of meiotic euploidy, and if it has any impact on mitotic chromosome segregation.
Because maternal age does not always account for the risk of producing aneuploid embryos (Franasiak et al., 2014), an important task is to investigate the genetic contribution to this phenomenon. To date, only variants of polo-like kinase 4 (PLK4) (McCoy et al., 2015) and tubulin (TUBB8) (Feng et al., 2016) have been correlated with increased embryonic aneuploidy rates. Here, we identified two variants within genes required for cell cycle regulation and evaluated their biological significance during mouse oocyte meiosis. While our results indicate that the AURKC variant does not seem to alter this protein's function, we identified a single amino acid in the N-terminus of AURKB that increases activities that may protect euploidy in mammalian female meiosis. Further studies characterizing the biological significance of mutations possibly responsible for reproductive impairments will help us understand why some individuals are prone to generate genetically imbalanced embryos. Likewise, whole genome sequencing approaches should facilitate the discovery and characterization of genetic variants that could become biomarkers for a patients’ embryonic aneuploidy risk, which will be invaluable in personalized infertility treatments.
Supplementary data
Supplementary data are available at Molecular Human Reproduction online.
Authors’ roles
D.M., A.L.N., K.S., N.T. and J.X. conceived and designed the experiments. D.M., A.L.N., A.F., A.S.G., E.M.S., Y.W. and Z.C. performed the experiments. D.M., A.L.N., E.M.S., A.Z., D.T., J.X. and N.T. analyzed the data. K.S., J.X., R.T.S. and N.T. contributed reagents, materials or analysis tools. D.M., A.L.N., A.Z., J.X. and K.S. wrote the paper.
Funding
Research Grant from the American Society for Reproductive Medicine and support from the Charles and Johanna Busch Memorial Fund at Rutgers, the State University of NJ to K.S. and the Foundation for Embryonic Competence, Inc. to N.T.
Conflict of interest
None declared.
Supplementary Material
Acknowledgements
We thank Dr Jessica Fellmeth and Mustafa Ladha for cloning and mutagenesis of the AURKB variant.
Contributor Information
Alexandra L. Nguyen, Department of Genetics, Rutgers, The State University of New Jersey, 145 Bevier Rd. Piscataway, NJ 08854, USA
Diego Marin, Department of Genetics, Rutgers, The State University of New Jersey, 145 Bevier Rd. Piscataway, NJ 08854, USA; Reproductive Medicine Associates of New Jersey, 140 Allen Rd, Basking Ridge, NJ 07920, USA.
Anbo Zhou, Department of Genetics, Rutgers, The State University of New Jersey, 145 Bevier Rd. Piscataway, NJ 08854, USA.
Amanda S. Gentilello, Department of Genetics, Rutgers, The State University of New Jersey, 145 Bevier Rd. Piscataway, NJ 08854, USA
Evan M. Smoak, Department of Genetics, Rutgers, The State University of New Jersey, 145 Bevier Rd. Piscataway, NJ 08854, USA
Zubing Cao, Department of Genetics, Rutgers, The State University of New Jersey, 145 Bevier Rd. Piscataway, NJ 08854, USA.
Anastasia Fedick, Department of Genetics, Rutgers, The State University of New Jersey, 145 Bevier Rd. Piscataway, NJ 08854, USA; Reproductive Medicine Associates of New Jersey, 140 Allen Rd, Basking Ridge, NJ 07920, USA.
Yujue Wang, Department of Genetics, Rutgers, The State University of New Jersey, 145 Bevier Rd. Piscataway, NJ 08854, USA; Reproductive Medicine Associates of New Jersey, 140 Allen Rd, Basking Ridge, NJ 07920, USA.
Deanne Taylor, Reproductive Medicine Associates of New Jersey, 140 Allen Rd, Basking Ridge, NJ 07920, USA; Present address: Department of Biomedical and Health Informatics, The Children's Hospital of Philadelphia; Department of Pediatrics, Perelman School of Medicine, University of Pennsylvania, 3501 Civic Center Blvd, Philadelphia, PA 19104, USA.
Richard T. Scott, Jr, Reproductive Medicine Associates of New Jersey, 140 Allen Rd, Basking Ridge, NJ 07920, USA.
Jinchuan Xing, Department of Genetics, Rutgers, The State University of New Jersey, 145 Bevier Rd. Piscataway, NJ 08854, USA.
Nathan Treff, Reproductive Medicine Associates of New Jersey, 140 Allen Rd, Basking Ridge, NJ 07920, USA.
Karen Schindler, Department of Genetics, Rutgers, The State University of New Jersey, 145 Bevier Rd. Piscataway, NJ 08854, USA.
References
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature 2010;467:1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agresti A, Coull BA. Approximate is better than ‘Exact’ for interval estimation of binomial proportions. Am Stat 1998;52:119–126. [Google Scholar]
- Ahituv N, Kavaslar N, Schackwitz W, Ustaszewska A, Martin J, Hebert S, Doelle H, Ersoy B, Kryukov G, Schmidt Set al. Medical sequencing at the extremes of human body mass. Am J Hum Genet 2007;80:779–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anger M, Stein P, Schultz RM. CDC6 requirement for spindle formation during maturation of mouse oocytes. Biol Reprod 2005;72:188–194. [DOI] [PubMed] [Google Scholar]
- Avo Santos M, van de Werken C, de Vries M, Jahr H, Vromans MJ, Laven JS, Fauser BC, Kops GJ, Lens SM, Baart EB. A role for Aurora C in the chromosomal passenger complex during human preimplantation embryo development. Hum Reprod 2011;26:1868–1881. [DOI] [PubMed] [Google Scholar]
- Balboula AZ, Schindler K. Selective disruption of aurora C kinase reveals distinct functions from aurora B kinase during meiosis in mouse oocytes. PLoS Genet 2014;10:e1004194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Khelifa M, Coutton C, Blum MG, Abada F, Harbuz R, Zouari R, Guichet A, May-Panloup P, Mitchell V, Rollet Jet al. Identification of a new recurrent aurora kinase C mutation in both European and African men with macrozoospermia. Hum Reprod 2012;27:3337–3346. [DOI] [PubMed] [Google Scholar]
- Ben Khelifa M, Zouari R, Harbuz R, Halouani L, Arnoult C, Lunardi J, Ray PF. A new AURKC mutation causing macrozoospermia: implications for human spermatogenesis and clinical diagnosis. Mol Hum Reprod 2011;17:762–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet 2008;40:695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg LM, Stone G, Butler MK, Hugenholtz P, Tyson GW. Shining a light on dark sequencing: characterising errors in Ion Torrent PGM data. PLoS Comput Biol 2013;9:e1003031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruse S, Moreau M, Bromberg Y, Jang JH, Wang N, Ha H, Picchi M, Lin Y, Langley RJ, Qualls Cet al. Whole exome sequencing identifies novel candidate genes that modify chronic obstructive pulmonary disease susceptibility. Hum Genomics 2016;10:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M, Ruchaud S, Earnshaw WC. Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr Opin Cell Biol 2009;21:796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil 1989;86:679–688. [DOI] [PubMed] [Google Scholar]
- Cimini D, Wan X, Hirel CB, Salmon ED. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol 2006;16:1711–1718. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science 2004;305:869–872. [DOI] [PubMed] [Google Scholar]
- Dieterich K, Soto Rifo R, Faure AK, Hennebicq S, Ben Amar B, Zahi M, Perrin J, Martinez D, Sele B, Jouk PSet al. Homozygous mutation of AURKC yields large-headed polyploid spermatozoa and causes male infertility. Nat genetics 2007. a;39:661–665. [DOI] [PubMed] [Google Scholar]
- Dieterich K, Soto Rifo R, Faure AK, Hennebicq S, Ben Amar B, Zahi M, Perrin J, Martinez D, Sele B, Jouk PSet al. Homozygous mutation of AURKC yields large-headed polyploid spermatozoa and causes male infertility. Nat Genet 2007. b;39:661–665. [DOI] [PubMed] [Google Scholar]
- Dieterich K, Zouari R, Harbuz R, Vialard F, Martinez D, Bellayou H, Prisant N, Zoghmar A, Guichaoua MR, Koscinski Iet al. The Aurora Kinase C c.144delC mutation causes meiosis I arrest in men and is frequent in the North African population. Hum Mol Genet 2009;18:1301–1309. [DOI] [PubMed] [Google Scholar]
- Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol 2003;161:267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmeth JE, Ghanaim EM, Schindler K. Characterization of macrozoospermia-associated AURKC mutations in a mammalian meiotic system. Hum Mol Genet 2016;25:2698–2711. [DOI] [PubMed] [Google Scholar]
- Fellmeth JE, Gordon D, Robins CE, Scott RT Jr., Treff NR, Schindler K. Expression and characterization of three Aurora kinase C splice variants found in human oocytes. Mol Hum Reprod 2015;21:633–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng R, Yan Z, Li B, Yu M, Sang Q, Tian G, Xu Y, Chen B, Qu R, Sun Zet al. Mutations in TUBB8 cause a multiplicity of phenotypes in human oocytes and early embryos. J Med Genet 2016;53:662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragouli E, Alfarawati S, Spath K, Jaroudi S, Sarasa J, Enciso M, Wells D. The origin and impact of embryonic aneuploidy. Hum Genet 2013;132:1001–1013. [DOI] [PubMed] [Google Scholar]
- Franasiak JM, Forman EJ, Hong KH, Werner MD, Upham KM, Treff NR, Scott RT Jr. The nature of aneuploidy with increasing age of the female partner: a review of 15,169 consecutive trophectoderm biopsies evaluated with comprehensive chromosomal screening. Fertil Steril 2014;101:656–663.e651. [DOI] [PubMed] [Google Scholar]
- Harbuz R, Zouari R, Dieterich K, Nikas Y, Lunardi J, Hennebicq S, Ray PF. Function of aurora kinase C (AURKC) in human reproduction. Gynecol Obstet Fertil 2009;37:546–551. [DOI] [PubMed] [Google Scholar]
- Hassold T, Hall H, Hunt P. The origin of human aneuploidy: where we have been, where we are going. Hum Mol Genet 2007;16:R203–R208. [DOI] [PubMed] [Google Scholar]
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2001;2:280–291. [DOI] [PubMed] [Google Scholar]
- Hassold T, Jacobs PA, Leppert M, Sheldon M. Cytogenetic and molecular studies of trisomy 13. J Med Genet 1987;24:725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold TJ, Pettay D, Freeman SB, Grantham M, Takaesu N. Molecular studies of non-disjunction in trisomy 16. J Med Genet 1991;28:159–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi H, Knott JG, Schultz RM, Williams CJ. Alterations of PLCbeta1 in mouse eggs change calcium oscillatory behavior following fertilization. Dev Biol 2007;312:321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KT. Meiosis in oocytes: predisposition to aneuploidy and its increased incidence with age. Hum Reprod Update 2008;14:143–158. [DOI] [PubMed] [Google Scholar]
- Kabisch M, Lorenzo Bermejo J, Dunnebier T, Ying S, Michailidou K, Bolla MK, Wang Q, Dennis J, Shah M, Perkins BJet al. Inherited variants in the inner centromere protein (INCENP) gene of the chromosomal passenger complex contribute to the susceptibility of ER-negative breast cancer. Carcinogenesis 2015;36:256–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio MJ, McCleland ML, Stukenberg PT, Gorbsky GJ. Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr Biol 2002;12:900–905. [DOI] [PubMed] [Google Scholar]
- Kimmins S, Crosio C, Kotaja N, Hirayama J, Monaco L, Hoog C, van Duin M, Gossen JA, Sassone-Corsi P. Differential functions of the Aurora-B and Aurora-C kinases in mammalian spermatogenesis. Mol Endocrinol 2007;21:726–739. [DOI] [PubMed] [Google Scholar]
- Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol 2004;6:232–237. [DOI] [PubMed] [Google Scholar]
- Lane SI, Yun Y, Jones KT. Timing of anaphase-promoting complex activation in mouse oocytes is predicted by microtubule-kinetochore attachment but not by bivalent alignment or tension. Development 2012;139:1947–1955. [DOI] [PubMed] [Google Scholar]
- Lejeune J, Gautier M, Turpin R. Study of somatic chromosomes from 9 mongoloid children. C R Hebd Seances Acad Sci 1959;248:1721–1722. [PubMed] [Google Scholar]
- Lek M, Karczewski K, Minikel E, Samocha K, Banks E, Fennell T, O'Donnell-Luria A, Ware J, Hill A, Cummings Bet al. Analysis of protein-coding genetic variation in 60,706 humans. bioRxiv 2015;536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Carrasco A, Oltra S, Monfort S, Mayo S, Rosello M, Martinez F, Orellana C. Mutation screening of AURKB and SYCP3 in patients with reproductive problems. Mol Hum Reprod 2013;19:102–108. [DOI] [PubMed] [Google Scholar]
- May KM, Jacobs PA, Lee M, Ratcliffe S, Robinson A, Nielsen J, Hassold TJ. The parental origin of the extra X chromosome in 47,XXX females. Am J Hum Genet 1990;46:754–761. [PMC free article] [PubMed] [Google Scholar]
- McCoy RC, Demko Z, Ryan A, Banjevic M, Hill M, Sigurjonsson S, Rabinowitz M, Fraser HB, Petrov DA. Common variants spanning PLK4 are associated with mitotic-origin aneuploidy in human embryos. Science 2015;348:235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka SI, Hassold TJ, Hunt PA. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet 2012;13:493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res 2001;11:863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikonova AS, Astsaturov I, Serebriiskii IG, Dunbrack RL Jr, Golemis EA. Aurora A kinase (AURKA) in normal and pathological cell division. Cell Mol Life Sci 2013;70:661–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton N, Robertson PD, Rieder MJ, Zuchner S, Rampersaud E, Martin E, Li D, Nickerson DA, Hershberger RE. Evaluating pathogenicity of rare variants from dilated cardiomyopathy in the exome era. Circ Cardiovasc Genet 2012;5:167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penrose LS. The relative effects of paternal and maternal age in mongolism. 1933. J Genet 2009;88:9–14. [DOI] [PubMed] [Google Scholar]
- Pritchard JK. Are rare variants responsible for susceptibility to complex diseases. Am J Hum Genet 2001;69:124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quartuccio SM, Schindler K. Functions of Aurora kinase C in meiosis and cancer. Front Cell Dev Biol 2015;3:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler K, Davydenko O, Fram B, Lampson MA, Schultz RM. Maternally recruited Aurora C kinase is more stable than Aurora B to support mouse oocyte maturation and early development. Proc Natl Acad Sci USA 2012;109:E2215–E2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif B, Na J, Lykke-Hartmann K, McLaughlin SH, Laue E, Glover DM, Zernicka-Goetz M. The chromosome passenger complex is required for fidelity of chromosome transmission and cytokinesis in meiosis of mouse oocytes. J Cell Sci 2010;123:4292–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001;29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuda K, Schindler K, Ma J, Schultz RM, Donovan PJ. Aurora kinase B modulates chromosome alignment in mouse oocytes. Mol Reprod Dev 2009;76:1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solc P, Baran V, Mayer A, Bohmova T, Panenkova-Havlova G, Saskova A, Schultz RM, Motlik J. Aurora kinase A drives MTOC biogenesis but does not trigger resumption of meiosis in mouse oocytes matured in vivo. Biol Reprod 2012;87:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med 2010;61:437–455. [DOI] [PubMed] [Google Scholar]
- Swain JE, Ding J, Wu J, Smith GD. Regulation of spindle and chromatin dynamics during early and late stages of oocyte maturation by aurora kinases. Mol Hum Reprod 2008;14:291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaesu N, Jacobs PA, Cockwell A, Blackston RD, Freeman S, Nuccio J, Kurnit DM, Uchida I, Freeman V, Hassold T. Nondisjunction of chromosome 21. Am J Med Genet Suppl 1990;7:175–181. [DOI] [PubMed] [Google Scholar]
- Tang CJ, Lin CY, Tang TK. Dynamic localization and functional implications of Aurora-C kinase during male mouse meiosis. Dev Biol 2006;290:398–410. [DOI] [PubMed] [Google Scholar]
- Tchatchou S, Wirtenberger M, Hemminki K, Sutter C, Meindl A, Wappenschmidt B, Kiechle M, Bugert P, Schmutzler RK, Bartram CRet al. Aurora kinases A and B and familial breast cancer risk. Cancer Lett 2007;247:266–272. [DOI] [PubMed] [Google Scholar]
- Thoma ME, McLain AC, Louis JF, King RB, Trumble AC, Sundaram R, Buck Louis GM. Prevalence of infertility in the United States as estimated by the current duration approach and a traditional constructed approach. Fertil Steril 2013;99:1324–1331.e1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsafriri A, Chun SY, Zhang R, Hsueh AJ, Conti M. Oocyte maturation involves compartmentalization and opposing changes of cAMP levels in follicular somatic and germ cells: studies using selective phosphodiesterase inhibitors. Dev Biol 1996;178:393–402. [DOI] [PubMed] [Google Scholar]
- van der Horst A, Vromans MJ, Bouwman K, van der Waal MS, Hadders MA, Lens SM. Inter-domain cooperation in INCENP promotes Aurora B relocation from centromeres to microtubules. Cell Rep 2015;12:380–387. [DOI] [PubMed] [Google Scholar]
- Vogt E, Kipp A, Eichenlaub-Ritter U. Aurora kinase B, epigenetic state of centromeric heterochromatin and chiasma resolution in oocytes. Reprod Biomed Online 2009;19:352–368. [DOI] [PubMed] [Google Scholar]
- Vogt E, Sanhaji M, Klein W, Seidel T, Wordeman L, Eichenlaub-Ritter U. MCAK is present at centromeres, midspindle and chiasmata and involved in silencing of the spindle assembly checkpoint in mammalian oocytes. Mol Hum Reprod 2010;16:665–684. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Kaido M, Kitajima TS. Inherent instability of correct Kinetochore-microtubule attachments during Meiosis I in oocytes. Dev Cell 2015;33:589–602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.