Abstract
The SUPERFAMILY database contains a library of hidden Markov models representing all proteins of known structure. The database is based on the SCOP ‘superfamily’ level of protein domain classification which groups together the most distantly related proteins which have a common evolutionary ancestor. There is a public server at http://supfam.org which provides three services: sequence searching, multiple alignments to sequences of known structure, and structural assignments to all complete genomes. Given an amino acid or nucleotide query sequence the server will return the domain architecture and SCOP classification. The server produces alignments of the query sequences with sequences of known structure, and includes multiple alignments of genome and PDB sequences. The structural assignments are carried out on all complete genomes (currently 59) covering approximately half of the soluble protein domains. The assignments, superfamily breakdown and statistics on them are available from the server. The database is currently used by this group and others for genome annotation, structural genomics, gene prediction and domain-based genomic studies.
INTRODUCTION
The SUPERFAMILY database is based on the SCOP (1) classification of protein domains. SCOP is a structural domain-based heirarchical classification with several levels including the ‘superfamily’ level. Proteins grouped together at the superfamily level are defined as having structural, functional and sequence evidence for a common evolutionary ancestor. It is at this level, as the name suggests, that SUPERFAMILY operates because it is the level with the most distantly related protein domains. The level below is the ‘family’ level which groups more closely related domains, and the level above is the ‘fold’ level which groups domains with similar topology which are not necessarily related.
The database uses hidden Markov models (HMMs) which are profiles based on multiple sequence alignments designed to represent a protein family (or superfamily) which can be used to search sequence databases for homologues. The SAM-T99 HMM software (2) is one of the best methods for the detection of remote protein homologues. The SAM software was used to build a library of models (3) representing all proteins of known structure, which forms the core of the SUPERFAMILY database. These models have added value by expert curation and tuning designed to detect and classify SCOP domains at the superfamily level.
There are existing databases which use HMMs representing protein domains such as Pfam (4), SMART (5) and others. There are also unifying databases which have several of these methods included, e.g. InterPro (6) and CDD (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). There are two main differences to SUPERFAMILY: these other databases span all proteins whereas SUPERFAMILY only covers those with a known structural representative, and they also group domains into families based on sequence similarity alone leading to a level of classification more similar to the family than the superfamily level. Structural assignments have been carried out using PSI-BLAST (7) based on the CATH (8) database but are much less extensive (http://www.biochem.ucl.ac.uk/bsm/cath_new/Gene3D).
DATABASE CONTENTS
The database may be accessed directly via a public server at http://stash.mrc-lmb.cam.ac.uk/SUPERFAMILY or via a link from each domain entry in SCOP at http://scop.mrc-lmb.cam.ac.uk/scop. There are also links from some genome databases, for example, Ensembl at http://www.ensembl.org. The underlying machinery of the database consists of a library of HMMs, a relational database and some programs. All of these are also available for download.
Structural assignments to sequences
The HMM library representing all proteins of known structure may be used to assign structural domains to sequences of unknown structure. An amino acid or nucleotide sequence may be queried against the library, and then the domain architecture and SCOP classification is returned (Fig. 1). The procedure has been optimised for remote homology detection retaining an estimated error rate of <1%. Three-dimensional models can be generated and these have been used to compare the method to other automatic structure prediction servers at LiveBench (http://bioinfo.pl). The server’s specificity is one of the best, especially for hard targets.
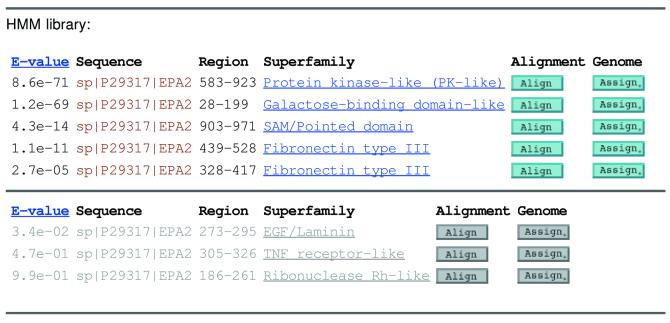
Figure 1.
An example of the result of a sequence query. The protein (sp│P2931│EPA2_HUMAN) is a multi-domain protein with five structural domains predicted with confidence, and shown in grey, three non-significant predictions. Each domain covers a different region of the query sequence and may be classified in a different SCOP superfamily with a different score. The ‘Align’ button links to a sequence alignment, and the ‘Assign.’ button links to all genome assignments for the given superfamily.
Nucleotide searches are carried out by translating sequences into the six reading frames and splitting across stop codons. Thus, the resulting structural assignments do not require any prior gene prediction and can be used to locate possible genes from raw DNA (Fig. 2). This does not provide gene prediction on its own, but is useful if no gene prediction is available and may suggest possible coding regions which gene prediction algorithms may not have identified.
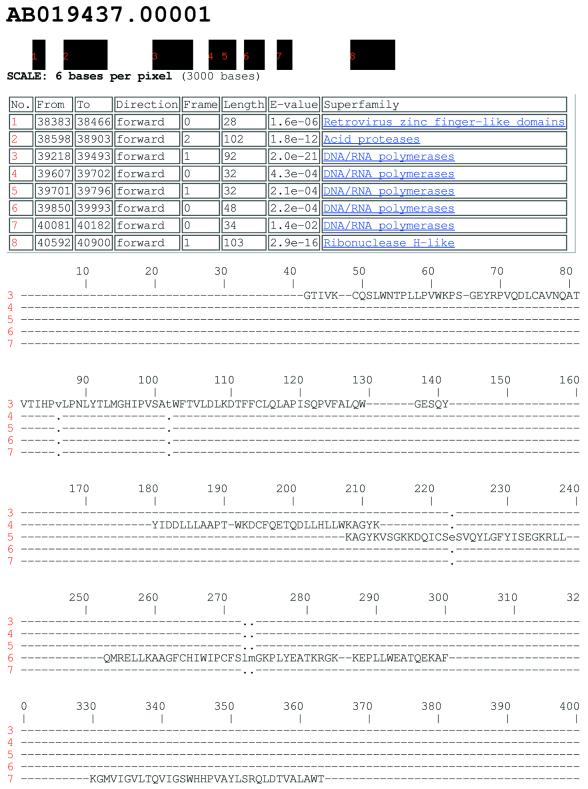
Figure 2.
A section of a result of a nucleotide search of human contig AB019437.00001 clearly showing a DNA/RNA polymerase domain consisting of exons 3–7. The alignment shows how the exons combine in order to make up a complete domain.
Multiple sequence alignments
The models are used to generate multiple sequence alignments to sequences of known structure. A sequence with structural domains assigned (as above) can be aligned to a known sequence of the structural domain in question. On the public server there is a link to the alignment from the result page from a sequence query (Fig. 1). The server contains all PDB sequences and all complete genome sequences, which can be added to obtain a multiple alignment; users can also upload their own sequences for addition to the multiple alignment.
In the absence of a sequence query, the multiple alignments can be reached via links from SCOP or a keyword search on the server.
Genome assignments
The SUPERFAMILY procedure has been used to carry out structural assignments to all complete genomes (Tables 1 and 2). The assignments cover ∼35% of eukaryote and 45% of prokaryote sequence, which is estimated as half of the soluble protein domains. This coverage is expected to increase as structural genomics projects solve more novel structures, giving a more complete structural picture of the genomes.
Table 1. The genome assignments for 56 genomes using the model library and assignment procedure.
| Genome | A | B | C | D | E | F | G | H |
|---|---|---|---|---|---|---|---|---|
| Arabidopsis thaliana | at | E | 25 470 | 13 320 | 52 | 38 | 17 957 | 564 |
| Homo sapiens | hs | E | 23 867 | 11 661 | 49 | 37 | 21 201 | 595 |
| Caenorhabditis elegans | ce | E | 19 705 | 7851 | 40 | 29 | 12 628 | 537 |
| Drosophila melanogaster | dm | E | 14 331 | 6851 | 48 | 34 | 11 479 | 554 |
| Mesorhizobium loti | mk | B | 6752 | 3552 | 53 | 44 | 4631 | 433 |
| Saccharomyces cerevisiae | sc | E | 6297 | 2770 | 44 | 33 | 3760 | 461 |
| Pseudomonas aeruginosa | pa | B | 5570 | 3079 | 55 | 45 | 4261 | 439 |
| Escherichia coli o157 | eo | B | 5283 | 2502 | 47 | 41 | 3346 | 454 |
| Escherichia coli | ec | B | 4289 | 2292 | 53 | 45 | 3097 | 453 |
| Mycobacterium tuberculosis CDC1551 | mu | B | 4187 | 1911 | 46 | 41 | 2594 | 391 |
| Bacillus subtilis | bs | B | 4100 | 2027 | 49 | 44 | 2754 | 417 |
| Bacillus halodurans | bh | B | 4066 | 2000 | 49 | 43 | 2688 | 415 |
| Mycobacterium tuberculosis | mb | B | 3918 | 1959 | 50 | 41 | 2650 | 392 |
| Vibrio cholerae | vc | B | 3835 | 1852 | 48 | 42 | 2527 | 424 |
| Caulobacter crescentus | cc | B | 3737 | 1997 | 53 | 46 | 2663 | 404 |
| Clostridium acetobutylicum | ca | B | 3672 | 1819 | 50 | 41 | 2382 | 401 |
| Cyanobacterium synechocystis | cs | B | 3169 | 1589 | 50 | 42 | 2164 | 379 |
| Deinococcus radiodurans | dr | B | 3102 | 1561 | 50 | 42 | 2007 | 379 |
| Sulfolobus solfataricus | ss | A | 2977 | 1412 | 47 | 40 | 1790 | 323 |
| Xylella fastidiosa | xf | B | 2766 | 1097 | 40 | 41 | 1477 | 359 |
| Aeropyrum pernix | ap | A | 2694 | 836 | 31 | 33 | 1067 | 289 |
| Staphylococcus aureus | sa | B | 2594 | 1313 | 51 | 43 | 1728 | 368 |
| Archaeoglobus fulgidus | af | A | 2407 | 1238 | 51 | 45 | 1664 | 320 |
| Lactococcus lactis | ll | B | 2266 | 1170 | 52 | 43 | 1514 | 334 |
| Streptococcus pneumoniae | sr | B | 2094 | 1044 | 50 | 43 | 1351 | 330 |
| Neisseria meningitidis A | nn | B | 2065 | 958 | 46 | 42 | 1266 | 342 |
| Pyrococcus horikoshii | ph | A | 2064 | 904 | 44 | 40 | 1175 | 294 |
| Halobacterium | hb | A | 2058 | 1023 | 50 | 42 | 1351 | 306 |
| Neisseria meningitidis | nm | B | 2025 | 941 | 46 | 43 | 1264 | 342 |
| Pasteurella multocida | pm | B | 2014 | 1112 | 55 | 46 | 1467 | 359 |
| Methanobacterium thermoautotrophicum | mt | A | 1869 | 971 | 52 | 44 | 1297 | 307 |
| Thermotoga maritima | tm | B | 1846 | 1003 | 54 | 46 | 1335 | 343 |
| Pyrococcus abyssi | pb | A | 1765 | 957 | 54 | 45 | 1231 | 298 |
| Methanococcus jannaschii | mj | A | 1715 | 872 | 51 | 45 | 1132 | 288 |
| Haemophilus influenzae | hi | B | 1709 | 943 | 55 | 48 | 1243 | 341 |
| Streptococcus pyogenes | sq | B | 1696 | 887 | 52 | 44 | 1189 | 328 |
| Campylobacter jejuni | cj | B | 1634 | 845 | 52 | 43 | 1095 | 329 |
| Mycobacterium leprae | ml | B | 1605 | 844 | 53 | 48 | 1215 | 327 |
| Helicobacter pylori | hp | B | 1553 | 670 | 43 | 38 | 882 | 295 |
| Aquifex aeolicus | aa | B | 1522 | 902 | 59 | 49 | 1203 | 334 |
| Thermoplasma volcanium | tv | A | 1499 | 795 | 53 | 45 | 1034 | 284 |
| Helicobacter pylori J99 | hq | B | 1491 | 681 | 46 | 38 | 896 | 287 |
| Thermoplasma acidophilum | ta | A | 1478 | 795 | 54 | 45 | 1051 | 286 |
| Chlamydophila pneumoniae AR39 | cq | B | 1110 | 443 | 40 | 36 | 625 | 243 |
| Chlamydophila pneumoniae J138 | cp | B | 1070 | 446 | 42 | 36 | 628 | 243 |
| Chlamydophila pneumoniae | cr | B | 1052 | 443 | 42 | 36 | 625 | 242 |
| Treponema pallidum | tp | B | 1031 | 467 | 45 | 38 | 655 | 235 |
| Chlamydia muridarum | cm | B | 909 | 423 | 47 | 39 | 604 | 234 |
| Chlamydia trachomatis | ct | B | 894 | 419 | 47 | 40 | 597 | 235 |
| Borrelia burgdorferi | bb | B | 850 | 415 | 49 | 42 | 574 | 225 |
| Rickettsia prowazekii | rp | B | 834 | 437 | 52 | 44 | 605 | 248 |
| Mycoplasma pulmonis | mq | B | 782 | 363 | 46 | 34 | 485 | 186 |
| Mycoplasma pneumoniae | mp | B | 677 | 308 | 45 | 35 | 414 | 179 |
| Ureaplasma urealyticum | uu | B | 611 | 267 | 44 | 33 | 367 | 170 |
| Buchnera sp. | bn | B | 564 | 380 | 67 | 56 | 560 | 248 |
| Mycoplasma genitalium | mg | B | 480 | 261 | 54 | 41 | 362 | 172 |
For each genome the table shows in order: the name of the species of the genome; a two-character code (A); the domain, where ‘E’ is for eukaryota, ‘A’ is for archea and ‘B’ is for bacteria (B); the number of genes comprising the genome (C); the number of genes which have at least one SCOP domain assigned (D); the percentage of genes with at least one domain assigned (E); the percentage of the actual sequence covered by SCOP domains because multi-domain genes may have some domains assigned but not others (F); the total number of domains assigned (G); the total number (out of a possible 859) of superfamilies represented by at least one domain in the genome (H).
Table 2. The assignments for 11 miscellaneous sequence sets including, amongst other things, five alternative human gene sets and some incomplete genomes.
| Genome | A | B | C | D | E | F | G | H |
|---|---|---|---|---|---|---|---|---|
| Viridiplantae sequences from GenPept | sp | E | 46 369 | 31 232 | 67 | 58 | 64 711 | 546 |
| Softberry human gene predictions | hv | E | 38 170 | 15 235 | 40 | 31 | 28 223 | 613 |
| Ensembl 0.8 human gene predictions | hx | E | 29 303 | 14 437 | 49 | 39 | 25 558 | 597 |
| Ensembl 1.0 human gene predictions | hs | E | 27 615 | 13 210 | 48 | 37 | 23 402 | 595 |
| Affymetrix human gene predictions | hu | E | 21 111 | 10 339 | 49 | 37 | 19 876 | 581 |
| Mus musculus cDNAs | mm | E | 21 076 | 6223 | 30 | 29 | 8047 | 496 |
| Human known genes | ht | E | 8243 | 4995 | 61 | 41 | 9769 | 531 |
| Mus musculus incomplete genome | mn | E | 6978 | 3463 | 50 | 39 | 5599 | 391 |
| Oryza sativa incomplete genome | os | E | 2425 | 759 | 31 | 28 | 987 | 177 |
| Guillardia theta nucleomorph genome | gt | E | 485 | 203 | 42 | 33 | 261 | 92 |
| Rhizobium plasmid | pn | P | 417 | 202 | 48 | 40 | 250 | 77 |
In Table 1 the current Ensembl (version 1.1) is used for Homo sapiens.
The SCOP classification of the structural domains in genomes provides a framework for comparing superfamilies within and across genomes. The public server provides statistics, and the breakdown of the genomes into superfamilies of different sizes. Within each superfamily of a given genome the individual genes may be displayed, with links to their domain architecture and sequence alignments.
A growing number of genome assignments are served via a distributed annotation system (DAS) server at http://supfam.org:8080/das allowing people to view the annotation from different sources in a single browser. To use this service a DAS client is required which can be obtained from http://www.biodas.org.
APPLICATIONS
The most straightforward application is a simple sequence search, of which the public server currently (pre-publication) receives over 1000 per month. Many larger sets of sequences have been run as special requests for specific studies; the database is used on several structural genomics projects’ targets (e.g. SPiNE at http://spine.mbb.yale.edu/spine).
Although the assignments to nucleotide sequence do not provide complete gene predictions, they can be used as information contributing to a gene prediction. Current work is generating the data for the human genome for this purpose.
The genome assignments provide annotation of the genes, much of which is novel. This information is not just accessed by users of the database but is also used by several genome projects (including all completed large eukaryotes) to aid their annotation efforts, or verbatim as annotation in its own right.
The SUPERFAMILY data provides a framework which already forms the basis of several ongoing genomic studies (9,10). The data is also used by the HIGH database (http://genomesapiens.org) of immunoglobulin genes in human.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Thomas Down for help setting up the DAS server, Matthew Bashton for contributing to web design, and the SCOP authors for helpful discussions.
REFERENCES
- 1.Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- 2.Karplus K., Barrett,C. and Hughey,R. (1998) Hidden Markov models for detecting remote protein homologies. Bioinformatics, 14, 846–856. [DOI] [PubMed] [Google Scholar]
- 3.Gough J., Karplus,K., Hughey,R. and Chothia,C. (2001) Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol., 313, 903–919. [DOI] [PubMed] [Google Scholar]
- 4.Bateman A., Birney,E., Durbin,R., Eddy,S.E., Lowe,K.L. and Sonnhammer,E.L. (2000) The Pfam protein families database. Nucleic Acids Res., 28, 263–266. Updated article in this issue: Nucleic Acids Res. (2002), 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schultz J., Copley,R.R., Doerks,T., Ponting,C.P. and Bork,P. (2000) SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res., 28, 231–234. Updated article in this issue: Nucleic Acids Res. (2002), 30, 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apweiler R., Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,L., Corpet,F., Croning,M.D.R. et al. (2000) InterPro—an integrated documentation resource for protein families, domains and functional sites. Bioinformatics, 16, 1145–1150. [DOI] [PubMed] [Google Scholar]
- 7.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orengo C.A., Michie,A.D., Jones,S., Jones,D.T., Swindells,M.B. and Thornton,J.M. (1997) CATH—a hierarchic classification of protein domain structures. Structure, 5, 1093–1108. [DOI] [PubMed] [Google Scholar]
- 9.Apic G., Gough,J. and Teichmann,S.A. (2001) Domain combinations in archael, eubacterial and eukaryotic proteins. J. Mol. Biol., 310, 311–325. [DOI] [PubMed] [Google Scholar]
- 10.Teichmann S.A., Rison,S.C.G., Thornton,J.M., Riley,M., Gough,J. and Chothia,C. (2001) The evolution and structural anatomy of the small molecule metabolic pathways in Escherichia coli. J. Mol. Biol., 311, 693–708. [DOI] [PubMed] [Google Scholar]