Abstract
NaV1.7, a membrane-bound voltage-gated sodium channel, is preferentially expressed along primary sensory neurons, including their peripheral & central nerve endings, axons, and soma within the dorsal root ganglia and plays an integral role in amplifying membrane depolarization and pain neurotransmission. Loss- and gain-of-function mutations in the gene encoding NaV1.7, SCN9A, are associated with a complete loss of pain sensation or exacerbated pain in humans, respectively. As an enticing pain target supported by human genetic validation, many compounds have been developed to inhibit NaV1.7 but have disappointed in clinical trials. The underlying reasons are still unclear, but recent reports suggest that inhibiting NaV1.7 in central terminals of nociceptor afferents is critical for achieving pain relief by pharmacological inhibition of NaV1.7. We report for the first time that NaV1.7 mRNA is expressed in putative projection neurons (NK1R+) in the human spinal dorsal horn, predominantly in lamina 1 and 2, as well as in deep dorsal horn neurons and motor neurons in the ventral horn. NaV1.7 protein was found in the central axons of sensory neurons terminating in lamina 1–2, but also was detected in the axon initial segment of resident spinal dorsal horn neurons and in axons entering the anterior commissure. Given that projection neurons are critical for conveying nociceptive information from the dorsal horn to the brain, these data support that dorsal horn NaV1.7 expression may play an unappreciated role in pain phenotypes observed in humans with genetic SCN9A mutations, and in achieving analgesic efficacy in clinical trials.
Keywords: NaV1.7, spinal cord, dorsal horn
Classification: Biological Sciences
Introduction
In the mid-2000s, it was found that loss- and-gain-of-function mutations in the gene encoding voltage-gated sodium channel (NaV) 1.7 (Baker and Nassar, 2020), SCN9A, resulted in congenital pain insensitivity (Cox et al., 2006) or in extreme pain disorders like primary erythromelalgia (Yang et al., 2004; Mann et al., 2019), paroxysmal extreme pain disorder (Fertleman et al., 2006; Hua et al., 2022), and idiopathic small fiber neuropathy (Faber et al., 2012), respectively. The rarity of finding a gene that is critical for sensory pathophysiology and that is backed by human genetics framed NaV1.7 as a prime therapeutic target for pain treatment. Several NaV1.7 inhibitors have been developed, and a series of clinical trials have been conducted with mixed reports of success (Price et al., 2017; McDonnell et al., 2018; Kingwell, 2019; Alles and Smith, 2021; Biogen, 2021; Eagles et al., 2022). While these outcomes could be due to poor drug pharmacokinetics and/or limited selectivity towards other sodium channels, one hypothesis is that peripherally restricted drugs lack significant efficacy because spinal NaV1.7 must be targeted to achieve analgesia. This idea has been supported by mouse studies which suggest that NaV1.7 in the central terminals of nociceptors is critical for nociceptive neurotransmission in the dorsal horn (MacDonald et al., 2021). However, it is also notable that mouse genetic studies generating gain-of-function mutations in Scn9a have failed to recapitulate human phenotypes (Chen et al., 2021).
NaV1.7 is predominantly expressed in the peripheral nervous system in sympathetic neurons and nociceptive sensory neurons in the dorsal root ganglia (DRG) and trigeminal ganglia (Hameed, 2019a). Subcellularly, it is localized to the membrane of sensory neurons including their soma, peripheral axons that innervate the skin, muscle and other organs, and central axons that cross the blood brain barrier and terminate in the dorsal horn of the spinal cord (Black et al., 2012; Shiers et al., 2020; Shiers et al., 2021). It is a key regulator of neuronal excitability as it mediates Na currents during membrane depolarization and action potential firing (McDermott et al., 2019; Middleton et al., 2022) and is dysregulated in pathological pain conditions in both rodents and humans (Black et al., 2008; Li et al., 2018; Sun et al., 2018; Hameed, 2019b; Alvarez et al., 2021; Liu et al., 2021; Nascimento et al., 2022). For this reason, the peripheral nociceptive system has been considered as the primary site of action for therapeutic targeting. However, peripherally restricted NaV1.7 inhibitors like Pfizer’s small molecule inhibitor, PF-05089771, and Teva/Xenon’s topical small-molecule inhibitor, TV-45070, failed to show significant analgesic efficacy in clinical trials (Price et al., 2017; McDonnell et al., 2018), suggesting that central nervous system-targeting of NaV1.7 may be critical for analgesia.
While absent in the rodent brain(Lein et al., 2007; Allen Institute for Brain Science, 2022a) and human cortex,(Allen Institute for Brain Science, 2022b) NaV1.7 protein appears to be entirely localized to presynaptic axons and fibers in both the rodent and human spinal dorsal horn (Black et al., 2012; Shiers et al., 2021). A central analgesic mechanism for NaV1.7 was recently proposed wherein NaV1.7 -null mice that were pain insensitive, retained nociceptor firing properties but displayed opioid-dependent impaired synaptic transmission and neurotransmitter release at sensory neuron presynaptic terminals in the spinal dorsal horn (MacDonald et al., 2021). Indeed, this could be an important mechanism of action for NaV1.7’s key role in nociception but there is some evidence, although inconsistent, for NaV1.7 expression post-synaptically within the spinal cord. NaV1.7 mRNA has been identified in several subsets of mouse spinal cord neurons using single-cell RNA sequencing (Russ et al., 2021), but only its motor neuron expression could be confirmed with in situ hybridization (Alles et al., 2020; Allen Institute for Brain Science, 2022a). However, electron microscopy revealed a substantial proportion of NaV1.7 immunoreactivity was localized to dendrites of mouse spinal dorsal horn neurons, but these authors proposed that the protein originated from presynaptic fibers and was transferred to post-synaptic sites through a mechanism that remains to be elucidated (Alles et al., 2020).
We hypothesized that SCN9A mRNA and NaV1.7 protein might be expressed by human dorsal horn projection neurons. If post-synaptic NaV1.7 exists, especially within the pain neurocircuitry in the spinal dorsal horn such as projection neurons, this would provide important evidence to explain the lack of efficacy of peripherally restricted NaV1.7 inhibitors and invigorate research towards investigating how NaV1.7 expression in intrinsic dorsal horn neurons regulates nociception in addition to its well-described role in nociceptors.
Methods
Tissue preparation
All human tissue procurement procedures were approved by the Institutional Review Boards at the University of Texas at Dallas. Human lumbar spinal cords were surgically extracted using a ventral approach (Valtcheva et al., 2016) from organ donors within 4 hours of cross-clamp, frozen immediately in dry ice, and stored in a −80°C freezer. All tissues were recovered in the Dallas area via a collaboration with the Southwest Transplant Alliance. Donor information is provided in Table 1. All tissues were collected from neurologic determination of death donors. Human spinal cords were gradually embedded in OCT in a cryomold by adding small volumes of OCT over dry ice to avoid thawing. All tissues were cryostat sectioned at 20 μm onto SuperFrost Plus charged slides.
Table 1.
Donor Demographic Information
| Donor ID | Age | Sex | Race | Cause of Death | Experiment used |
|---|---|---|---|---|---|
| AICS171 | 19 | M | White | Anoxia/Drug Overdose | RNAscope SCN9A/TACR1 |
| AIAG131 | 61 | F | White | Anoxia/Cardiac Arrest | RNAscope SCN9A/TACR1 |
| AHLC292 | 51 | M | White | CVA/Stroke | RNAscope SCN9A/TACR1 |
| AIBR013 | 18 | F | Black | Anoxia/Seizure | IHC NaV1.7/CGRP/Bassoon |
| AHJZ471 | 24 | M | White | Head Trauma/MVA | IHC NaV1.7/CGRP/Bassoon |
| AHJD193 | 53 | F | White | CVA/Stroke | IHC NaV1.7/CGRP/Bassoon |
| AIEJ414 | 69 | M | Black | CVA/Stroke | IHC NaV1.7/CGRP/Bassoon |
| AJD2236 | 56 | M | White | Head Trauma/GSW | IHC NaV1.7/Ankyrin-G/MAP2 |
| AIFN459 | 44 | F | Hispanic | CVA/Stroke | IHC NaV1.7/Ankyrin-G/MAP2 |
| AJEK100 | 22 | M | Black | Overdose/Cardiovascular | IHC NaV1.7/Ankyrin-G/MAP2 |
| AIGD395 | 45 | F | Black | CVA/Stroke | IHC NaV1.7/Ankyrin-G/MAP2 |
Sections were only briefly thawed in order to adhere to the slide but were immediately returned to the −20°C cryostat chamber until completion of sectioning. The slides were then immediately utilized for histology. 3 sections from each donor were stained in each experiment, and 3–4 donors were used in each experiment. Donor tissues used in each experiment are also indicated in Table 1.
RNAscope in situ hybridization
RNAscope (multiplex fluorescent v2 kit, Cat 323100) was performed as instructed by Advanced Cell Diagnostics (ACD). The protease IV digestion (2 minute incubation) was optimized for human spinal cord. The probe combination used was human specific SCN9A (Cat 562251, ACD) paired with Cy3 (Cat NEL744001KT, Akoya Biosciences) and TACR1 (Cat 310701–3, ACD) paired with Cy5 (Cat NEL745001KT, Akoya Biosciences). The third channel (green, 488) was left unstained to detect background lipofuscin. All tissues were checked for RNA quality by using a positive control probe cocktail (Cat 320861, ACD) which contains probes for high, medium and low-expressing mRNAs that are present in all cells (ubiquitin C > Peptidyl-prolyl cis-trans isomerase B > DNA-directed RNA polymerase II subunit RPB1). A negative control probe against the bacterial DapB gene (Cat 310043, ACD) was used to reference non-specific/background label. Tissues were coverslipped with Prolong Gold Antifade Reagent (Cat P36930, Fisher Scientific).
Immunohistochemistry (IHC)
Slides were removed from the cryostat and immediately transferred to cold 10% formalin (4°C; pH 7.4) for 15 minutes. The tissues were then dehydrated in 50% ethanol (5 min), 70% ethanol (5 min), 100% ethanol (5 min), 100% ethanol (5 min) at room temperature. The slides were air dried briefly and then boundaries were drawn around each section using a hydrophobic pen (ImmEdge PAP pen, Vector Labs). When hydrophobic boundaries had dried, the slides were submerged in blocking buffer (10% Normal Goat Serum, 0.3% Triton-X 100 in 0.1M Phosphate Buffer (PB) for 1 hour at room temperature. Slides were then rinsed in 0.1M PB, placed in a light-protected humidity-controlled tray and incubated in primary antibodies diluted in blocking buffer overnight at 4°C. The next day, slides were washed in 0.1M PB and then incubated in their respective secondary antibody diluted at 1:2000 with DAPI (1:5000; Cayman Chemical; Cat 14285) in blocking buffer for 1 hour at room temperature. The antibodies used are provided in Table 2. The sections were washed in 0.1M PB and then covered with True Black (20% diluted in 70% Ethanol), a blocker of lipofuscin, for 1 minute. Sections were then washed in water, air dried and coverslipped with Prolong Gold Antifade reagent (Cat P36930, Fisher Scientific).
Table 2.
Antibodies used for immunohistochemistry.
| Antibody | Vendor | Catalog # or Clone # | RRID | Dilution/Conc. |
|---|---|---|---|---|
| Mouse-anti- NaV1.7 | NeuroMab | N68/6 | AB_2184355 | 2μg/mL |
| Rabbit-anti-CGRP | ImmunoStar | 24112 | AB_572217 | 1:1000 |
| Mouse-anti-Bassoon | Enzo Life Sci | SAP7F407 | AB_11181058 | 2μg/mL |
| Mouse-anti-Ankyrin G | NeuroMab | N106/36 | AB_10673030 | 2μg/mL |
| Chicken-anti-MAP2 | Novus Bio | NB300–213 | AB_2138178 | 2μg/mL |
| Goat-anti-rabbit H&L 647 | ThermoFisher | A21245 | AB_2535813 | 1:2000 |
| Goat-anti-chicken H&L 647 | ThermoFisher | A21449 | AB_2535866 | 1:2000 |
| Goat-anti-mouse IgG2a 555 | ThermoFisher | A21137 | AB_2535776 | 1:2000 |
| Goat-anti-mouse IgG2a 488 | ThermoFisher | AB21131 | AB_2535771 | 1:2000 |
| Goat-anti-mouse IgG1 488 | ThermoFisher | A21121 | AB_2535764 | 1:2000 |
| Goat-anti-mouse IgG1 555 | ThermoFisher | A21127 | AB_2535769 | 1:2000 |
NaV1.7 antibody information and validation
The NaV1.7 mouse monoclonal antibody has been knockout validated using immunocytochemistry on mouse cultured DRG neurons and using IHC on rat brain (Grubinska et al., 2019). We have also recently shown that this antibody robustly stains human DRG and shows a specific and similar expression pattern to its mRNA (Shiers et al., 2020; Shiers et al., 2021; Tavares-Ferreira et al., 2022).
Imaging
All sections were imaged on an Olympus FV3000 confocal microscope. Acquisition parameters were set based on guidelines for the FV3000 provided by Olympus. In particular, the gain was kept at the default setting 1, HV ≤ 600, offset = 4, and laser power ≤ 10%. For the MAP2/NaV1.7 experiment, an epifluorescent mosaic image of the entire spinal cord section was captured on an Olympus vs120 slide scanner as a means to visualize the precise anatomical location of the confocal images. Laminar boundaries were drawn using a reference atlas (Sengul et al., 2013; Shiers et al., 2021).
For human spinal cord RNAscope, multiple 20X confocal images with overlapping lipofuscin signal were acquired of adjacent regions of the spinal dorsal horn (mosaic imaging). The images were manually stitched together in Adobe Photoshop (v21.2.12, 2020) by overlaying the overlapping lipofuscin in each image. Once the entire spinal dorsal horn was visualized, laminar boundaries were drawn in Adobe Illustrator using a reference atlas (Sengul et al., 2013; Shiers et al., 2021). This imaging was performed on one section from 3 donors.
Images Analysis
Raw images were brightened and contrasted in Olympus CellSens (v1.18). Images were pseudocolored for visualization purposes.
For RNAscope, the number of nuclei expressing SCN9A-only, TACR1-only, and SCN9A-and-TACR1 were counted in each laminar region. For density analysis, the number of RNAscope positive nuclei (neurons) was divided by the area of each laminar subregion (neuron / μm2). One section from three donors was analyzed. Graphs were generated using GraphPad Prism version 8.4.3 (GraphPad Software, Inc. San Diego, CA USA).
Results
SCN9A mRNA is expressed in TACR1+ putative projection neurons in the human spinal dorsal horn
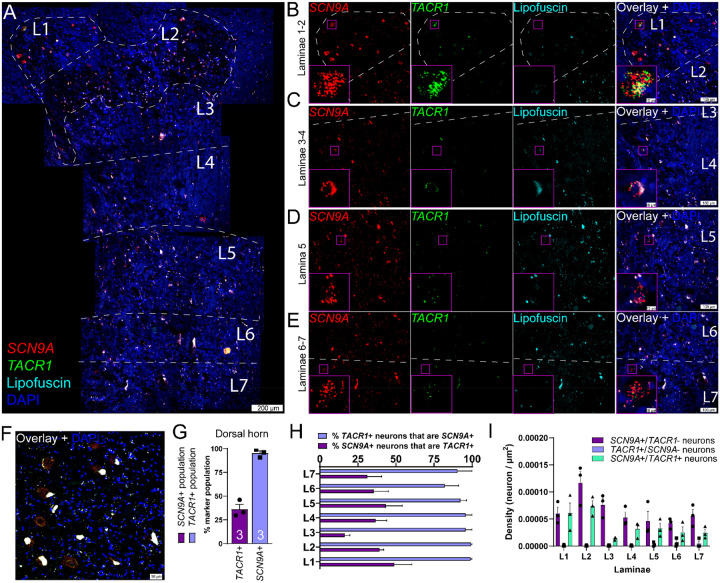
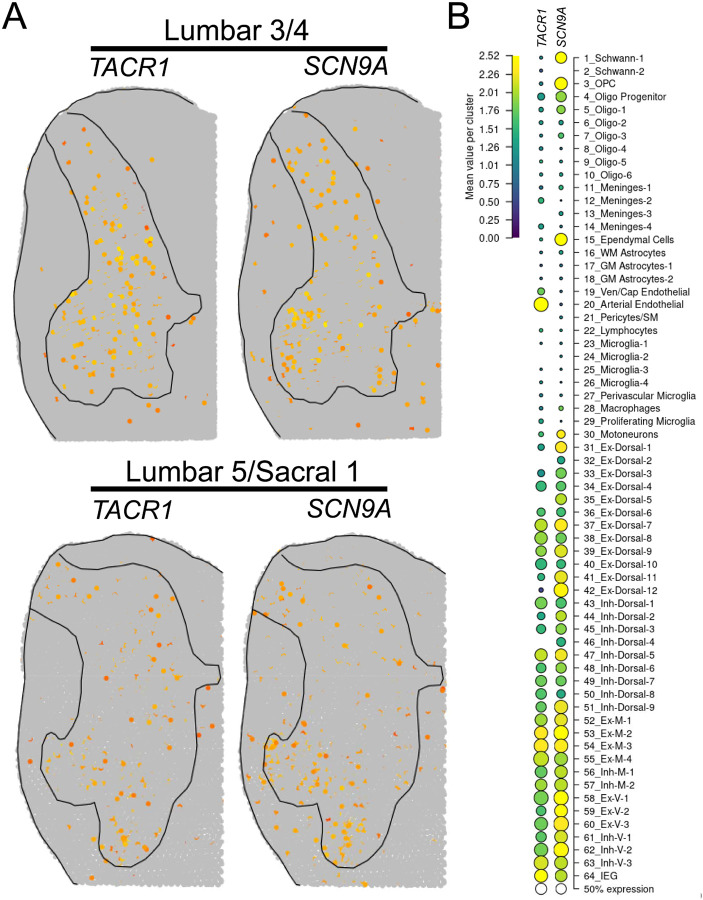
The substance P receptor (protein: Nk1r, mRNA: Tacr1) labels a subset of projection neurons in the rodent spinal dorsal horn that transmit pain and itch information to the parabrachial nucleus in the brainstem (Blomqvist and Mackerlova, 1995; Mantyh et al., 1997; Carstens et al., 2010). These neurons are localized to the superficial laminae, primarily lamina 1 (Mantyh et al., 1997; Carstens et al., 2010), but they are also found in the deep dorsal horn, mostly in lamina V (Brown et al., 1995). We assessed the distribution of SCN9A (NaV1.7) and TACR1 (Nk1r) mRNA in the human spinal dorsal horn using RNAscope. We found that SCN9A and TACR1 mRNAs were detected in cells throughout all laminae in the human spinal dorsal horn (FIG 1A–E), and in motor neurons in the ventral horn (FIG 1F). Large SCN9A/TACR1 co-expressing neurons were prevalent in lamina 1 and were ~30–40μm in diameter (FIG 1B). In the dorsal horn (laminae 1–7), virtually all of the TACR1+ cells were SCN9A+ (95%), but only 36% of SCN9A+ cells were TACR1+ (FIG 1G–H). SCN9A+ and TACR1+ co-expressing neurons were more densely populated in laminae 1–2 (FIG 1I). This distribution closely resembles recently published human spinal cord spatial transcriptomic data (Yadav et al., 2023) in which both SCN9A and TACR1 mRNAs were found throughout the dorsal and ventral horns (FIG 2A). SCN9A was detected in a broad population of excitatory and inhibitory dorsal horn neurons using single-nucleus RNA sequencing of human spinal cord (Yadav et al., 2023), many of which co-expressed TACR1 (FIG 2B).
Figure 1. Distribution of SCN9A (NaV1.7) and TACR1 (NK1R) mRNAs in the human spinal dorsal horn using RNAscope.
A) Representative stitched mosaic image of human lumbar spinal cord labeled with RNAscope in situ hybridization for SCN9A (red) and TACR1 (green) mRNAs and co-stained with DAPI (cyan). The 488 channel was left unstained (pseudocolored to cyan) to reveal background autofluorescence and lipofuscin which is present in all human neurons. 20X images for each channel are shown for B) laminae 1–2, C) laminae 3–4, D) lamina 5, and E) laminae 6–7. The inset images are a zoomed-in, cropped image of a single SCN9A/TACR1 co-positive cell. F) SCN9A and TACR1 mRNAs were coexpressed by motor neurons in the ventral horn. G) Percentage of SCN9A+ neurons in the dorsal horn that coexpressed TACR1 (purple bar), and the percentage of TACR1+ neurons that coexpressed SCN9A (blue bar). H) Percentage of TACR1+ neurons that were copositive for SCN9A, and the percentage of SCN9A+ neurons that were copositive for TACR1 for each lamina (L1-L7) of the spinal dorsal horn. I) Density of each neuronal subpopulation within each lamina (neuron/μm2). Scales bar = panel A: 200 μm; panels B-E: 100 μm and inset is 10 μm; panel F: 50 μm.
Figure 2. Spatial and single-nuclear RNA-sequencing detection of SCN9A and TACR1 in the human spinal dorsal horn.
A) Normalized spatial transcriptomic gene expression for SCN9A and TACR1 per barcoded spot on aggregated human lumbar spinal cord sections (Yadav et al., 2023). Solid lines mark grey matter boundaries. B) Dot plot showing the average gene expression for SCN9A and TACR1 for each human spinal cord cluster identified using single-nucleus RNA sequencing.
Data found at https://vmenon.shinyapps.io/humanspinalcord/
NaV1.7 protein is localized pre-synaptically in the human spinal cord.
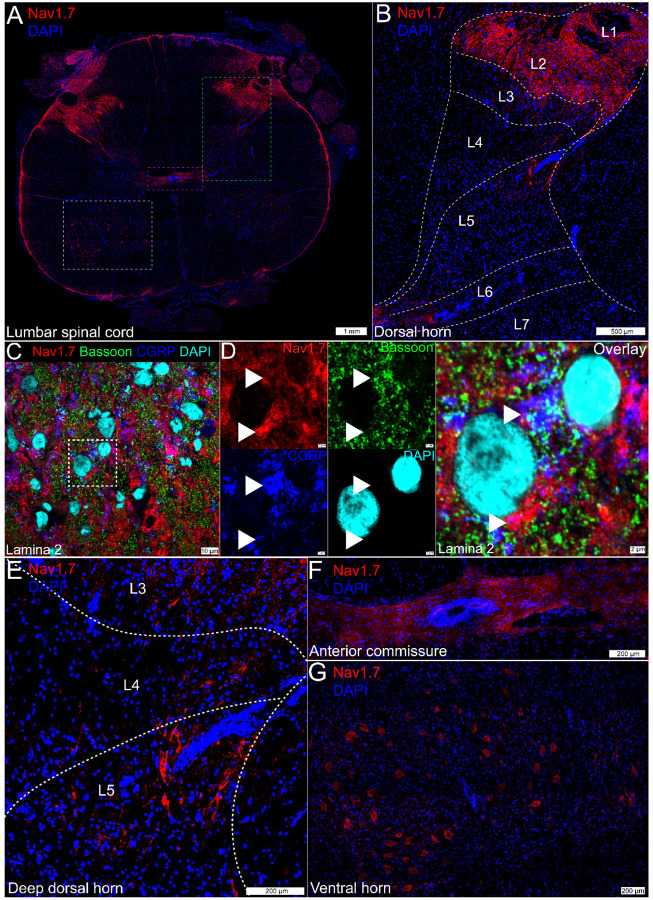
NaV1.7 protein staining in the human spinal cord was robustly found in the dorsal rootlet and substantia gelatinosa which comprises lamina 1 and 2, as we have previously reported (Shiers et al., 2021) (FIG 3A–3B).
Figure 3. NaV1.7 protein expression in the human lumbar spinal cord.
A) Mosaic image of NaV1.7 (red) protein staining in the human lumbar spinal cord, costained with DAPI (blue). The green box represents B) the dorsal horn from L1-L6 where robust NaV1.7 neuropil staining was observed in lamina 1-lamina 2. C) Representative 100X image of NaV1.7 (red) protein co-stained with the nociceptive presynaptic marker, CGRP (blue), the presynaptic active zone protein, Bassoon (green), and DAPI (cyan) in lamina 2 of the spinal dorsal horn. A cropped, zoomed in area outlined in white is shown in D) where NaV1.7 signal is colocalized with CGRP and/or Bassoon (white arrows) or is in close proximity to these proteins. However, not all NaV1.7 signal appeared to be localized to the presynaptic compartment. E) Axonal NaV1.7 was observed in the deeper laminae, most prominently in lamina 5. The magenta box in panel A represents F) the anterior commissure where NaV1.7 was also detected in axons. The white box in panel A represents G) the ventral horn where NaV1.7 was localized to the cytoplasm of large motor neurons. Scales bar = panel A: 1 mm; panel B: 500 μm; panels C: 10 μm; panel D: 2 μm; panels E-G: 200 μm.
NaV1.7 staining gave a neuropil-like pattern, potentially indicative of synaptic staining, throughout laminae 1–2 (FIG 3B). When colabeled with the nociceptive presynaptic marker, CGRP, and the presynaptic active-zone protein, Bassoon (FIG 3C), we observed NaV1.7 signal that co-localized or was in close proximity to these presynaptic markers (Fig 3D). However, not all of the NaV1.7 signal in the human spinal cord appeared to be localized to the presynaptic compartment.
NaV1.7 was sparsely detected in axons in deeper lamina, with the highest abundance in lamina 5 (FIG 3E) and also in axons entering the anterior commissure, where dorsal horn projection neurons cross hemispheres to enter the contralateral spinal thalamic tract (FIG 3F). As tracing studies in cats and macaques have demonstrated that neurons projecting to the spinal thalamic tract originate in lamina 1 and lamina 5, it is possible that the non-presynaptic signal in the superficial lamina as well as the axonal signal in the deep lamina and anterior commissure could represent NaV1.7 protein expressed by projection neurons (Willis et al., 1979; Jones et al., 1987; Craig, 2006), a hypothesis we explored with additional experiments. Additionally, we observed NaV1.7 protein staining in the cytoplasm of motor neurons in the ventral horn (FIG 3G).
Evidence for post-synaptic NaV1.7 protein in the human spinal cord.
In an effort to identify post-synaptic NaV1.7 immunoreactivity in the spinal dorsal horn, we co-stained NaV1.7 with a variety of subcellular protein markers. First, we assessed NaV1.7 with the cytoskeletal marker, MAP2, which is primarily localized to dendrites (FIG 4A). We assessed the dorsal horn from 3 organ donors but observed no convincing evidence for any NaV1.7 signal that was localized to the dendritic compartment of resident dorsal horn neurons, nor their soma in neither lamina 1 (FIG 4B), nor lamina 2 (FIG 4C), nor lamina 4–5 (FIG 4D). In the anterior commissure, intensely labeled NaV1.7 axonal fibers could be observed crossing hemispheres (FIG 4E), but we could not determine what neurons these axons originated from; however, it is likely they are projection neurons as this is a major hub for cross-hemispheric spinal neurotransmission.
Figure 4. Evidence for post-synaptic NaV1.7 expression in the human spinal cord.
A) Representative 10X image of NaV1.7 (red), MAP2 (green), and DAPI (blue) staining in the human lumbar dorsal horn. The white outline demarcates the substantia gelatinosa which comprises lamina 1 and lamina 2. B) Representative 40X images of NaV1.7 (red), MAP2 (green), and DAPI (blue) in lamina 1, and C) lamina 2. White arrows point to MAP2 signal that is localized around resident neurons (appears to be the plasma membrane) but are absent of NaV1.7 signal. The image inset in panel C shows a 100X image of a large, L2 neuron with a large apical dendrite that is devoid of NaV1.7 signal. D) 20X image of NaV1.7-positive axonal fibers in the deeper lamina around L4-L5. The white arrow points to NaV1.7 and MAP2 copositive signal (yellow in overlay) that does not have a nucleus and is not a cell body. E) 20X image of NaV1.7 staining in the anterior commissure (ac) where intensely labeled NaV1.7-positive axons are highlighted (white arrow). F) A 100X image of NaV1.7 (red), Ankyrin-G (green), and DAPI (blue) staining in a motor neuron in the ventral horn. G) A cropped, zoomed-in image of NaV1.7 (red), Ankyrin-G (green), and DAPI (blue) signal in a lamina 2 dorsal horn neuron. Scales bar = panel A: 100 μm; panels B-C: 20 μm; panel C inset: 10 μm; panel D-E: 50 μm; panel F: 10 μm, panel G: 2 μm.
Interestingly, we noted that NaV1.7 appeared to be localized to the cytoplasm of motor neurons, including their soma and what appeared to be their axon initial segment (AIS). Recent work with NaV1.7 in the rodent DRG found that sensory neurons contain an AIS that it is enriched with NaV1.7 and that its localization there is critical for spontaneous activity in neuropathic pain (Nascimento et al., 2022). Other Navs are also known to be enriched in the AIS in the CNS (Leterrier, 2018). To confirm our observation in motor neurons, we co-stained NaV1.7 with the AIS marker, Ankyrin-G, and found that, indeed, NaV1.7 colocalized with Ankyrin-G at the AIS of motor neurons in the human ventral horn (Fig 4D). In the dorsal horn, we also found evidence for NaV1.7 localized to the AIS of resident neurons (Fig 4E), but this was sparse, and we only observed this pattern in a few neurons across 3 donors. As we see robust NaV1.7 mRNA expression in virtually all of the putative (TACR1+) projection neurons, its low prevalence in the AIS and absence in the soma and cytoplasm of resident neurons suggests that either NaV1.7 is not translated into protein in these neurons, or which is more likely, that it is localized to the membrane of post-synaptic axons. This is further supported by NaV1.7 labeling in the anterior commissure.
Discussion
In the present study, we found evidence for the presence of NaV1.7 expression in intrinsic neurons of the human spinal cord, including in the dorsal and ventral horns. First, we identified that SCN9A (protein: NaV1.7) mRNA was detected in virtually all TACR1+ (protein: Nk1r) neurons and that these neurons were more densely populated in lamina 1, 2, 4 and 5, with their lowest density in lamina 3, 6, and 7. In rodents, the Nk1r is expressed by a subset of large-diameter projection neurons that send projections to higher brain regions like the parabrachial nucleus, and are found predominantly in lamina 1 and in the deeper dorsal horn around lamina 5 (Brown et al., 1995; Marshall et al., 1996; Todd et al., 2000). It is important to note that not all rodent projection neurons are Tacr1+ (Nk1r+), and interneurons as well as motor neurons express this gene (Russ et al., 2021). Single-nucleus RNA sequencing recapitulates this expression pattern as SCN9A and TACR1 mRNAs were co-expressed in a wide range of human spinal neuronal populations, including motor neurons (Yadav et al., 2023). The high density of the signal in known projection-neuron enriched lamina, as well as the large size profile of the neurons supports that a subset of these SCN9A/TACR1 co-expressing neurons are likely projection neurons.
Similarly, Scn9a mRNA was also detected in a variety of Tacr1 co-expressing neuron populations in mouse spinal cord using single cell sequencing (Russ et al., 2021). However, only its motor neuron expression could be validated with in situ hybridization (Alles et al., 2020; Allen Institute for Brain Science, 2022a). While sensitivity issues could underlie these technical differences, detection of NaV1.7 mRNA and protein in rodent spinal dorsal horn neurons has also been inconclusive likely due to its unique subcellular localization to the neuronal membrane which could comprise the soma, dendrites, axon initial segment, nodes of Ranvier, and/or synapse. Indeed, most reports suggest that spinal NaV1.7 protein expression is entirely presynaptic due to its robust neuropil staining pattern within lamina 1–2 (Black et al., 2012; Shiers et al., 2021). However, immuno-electron microscopy detected NaV1.7 protein localized to dendrites of mouse dorsal horn neurons, but it was hypothesized that these proteins originated in the presynaptic compartment and were transferred to post-synaptic sites through an unknown mechanism (Alles et al., 2020).
While we also detected intense presynaptic NaV1.7 labeling in lamina 1–2 of the human spinal cord, not all of its expression was colocalized with presynaptic markers like CGRP and Bassoon. NaV1.7 protein was also localized to axons in the deeper lamina, particularly lamina 4 and 5, and to axons in the anterior commissure, the white matter tract connecting the two spinal hemispheres and an important relay for dorsal horn projection neurons transmitting nociceptive information to the contralateral spinothalamic tract (Ku and Morrison, 2022). Because axon tracing methods cannot be employed in the human spinal cord, we could not identify if these were primary afferents or the axons of intrinsic dorsal horn neurons; however, ascending projections of primary afferents ascend in the dorsal columns and do not cross the midline, so it is exceedingly unlikely that these commissural axons are contributed by sensory neurons.
Interestingly, we did not detect NaV1.7 localized to dendrites, but instead identified its expression in the AISs of some dorsal horn neurons in lamina1-2 and also in motor neurons. Other Nav family members are highly concentrated at the AIS and their localization there is critical for integration of synaptic currents into action potential generation (Grubb and Burrone, 2010; Leterrier, 2018). Importantly, neurons are known to increase or decrease the size of their AIS as a means to augment or depress their neuronal excitability in response to changing presynaptic input (Grubb and Burrone, 2010; Kuba et al., 2010; Grubb et al., 2011). As sensory neuron hyperexcitability and/or spontaneous activity is a major driver of abnormal nociceptive signaling into the dorsal horn in chronic pain conditions, it is possible that NaV1.7 regulation at the AIS in resident dorsal horn neurons is critical for nociceptive processing in the dorsal horn.
While loss-of-function mutations in SCN9A result in pain insensitivity in both rodents and humans (Gingras et al., 2014; Shields et al., 2018; Grubinska et al., 2019), gain-of-function mutations engineered in mice to match human mutations that cause pain disorders do not recapitulate the human pain phenotype (Chen et al., 2021). One potential explanation for this is that the loss-of-function phenotype is dependent upon sensory neuron expression of NaV1.7 which is conserved across species and the loss of function leads to a conserved loss of action potential generation in nociceptors. On the other hand, the human gain-of-function pain phenotype may require both peripheral sensitization and spinal amplification. As there has been little convincing evidence for Nav1.7 expression in intrinsic neurons of the rodent spinal cord, it is probable that spinal amplification of nociceptive signals does not occur in rodents but may occur in humans with gain-of-function mutations given the broad expression of SCN9A in dorsal horn neurons, including putative projection neurons. Gain-of-function mutations in SCN9A could lead to increased excitability of projection neurons, which would be further amplified by increased nociceptive input from hyperexcitable nociceptors in the periphery. However, this mechanistic hypothesis may not be as straightforward as this because SCN9A mRNA was found in many neurons in the dorsal horn, including inhibitory interneurons by single-nuclear sequencing (Yadav et al., 2023). Nevertheless, dysregulated circuit dynamics in the dorsal horn caused by gain-of-function mutations in Nav1.7 potentially explain the difference in pain phenotype between humans and rodents.
In summary, we offer several compelling pieces of evidence to support the existence of NaV1.7 mRNA and protein expression by intrinsic neurons of the human spinal dorsal horn: 1) SCN9A mRNA is expressed by human resident dorsal horn neurons detected by both RNAscope in situ hybridization, spatial sequencing, and single-nucleus sequencing; 2) all TACR1+ human resident dorsal horn neurons express SCN9A, a subset of which are likely projection neurons; 3) many SCN9A+ and TACR1+ co-expressing neurons were large diameter and were most abundant in laminar regions (lamina 1 and lamina 5) that are known to be enriched with projection neurons; 4) not all NaV1.7 protein signal was limited to the presynaptic compartment as demonstrated with co-labeling with CGRP and Bassoon; 5) NaV1.7 was detected in the axon initial segment of some resident dorsal horn neurons; 6) NaV1.7+ axons were detected in the anterior commissure. The existence of NaV1.7 in dorsal horn neurons could explain the lack of analgesic efficacy of peripherally restricted NaV1.7 inhibitors and offer new insight into a centrally mediated NaV1.7 regulatory mechanism for nociceptive processing in the dorsal horn.
ACKNOWLEDGEMENTS:
The authors are grateful to the organ donors and their families for the gift of life and research provided by their organ donation.
FUNDING:
This work was supported by NIH grant U19NS130608 and a research grant from Grunenthal.
The data that support the findings of this study are available from the corresponding author upon request.
Footnotes
The authors declare no conflicts of interest. SH is an employee of Grunenthal Gmbh.
References
- Allen Institute for Brain Science (2022a) Allen Mouse Brain Atlas [Scn9a]. In: Allen Institute for Brain Science [Google Scholar]
- Allen Institute for Brain Science (2022b) Allen Human Brain Atlas [SCN9A].
- Alles SRA, Smith PA (2021) Peripheral Voltage-Gated Cation Channels in Neuropathic Pain and Their Potential as Therapeutic Targets. Front Pain Res (Lausanne) 2:750583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alles SRA, Nascimento F, Lujan R, Luiz AP, Millet Q, Bangash MA, Santana-Varela S, Zhou X, Cox JJ, Okorokov AL, Beato M, Zhao J, Wood JN (2020) Sensory neuron-derived Na(V)1.7 contributes to dorsal horn neuron excitability. Sci Adv 6:eaax4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez P, Bogen O, Green PG, Levine JD (2021) Nociceptor Overexpression of Na(V)1.7 Contributes to Chronic Muscle Pain Induced by Early-Life Stress. J Pain 22:806–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MD, Nassar MA (2020) Painful and painless mutations of SCN9A and SCN11A voltage-gated sodium channels. Pflugers Arch 472:865–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biogen (2021) Biogen Announces Postive Topline Results from Phase 2 CONVEY Study in Small Fiber Neuropathy. In. biogen.com. [Google Scholar]
- Black JA, Frezel N, Dib-Hajj SD, Waxman SG (2012) Expression of Nav1.7 in DRG neurons extends from peripheral terminals in the skin to central preterminal branches and terminals in the dorsal horn. Mol Pain 8:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JA, Nikolajsen L, Kroner K, Jensen TS, Waxman SG (2008) Multiple sodium channel isoforms and mitogen-activated protein kinases are present in painful human neuromas. Ann Neurol 64:644–653. [DOI] [PubMed] [Google Scholar]
- Blomqvist A, Mackerlova L (1995) Spinal projections to the parabrachial nucleus are substance P-immunoreactive. Neuroreport 6:605–608. [DOI] [PubMed] [Google Scholar]
- Brown JL, Liu H, Maggio JE, Vigna SR, Mantyh PW, Basbaum AI (1995) Morphological characterization of substance P receptor-immunoreactive neurons in the rat spinal cord and trigeminal nucleus caudalis. J Comp Neurol 356:327–344. [DOI] [PubMed] [Google Scholar]
- Carstens EE, Carstens MI, Simons CT, Jinks SL (2010) Dorsal horn neurons expressing NK-1 receptors mediate scratching in rats. Neuroreport 21:303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wimalasena NK, Shim J, Han C, Lee SI, Gonzalez-Cano R, Estacion M, Faber CG, Lauria G, Dib-Hajj SD, Woolf CJ, Waxman SG (2021) Two independent mouse lines carrying the Nav1.7 I228M gain-of-function variant display dorsal root ganglion neuron hyperexcitability but a minimal pain phenotype. Pain 162:1758–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JJ, Reimann F, Nicholas AK, Thornton G, Roberts E, Springell K, Karbani G, Jafri H, Mannan J, Raashid Y, Al-Gazali L, Hamamy H, Valente EM, Gorman S, Williams R, McHale DP, Wood JN, Gribble FM, Woods CG (2006) An SCN9A channelopathy causes congenital inability to experience pain. Nature 444:894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AD (2006) Retrograde analyses of spinothalamic projections in the macaque monkey: input to ventral posterior nuclei. J Comp Neurol 499:965–978. [DOI] [PubMed] [Google Scholar]
- Eagles DA, Chow CY, King GF (2022) Fifteen years of Na(V) 1.7 channels as an analgesic target: Why has excellent in vitro pharmacology not translated into in vivo analgesic efficacy? Br J Pharmacol 179:3592–3611. [DOI] [PubMed] [Google Scholar]
- Faber CG, Hoeijmakers JG, Ahn HS, Cheng X, Han C, Choi JS, Estacion M, Lauria G, Vanhoutte EK, Gerrits MM, Dib-Hajj S, Drenth JP, Waxman SG, Merkies IS (2012) Gain of function Nanu1.7 mutations in idiopathic small fiber neuropathy. Ann Neurol 71:26–39. [DOI] [PubMed] [Google Scholar]
- Fertleman CR, Baker MD, Parker KA, Moffatt S, Elmslie FV, Abrahamsen B, Ostman J, Klugbauer N, Wood JN, Gardiner RM, Rees M (2006) SCN9A mutations in paroxysmal extreme pain disorder: allelic variants underlie distinct channel defects and phenotypes. Neuron 52:767–774. [DOI] [PubMed] [Google Scholar]
- Gingras J, Smith S, Matson DJ, Johnson D, Nye K, Couture L, Feric E, Yin R, Moyer BD, Peterson ML, Rottman JB, Beiler RJ, Malmberg AB, McDonough SI (2014) Global Nav1.7 knockout mice recapitulate the phenotype of human congenital indifference to pain. PLoS One 9:e105895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb MS, Burrone J (2010) Activity-dependent relocation of the axon initial segment fine-tunes neuronal excitability. Nature 465:1070–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb MS, Shu Y, Kuba H, Rasband MN, Wimmer VC, Bender KJ (2011) Short- and long-term plasticity at the axon initial segment. J Neurosci 31:16049–16055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubinska B, Chen L, Alsaloum M, Rampal N, Matson DJ, Yang C, Taborn K, Zhang M, Youngblood B, Liu D, Galbreath E, Allred S, Lepherd M, Ferrando R, Kornecook TJ, Lehto SG, Waxman SG, Moyer BD, Dib-Hajj S, Gingras J (2019) Rat NaV1.7 loss-of-function genetic model: Deficient nociceptive and neuropathic pain behavior with retained olfactory function and intra-epidermal nerve fibers. Mol Pain 15:1744806919881846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameed S (2019a) Na(v)1.7 and Na(v)1.8: Role in the pathophysiology of pain. Mol Pain 15:1744806919858801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameed S (2019b) Nav1.7 and Nav1.8: Role in the pathophysiology of pain. Mol Pain 15:1744806919858801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Cui D, Han L, Xu L, Mao S, Yang C, Gao F, Yuan Z (2022) A novel SCN9A gene variant identified in a Chinese girl with paroxysmal extreme pain disorder (PEPD): a rare case report. BMC Med Genomics 15:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MW, Apkarian AV, Stevens RT, Hodge CJ Jr. (1987) The spinothalamic tract: an examination of the cells of origin of the dorsolateral and ventral spinothalamic pathways in cats. J Comp Neurol 260:349–361. [DOI] [PubMed] [Google Scholar]
- Kingwell K (2019) Nav1.7 withholds its pain potential. Nat Rev Drug Discov. [DOI] [PubMed] [Google Scholar]
- Ku J, Morrison EH (2022) Neuroanatomy, Anterior White Commissure. In: StatPearls. Treasure Island; (FL: ). [PubMed] [Google Scholar]
- Kuba H, Oichi Y, Ohmori H (2010) Presynaptic activity regulates Na(+) channel distribution at the axon initial segment. Nature 465:1075–1078. [DOI] [PubMed] [Google Scholar]
- Lein ES et al. (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168–176. [DOI] [PubMed] [Google Scholar]
- Leterrier C (2018) The Axon Initial Segment: An Updated Viewpoint. J Neurosci 38:2135–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, North RY, Rhines LD, Tatsui CE, Rao G, Edwards DD, Cassidy RM, Harrison DS, Johansson CA, Zhang H, Dougherty PM (2018) DRG Voltage-Gated Sodium Channel 1.7 Is Upregulated in Paclitaxel-Induced Neuropathy in Rats and in Humans with Neuropathic Pain. J Neurosci 38:1124–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu BW, Zhang J, Hong YS, Li NB, Liu Y, Zhang M, Wu WY, Zheng H, Lampert A, Zhang XW (2021) NGF-Induced Nav1.7 Upregulation Contributes to Chronic Post-surgical Pain by Activating SGK1-Dependent Nedd4-2 Phosphorylation. Mol Neurobiol 58:964–982. [DOI] [PubMed] [Google Scholar]
- MacDonald DI, Sikandar S, Weiss J, Pyrski M, Luiz AP, Millet Q, Emery EC, Mancini F, Iannetti GD, Alles SRA, Arcangeletti M, Zhao J, Cox JJ, Brownstone RM, Zufall F, Wood JN (2021) A central mechanism of analgesia in mice and humans lacking the sodium channel Na(V)1.7. Neuron 109:1497–1512 e1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann N, King T, Murphy R (2019) Review of primary and secondary erythromelalgia. Clin Exp Dermatol 44:477–482. [DOI] [PubMed] [Google Scholar]
- Mantyh PW, Rogers SD, Honore P, Allen BJ, Ghilardi JR, Li J, Daughters RS, Lappi DA, Wiley RG, Simone DA (1997) Inhibition of hyperalgesia by ablation of lamina I spinal neurons expressing the substance P receptor. Science 278:275–279. [DOI] [PubMed] [Google Scholar]
- Marshall GE, Shehab SA, Spike RC, Todd AJ (1996) Neurokinin-1 receptors on lumbar spinothalamic neurons in the rat. Neuroscience 72:255–263. [DOI] [PubMed] [Google Scholar]
- McDermott LA, Weir GA, Themistocleous AC, Segerdahl AR, Blesneac I, Baskozos G, Clark AJ, Millar V, Peck LJ, Ebner D, Tracey I, Serra J, Bennett DL (2019) Defining the Functional Role of Na(V)1.7 in Human Nociception. Neuron 101:905–919 e908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell A, Collins S, Ali Z, Iavarone L, Surujbally R, Kirby S, Butt RP (2018) Efficacy of the Nav1.7 blocker PF-05089771 in a randomised, placebo-controlled, double-blind clinical study in subjects with painful diabetic peripheral neuropathy. Pain 159:1465–1476. [DOI] [PubMed] [Google Scholar]
- Middleton SJ et al. (2022) Nav1.7 is required for normal C-low threshold mechanoreceptor function in humans and mice. Brain 145:3637–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento AI, Da Silva TF, Fernandes EC, Luz LL, Mar FM, Safronov BV, Sousa MM (2022) Sensory neurons have an axon initial segment that initiates spontaneous activity in neuropathic pain. Brain 145:1632–1640. [DOI] [PubMed] [Google Scholar]
- Price N, Namdari R, Neville J, Proctor KJ, Kaber S, Vest J, Fetell M, Malamut R, Sherrington RP, Pimstone SN, Goldberg YP (2017) Safety and Efficacy of a Topical Sodium Channel Inhibitor (TV-45070) in Patients With Postherpetic Neuralgia (PHN): A Randomized, Controlled, Proof-of-Concept, Crossover Study, With a Subgroup Analysis of the Nav1.7 R1150W Genotype. Clin J Pain 33:310–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ DE, Cross RBP, Li L, Koch SC, Matson KJE, Yadav A, Alkaslasi MR, Lee DI, Le Pichon CE, Menon V, Levine AJ (2021) A harmonized atlas of mouse spinal cord cell types and their spatial organization. Nat Commun 12:5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengul G, Watson C, Tanaka I, Paxinos G (2013) Atlas of the spinal cord of the rat, mouse, marmoset, rhesus, and human, 1st Edition. London ; Boston: Elsevier Academic Press. [Google Scholar]
- Shields SD, Deng L, Reese RM, Dourado M, Tao J, Foreman O, Chang JH, Hackos DH (2018) Insensitivity to Pain upon Adult-Onset Deletion of Nav1.7 or Its Blockade with Selective Inhibitors. J Neurosci 38:10180–10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiers S, Klein RM, Price TJ (2020) Quantitative differences in neuronal subpopulations between mouse and human dorsal root ganglia demonstrated with RNAscope in situ hybridization. Pain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiers SI, Sankaranarayanan I, Jeevakumar V, Cervantes A, Reese JC, Price TJ (2021) Convergence of peptidergic and non-peptidergic protein markers in the human dorsal root ganglion and spinal dorsal horn. J Comp Neurol 529:2771–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Li N, Duan G, Liu Y, Guo S, Wang C, Zhu C, Zhang X (2018) Increased Na(v)1.7 expression in the dorsal root ganglion contributes to pain hypersensitivity after plantar incision in rats. Mol Pain 14:1744806918782323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares-Ferreira D, Shiers S, Ray PR, Wangzhou A, Jeevakumar V, Sankaranarayanan I, Cervantes AM, Reese JC, Chamessian A, Copits BA, Dougherty PM, Gereau RWt, Burton MD, Dussor G, Price TJ (2022) Spatial transcriptomics of dorsal root ganglia identifies molecular signatures of human nociceptors. Sci Transl Med 14:eabj8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd AJ, McGill MM, Shehab SA (2000) Neurokinin 1 receptor expression by neurons in laminae I, III and IV of the rat spinal dorsal horn that project to the brainstem. Eur J Neurosci 12:689–700. [DOI] [PubMed] [Google Scholar]
- Valtcheva MV, Copits BA, Davidson S, Sheahan TD, Pullen MY, McCall JG, Dikranian K, Gereau RWt (2016) Surgical extraction of human dorsal root ganglia from organ donors and preparation of primary sensory neuron cultures. Nat Protoc 11:1877–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis WD, Kenshalo DR Jr., Leonard RB (1979) The cells of origin of the primate spinothalamic tract. J Comp Neurol 188:543–573. [DOI] [PubMed] [Google Scholar]
- Yadav A et al. (2023) A cellular taxonomy of the adult human spinal cord. Neuron 111:328–344 e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang Y, Li S, Xu Z, Li H, Ma L, Fan J, Bu D, Liu B, Fan Z, Wu G, Jin J, Ding B, Zhu X, Shen Y (2004) Mutations in SCN9A, encoding a sodium channel alpha subunit, in patients with primary erythermalgia. J Med Genet 41:171–174. [DOI] [PMC free article] [PubMed] [Google Scholar]