Abstract
The Metalloprotein Database and Browser (MDB; http://metallo.scripps.edu) at The Scripps Research Institute is a web-accessible resource for metalloprotein research. It offers the scientific community quantitative information on geometrical parameters of metal-binding sites in protein structures available from the Protein Data Bank (PDB). The MDB also offers analytical tools for the examination of trends or patterns in the indexed metal-binding sites. A user can perform interactive searches, metal-site structure visualization (via a Java applet), and analysis of the quantitative data by accessing the MDB through a web browser without requiring an external application or platform-dependent plugin. The MDB also has a non-interactive interface with which other web sites and network-aware applications can seamlessly incorporate data or statistical analysis results from metal-binding sites. The information contained in the MDB is periodically updated with automated algorithms that find and index metal sites from new protein structures released by the PDB.
INTRODUCTION
The Metalloprotein Database and Browser (MDB, http://metallo.scripps.edu) is part of the Metalloprotein Structure, Bioinformatics and Design Program (http://www.scripps.edu/research/metallo) at The Scripps Research Institute (TSRI; http://www.scripps.edu). The main role of the MDB is to collect, and make viewable and easily accessible, key quantitative information on protein metal-binding sites from structures available at the Protein Data Bank (PDB) (1).
A major emerging challenge in structural biology is to develop a sufficient understanding of metalloproteins to allow their rational design with engineered metal-site geometries and properties. To achieve this, we need to comprehend the set of structural, environmental and functional requirements for metal-binding sites in existing metalloproteins. We need to know not only what types of metal ions are bound by proteins, but also the types of ligands that bind these metal ions (i.e. the first-shell ligands), the residues that contact the metal-binding ligands (i.e. the second-shell ligands) and what other geometrical or environmental effects may modulate the properties of the metal-binding site. This information is crucial whether the objective is to construct new metal-binding sites into a given protein scaffold or to modify an existing metal site.
We have created the MDB to address the need for quantitative information by biochemists, biologists, computational chemists, bioinformaticians and other metalloprotein researchers. The MDB contains key structural information that can be used not only for metal-binding site design, but also for obtaining parameters for the determination of X-ray structures of metal-containing proteins, developing constraints for computational modeling of metallosystems and statistical analyses of ligand geometries and ligating patterns.
The MDB is a bioinformatic web application designed for easy user access, which takes advantage of well known web technologies: a fast database engine (MySQL; http://www.mysql.com), a stable web server (Apache; http://www.apache.org) and a powerful web scripting language (PHP; http://www.php.net). With these Open Source tools, we have constructed interactive web query interfaces, functions to allow remote viewing and searching and a non-interactive web application program interface (API).
INDEXING PROTEIN METAL-SITE DATA FOR THE MDB
The metal-site structural data in the MDB are collected with automatic tools that periodically index protein structures with metal sites from the latest PDB release. The current publicly available version of the MDB reliably indexes first-shell data from mononuclear metal sites. To enhance the quality and usefulness of the data present in the MDB, we have developed the metal-binding site indexing tool (MSIT). The MSIT, an application written in Java, extracts first- and second-shell data, recognizes multinuclear metal sites and cluster-containing sites, classifies metal-binding sites according to several criteria (number of metal ions in the site, metal complexation geometry, type of metal ion, etc.) and determines non-covalent interactions within each indexed shell and among shells.
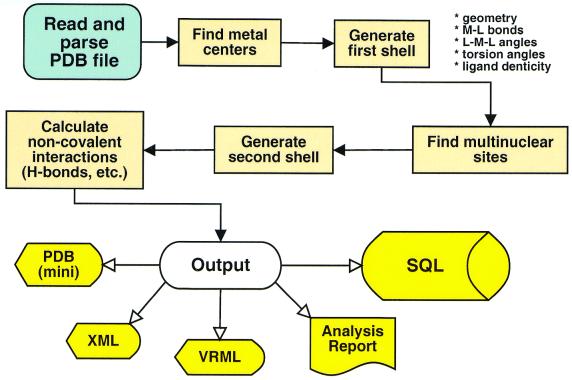
The MSIT uses a distance-dependent algorithm and table-based heuristics to recognize and extract metal-binding sites and to work around malformed PDB structure files (missing records, misaligned fields, etc.). The program reads and parses the input PDB file, recognizes metal centers in the structure and generates a first shell. It then executes a breadth-first search through all metal sites that have been found to search for sites that share common residues. Such sites are merged, creating a multinuclear site. Once the mono- and multinuclear sites have been defined, second-shell residues and non-covalent interactions are identified. The geometrical data are written to structure files (in PDB, XML/CML and VRML formats) and the calculated data are inserted into the SQL database (Fig. 1).
Figure 1.
Algorithm used by the MSIT to identify and extract information about metal-binding sites in metalloproteins.
INTERACTIVE AND NON-INTERACTIVE ACCESS TO THE MDB
Interactive interfaces
The MDB web site offers the researcher a series of query options, from the simple to the complex. Interactive interfaces are implemented either as simple HTML forms or as forms combined with an applet for real-time three-dimensional viewing of structures. In the MDB, you can search for metal-binding sites with HTML forms that allow you to:
Specify PDB codes of proteins (e.g. 2sod or 1fer), resulting in a list of all metal-binding sites found within the indicated proteins.
Select sites based on type of metal, number of ligands and other parameters. For example, you can search for zinc-containing sites with four to six ligands found in protein structures with resolutions <2.0 Å. A more extensive HTML form allows even more restricted searches. For example, a specified number of ligands can be restricted to be of certain type (‘one must be water’) or a range of resolutions may be specified instead of an upper limit (‘resolution between 1.5 and 2.8 Å’).
Perform a query using the SQL language, offering the user complete flexibility to examine any possible correlation among the data sets contained in the database. To assist the user, the database structure is documented (http://metallo.scripps.edu/sql_docs/structure.html and http://metallo.scripps.edu/sql_docs/table_descriptions.html). A possible complex query would be to select all copper-binding sites that contain exactly one Asp or Glu, two His residues and a water molecule as the metal-ligating pattern and then to display the copper–water distance, the resolution and the date when the structure was released for each site.
Use an interface with a lightweight (70 kB in size) Java applet to view and manipulate the metal-binding sites found by the queries. This viewer allows the user to perform geometrical measurements, including atom to atom distances, valence angles and torsion angles. This applet also allows simple superpositions, stereo visualization, display of atoms within a given distance of the one selected, independent selection and movement of structures when several are being displayed, the ability to detach or reattach the viewer from/to the page (to enlarge the window), etc.
Non-interactive interfaces
The non-interactive interfaces allow one to embed MDB data or visualization tools in any other web page or application. Currently there are three interfaces available.
The remote query/viewer interface, which uses some simple HTML code to allow transparent querying and visualization of metal-sites of interest (with the interactive Java viewer). Several sites are using this interface, including the IMB Jena Image Library of Biological Macromolecules (2), the PROMISE database (3) and the EF-Hand Calcium-Binding Proteins Data Library (http://structbio.vanderbilt.edu/cabp_database).
An SQL query interface (http://metallo.scripps.edu/api) that allows application developers to call a particular URL on the MDB site, pass the appropriate parameters and obtain the results in program parsable format, such as comma delimited records (for spreadsheet analysis) or WDDX packets (http://www.openwddx.org/), or in a format ready to embed into a web page (HTML tables).
An XML-RPC-based interface (http://www.xml-rpc.com) that accepts remote procedure calls from any application using the XML-RPC protocol, independent of the platform or programming language used by the application requesting the service. Added advantages are that the protocol supports introspection (i.e. an application can ask the server: ‘what procedures do you offer and how should I call them?’) and that there are XML-RPC libraries for most scripting and programming languages.
This last interface (XML-RPC), which is under vigorous development, will comprise a whole set of callable methods (API) for the MDB. For example, a program for metal-site design could use this interface to communicate directly with the MDB and request a list of observed ranges for a particular geometrical feature (distance, angle, etc.), and thus compare a proposed model value with those found in known metalloproteins.
DATA ANALYSIS WITH THE MDB
The MDB has been used to analyze the distribution of geometrical parameters such as metal–ligand bond distances and side-chain torsion angles, and also to obtain the frequencies for ligating patterns in metal sites. These analyses have been used to assist in the determination of X-ray crystallographic structures, to validate and compare designed metal-site candidates, and to find ligating tendencies (such as differences between Cu–S distances when Cys and Met are metal ligands).
These types of analyses have been very useful in our research, so we designed HTML forms to perform the analyses online. Using the forms, we can generate the distribution of the metal–ligand atom distances or the ligand patterns that are most common for a particular metal ion with a given coordination number.
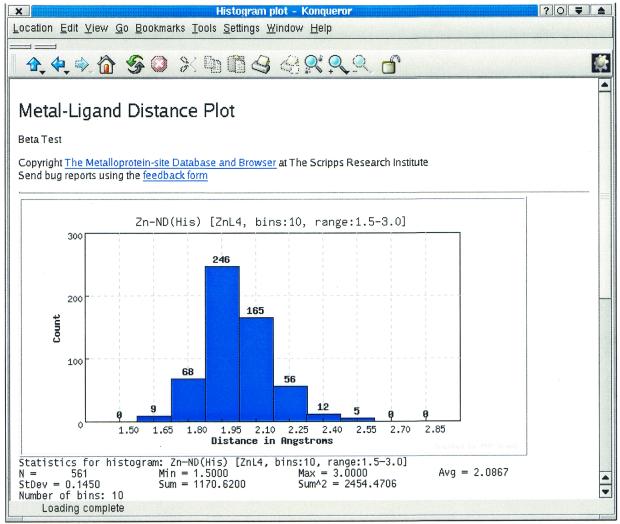
Figure 2 shows the result of analysis of the bond distance distribution for Zn-Nδ(His) (coordination number = 4 and fixed range of 1.5–3.0 Å). We find a normal distribution with an average Zn-Nδ(His) distance of 2.09 Å (SD 0.15) from 561 indexed distances. Not shown in Figure 2 is a table listing each of the plotted bins, their corresponding counts and a button allowing the researcher to obtain (in another window) a list of the metal-binding sites in which a particular distance occurs. From that list, the scientist can choose to download the structure of the site (in PDB format) or view it using our Java viewer. Usually, this tool takes between 2 and 10 s to process a distribution (including plot, table, etc.) and behaves linearly with the number of matching distances. Improvements planned for this tool include discriminating distances from mononuclear, multinuclear or cluster sites and making the analysis on a dehomologized list of protein structures.
Figure 2.
Distribution and statistics of the Zn-Nδ(His) distance (coordination number = 4, range 1.5–3.0) using the online histogram plotting tool.
A complementary analysis tool identifies ligand pattern tendencies of a particular metal ion in a specific coordination number. Using a simple form, we can choose what metal ion we are interested in and decide what coordination number it should present, and we will obtain a ranked list (by frequency count) of the ligand patterns that match those constraints. For example, if we were interested in designing a metal-binding site that matched the pattern CuL4, we would find that the three most frequent patterns are Cu(Cys)(Gly)(His)2, Cu(His)4 and Cu(His)3(H2O), and that the pattern Cu(Cys)4 does not appear in the data indexed in the MDB to date.
RELATIONSHIP OF THE MDB WITH OTHER DATABASES
The MDB currently includes, in the lists generated from queries, links to the appropriate structure page at the PDB site (1) and to the NIH Molecules R Us site (http://webasaurus.dcrt.nih.gov/cgi-bin/pdb). The MDB is used by other databases to provide three-dimensional viewing of specific metal-binding sites through the use of our remote query/viewer tool. Sites using the MDB in this manner include the IMB Jena Image Library of Biological Macromolecules (2), the PROMISE database (3) and the E-F Hand Calcium-Binding Proteins Data Library (http://structbio.vanderbilt.edu/cabp_database).
We also make use of data from the PDB’s Het group dictionary (http://pdb.rutgers.edu/het_dictionary.txt) and the lists of dehomologized structures from PDBSELECT (4) and from WHATIF SELECT (5). These lists have been manually converted into SQL tables so that they can be correlated with the other data being indexed in the MDB.
AVAILABILITY AND ACCESS STATISTICS
The MDB is available at http://metallo.scripps.edu/, which includes interactive interfaces for querying and browsing the MDB, requiring just a common web browser. The non-interactive interfaces are described at http://metallo.scripps.edu/api (MDB’s web API) and http://metallo.scripps.edu/remote/ (remote query/viewer tool). New analytical tools and features are available for testing at http://metallo.scripps.edu/beta/.
The use of the MDB by the research community has increased steadily since its inception back in early 1998. Table 1 summarizes some access statistics for the MDB from the third quarter of 1999 until the second quarter of 2001. In the last 2 years the number of users has increased almost 3.5-fold and the number of documents viewed (counted as full page displays, not as individual hits in a document) has increased 3-fold. More importantly, the number of queries performed and the number of structures of metal-binding sites downloaded have increased by a factor of 4.5 and 2, respectively.
Table 1. Access statistics of the MDB web site from July 1999–June 2001 (outside users only).
| Year | Quarter | Queriesa | Structuresb | Pagesc | Hostsd |
|---|---|---|---|---|---|
| 1999 | Jul–Sep | 1413 | 1327 | 8043 | 3431 |
| Oct–Dec | 2983 | 1679 | 12 257 | 4762 | |
| 2000 | Jan–Mar | 3711 | 2025 | 14 951 | 7522 |
| Apr–Jun | 2522 | 1914 | 13 026 | 7733 | |
| Jul–Sep | 5471 | 1573 | 15 772 | 7785 | |
| Oct–Dec | 5414 | 2080 | 18 662 | 9292 | |
| 2001 | Jan–Mar | 5716 | 2832 | 19 761 | 10 208 |
| Apr–Jun | 6379 | 2285 | 24 468 | 11 987 |
aNumber of searches performed.
bNumber of structure files downloaded or viewed using the interactive Java viewer.
cNumber of pages viewed (complete documents).
dEach host is counted only once per day.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to all our users for their valuable and continuous feedback that drives improvements to the MDB. We would also like to acknowledge the work of Marij van Gorkom. Her enthusiasm helped us complete a prototype of the MSIT tool during her summer stay at TSRI. The MDB is part of the Metalloprotein Structure and Design Program Project at TSRI, funded by NIH grant P01-GM48495. The macromolecular Java viewer was developed as part of the Computational Center for Macromolecular Structure (http://www.sdsc.edu/CCMS), funded by NSF grant BIO/DBI 99-04559.
REFERENCES
- 1.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. Updated article in this issue: Nucleic Acids Res. (2002), 30, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reichter J., Jabs,A., Slickers,P. and Sühnel ,J. (2000) The IMB Jena Image Library of Biological Macromolecules. Nucleic Acids Res., 28, 246–249. Updated article in this issue: Nucleic Acids Res. (2002), 30, 253–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Degtyarenko K.N., North,A.C.T. and Findlay,J.B.C (1999) PROMISE: a database of bioinorganic motifs. Nucleic Acids Res., 27, 233–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hobohm U. and Sander,C. (1994) Enlarged representative set of protein structures. Protein Sci., 3, 522–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooft R.W.W., Sander,C. and Vriend,G. (1996) Verification of protein structures: side-chain planarity. J. Appl. Cryst., 29, 714–716. [Google Scholar]