PURPOSE
Peripheral T-cell lymphoma (PTCL) includes heterogeneous clinicopathologic entities with numerous diagnostic and treatment challenges. We previously defined robust transcriptomic signatures that distinguish common PTCL entities and identified two novel biologic and prognostic PTCL-not otherwise specified subtypes (PTCL-TBX21 and PTCL-GATA3). We aimed to consolidate a gene expression–based subclassification using formalin-fixed, paraffin-embedded (FFPE) tissues to improve the accuracy and precision in PTCL diagnosis.
MATERIALS AND METHODS
We assembled a well-characterized PTCL training cohort (n = 105) with gene expression profiling data to derive a diagnostic signature using fresh-frozen tissue on the HG-U133plus2.0 platform (Affymetrix, Inc, Santa Clara, CA) subsequently validated using matched FFPE tissues in a digital gene expression profiling platform (nCounter, NanoString Technologies, Inc, Seattle, WA). Statistical filtering approaches were applied to refine the transcriptomic signatures and then validated in another PTCL cohort (n = 140) with rigorous pathology review and ancillary assays.
RESULTS
In the training cohort, the refined transcriptomic classifier in FFPE tissues showed high sensitivity (> 80%), specificity (> 95%), and accuracy (> 94%) for PTCL subclassification compared with the fresh-frozen–derived diagnostic model and showed high reproducibility between three independent laboratories. In the validation cohort, the transcriptional classifier matched the pathology diagnosis rendered by three expert hematopathologists in 85% (n = 119) of the cases, showed borderline association with the molecular signatures in 6% (n = 8), and disagreed in 8% (n = 11). The classifier improved the pathology diagnosis in two cases, validated by clinical findings. Of the 11 cases with disagreements, four had a molecular classification that may provide an improvement over pathology diagnosis on the basis of overall transcriptomic and morphological features. The molecular subclassification provided a comprehensive molecular characterization of PTCL subtypes, including viral etiologic factors and translocation partners.
CONCLUSION
We developed a novel transcriptomic approach for PTCL subclassification that facilitates translation into clinical practice with higher precision and uniformity than conventional pathology diagnosis.
INTRODUCTION
Peripheral T-cell lymphoma (PTCL) represents approximately 10%-15% of non-Hodgkin lymphoma1 with numerous challenges in diagnosis even for expert hematopathologists.2-4 The WHO classification identifies more than 25 different subtypes of PTCLs, with angioimmunoblastic T-cell lymphoma (AITL), anaplastic large cell lymphoma (ALCL), adult T-cell leukemia/lymphoma (ATLL), and extranodal natural killer (NK)/T-cell lymphoma of nasal type (ENKTCL) as the most frequent entities with geographic variations.5 However, 30% of PTCL cannot be classified into any of the specific entities in the WHO classification, and these are categorized as PTCL, not otherwise specified (PTCL-NOS).6,7 Tumor-defining abnormalities, such as translocations involving the ALK gene in ALK-positive ALCL (ALK+ ALCL),8 human T-lymphotropic virus infection in ATLL,9 Epstein-Barr virus (EBV) positivity in ENKTCL, and IDH2R172 mutations in AITL, are generally uncommon in PTCL. PTCLs generally have a poor prognosis with current therapies,2 and more intensive regimens have not been proven to be superior.10 However, novel targeted therapies are now being tested, with some remarkable results.11,12
CONTEXT
Key Objective
To evaluate the role of digital gene expression signatures for the classification of peripheral T-cell lymphoma (PTCL) using formalin-fixed, paraffin-embedded tissue and to develop a highly accurate and reproducible diagnostic assay applicable for routine clinical practice.
Knowledge Generated
Using digital quantitation of transcripts, we have defined robust transcriptomic signatures that can distinguish common PTCL subtypes according to the WHO classification, including two novel biologic and prognostic subgroups within PTCL-not otherwise specified. We refined the classification algorithm and standardized the assay procedure for a robust diagnostic assay that was validated in an independent PTCL cohort. This assay was reproducible across institutions and showed high classification accuracy.
Relevance (J.W. Friedberg)
-
The digital transcriptomic assay resulted in robust molecular classification of PTCL using mRNA from formalin-fixed, paraffin-embedded tissue. This classifier may enable development of precision medicine trials in PTCL and ultimately may be incorporated into diagnostic classification systems for lymphoma.*
*Relevance section written by JCO Editor-in-Chief Jonathan W. Friedberg, MD.
The subclassification is more challenging for PTCL compared with B-cell lymphomas because of the complexity of T-cell biology with numerous functional subsets and functional plasticity. Gene expression profiling (GEP) has aided in delineating novel biologic subtypes and in the identification of oncogenic pathways in several B-non–Hodgkin lymphomas.13-18 Similar approaches in PTCLs resulted in robust molecular classifiers for the common PTCL entities and identified two biologic and prognostic subgroups within PTCL-NOS (PTCL-GATA3 and PTCL-TBX21).19-21 However, these studies were performed on fresh-frozen (FF) samples with transcriptome-wide arrays, thus limiting the application to routine clinical practice.22,23 Formalin-fixed, paraffin-embedded (FFPE) tissue samples are widely used in routine diagnosis, but formalin fixation leads to fragmentation, cross-linking, and chemical modifications of RNA and DNA.24 Therefore, the effective translation of our highly accurate RNA-based PTCL diagnostic signatures to FFPE tissue is challenging but essential to implement an assay with wide clinical application.25 Using digital quantification of RNA, as in diffuse large B-cell lymphoma,26-28 we consolidated our PTCL diagnostic signatures from FF RNA into a single technical platform for the accurate diagnosis across major PTCL entities.19-21 We used a training (n = 105) and an independent validation (n = 140) PTCL cohort, representing the largest well-characterized PTCL series investigated on a single platform for subclassification. Several viral transcripts were added to improve diagnostic accuracy, and we report a diagnostic algorithm that attained high sensitivity, specificity, and accuracy in distinguishing PTCL entities, including the novel molecular biologic subtypes of PTCL-GATA3 and PTCL-TBX21.20,21
MATERIALS AND METHODS
Patient Information
We included 249 diagnostic PTCL cases from multiple institutions, which after the exclusion of 4 cases with poor RNA quality, were divided into a training cohort (105 cases) with previously generated GEP19-21 with matched FF and FFPE samples and a validation cohort (140 cases) that had not been previously analyzed (Table 1 and Fig 1A). The basic clinical and pathologic characteristics of the cases are shown in the Data Supplement (online only). Inclusion and exclusion criteria of PTCL cases are detailed in the Data Supplement.
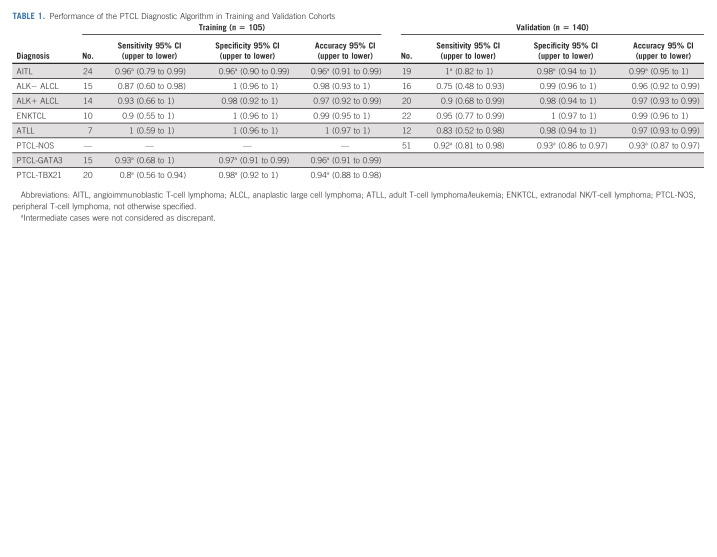
TABLE 1.
Performance of the PTCL Diagnostic Algorithm in Training and Validation Cohorts
FIG 1.
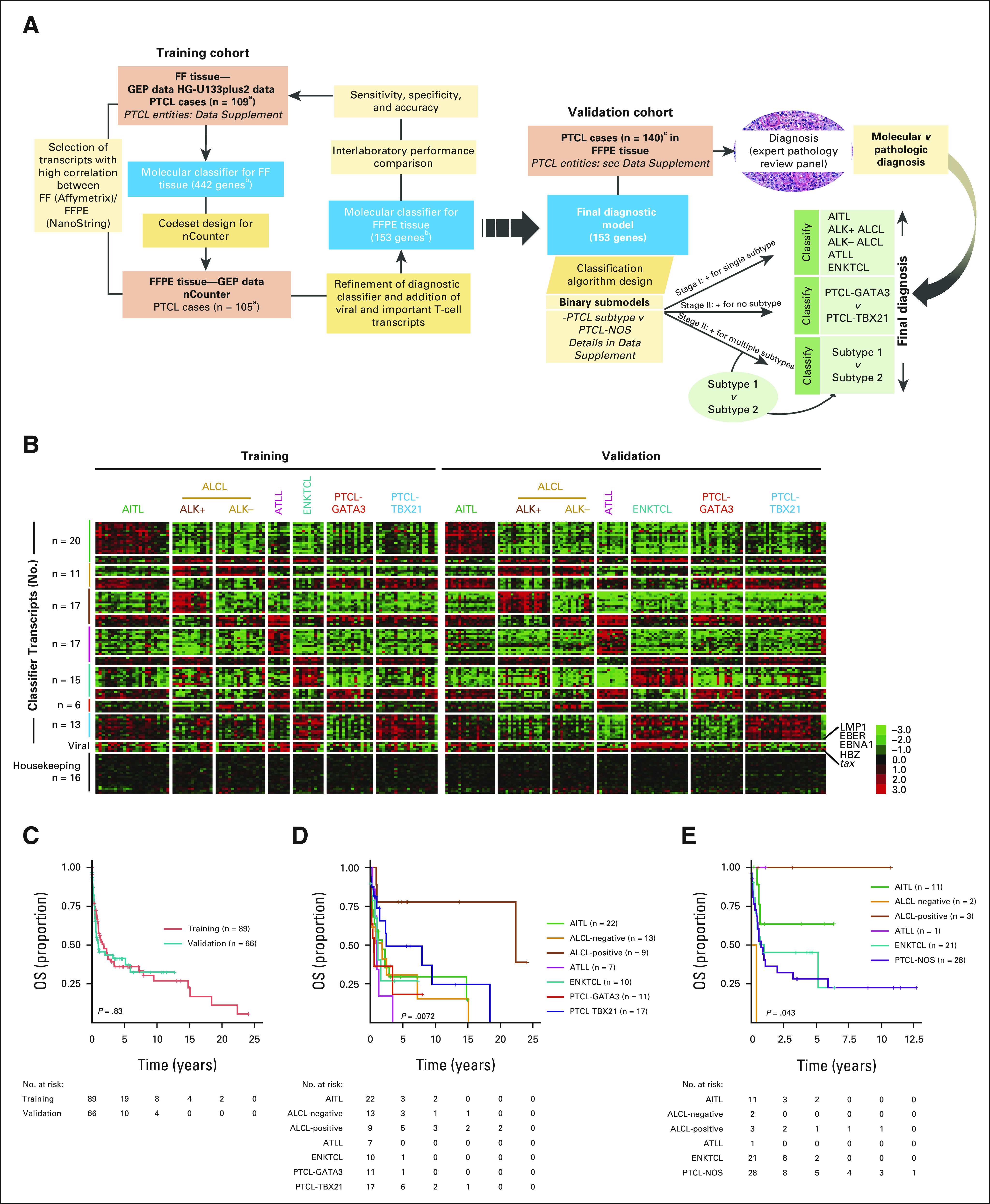
PTCL molecular classifier. (A) Study design and schematics of the molecular diagnosis of PTCL. A molecular classifier for PTCL subclassification was derived using HG-U133plus2.0 array data from PTCL with FF tissue (n = 109), designated as the training cohort. This molecular classifier had 442 distinct genes, including housekeeping and other genes involved in T-cell biology. This transcriptomic signature was considered for NanoString analysis using corresponding matched FFPE samples. Transcripts that showed a high correlation between FF and FFPE data were selected. The algorithm was further refined to have the minimum number of transcripts for subclassification and mimic the FF predictor score withthe nCounter platform (see the Methods and Materials for details). The final diagnostic model resulted in 153 transcripts (99 diagnostic, five viral, 16 housekeeping, and 33 T-cell biology–related) and was validated in an independent cohort of PTCL cases rigorously characterized by pathology and other ancillary methods. The classification algorithm was based on a series of several binary predictors to distinguish one entity from another, as detailed in the Data Supplement. (B) Heatmap of the finalized NanoString classifier in the training and validation cohorts. EBV and HTLV-1 viral transcripts commonly expressed in specific PTCL subtypes are shown, and housekeeping genes used for normalization and that do not vary between PTCL subtypes are displayed for comparison. (C) Kaplan-Meier curve of OS for 89 of the 105 training cohort cases and 66 of the 140 validation cases with available outcome data. (D) Kaplan-Meier curve of OS of PTCL subtypes included in the training cohort (molecular classification by the HG-U133plus2.0 array). (E) Kaplan-Meier curve of OS of PTCL entities included in the validation cohort (pathology classification). aFour cases excluded because of poor RNA quality. bIncludes classification, housekeeping, and important T-cell biology genes. cTwelve cases excluded because of reclassification as n-PTCL-TFH. AITL, angioimmunoblastic T-cell lymphoma; ALCL, anaplastic large cell lymphoma; ATLL, adult T-cell lymphoma/leukemia; EBV, Epstein-Barr virus; ENKTCL, extranodal natural killer/T-cell lymphoma; FF, fresh-frozen; FFPE, formalin-fixed, paraffin-embedded; GEP, gene expression profiling; HBZ, HTLV-1 bZIP factor; HTLV-1, human T-lymphotropic virus; NOS, not otherwise specified; OS, overall survival; PTCL, peripheral T-cell lymphoma; TFH, T follicular helper.
Histopathology/Immunomorphological Features of the PTCL Cohorts
PTCL cases were centrally reviewed and diagnosed according to the current WHO classification.6 The validation cohort was thoroughly re-evaluated by three hematopathologists (C.A., D.D.W., and W.C.C.) with a comprehensive immunostaining panel and T-cell receptor gene rearrangement analysis when needed. A consensus diagnosis was reached when there was unanimous agreement on the diagnosis (see the Data Supplement).
RNA Extraction and Digital Gene Expression for PTCL Subclassification
The details about RNA extraction protocols, quality control measures, NanoString assay (NanoString Technologies, Inc, Seattle, WA), data processing, cross-validation, and reproducibility assessment are given in the Data Supplement and in Figure 1A. The data analysis and normalization were designed to process samples individually, rather than in batches, so that the protocol would be suitable for processing patient samples on an as-needed basis. Class prediction was based on a series of binary comparisons that were combined for a final classification call for each sample (Fig 1A and Data Supplement). The detailed materials and methods are included in the Data Supplement.
Survival Analysis
The survival data were analyzed using the survival, survminer, and coin packages in R and are detailed in the Data Supplement.
RESULTS
Patient Characteristics in the Training and Validation Cohorts
The training cohort (n = 105), which had previously generated GEP data in FF19-21 and matched FFPE tissues, was used to select the classifier genes for the nCounter platform. The PTCL validation cohort (n = 140) without FF transcriptomic data was rigorously diagnosed by three expert hematopathologists using current WHO diagnostic criteria6 (Fig 1A and Table 1). Of note, PTCL-NOS cases were evaluated with T follicular helper (TFH) markers for exclusion of nodal PTCL-TFH cases and subsequently subclassified into PTCL-GATA3 and PTCL-TBX21 using the recently published IHC algorithm.29
The clinicopathologic characteristics of the training and validation cohorts are summarized in the Data Supplement. There was no significant difference in sex, age, and overall survival (OS; Fig 1C) between the validation and training cohorts. The median follow-up for survivors was 3.5 years (range, 0.01-24 years) for patients with available survival data. ALK+ ALCL cases showed a superior outcome than the other entities, consistent with published studies (Figs 1D and 1E).2,21
Development of Transcriptomic Signatures for FFPE Tissue
FFPE tissue blocks were selected on the basis of the presence of adequate tumor tissue and RNA quality assessed as shown in the Data Supplement. Transcriptomic signatures assessed between two platforms, HG-U133plus2.0 (Affymetrix, Inc, Santa Clara, CA) versus the nCounter platform, revealed high correlation (correlation coefficient r > 0.4) in the majority (approximately 60%) of signature-specified genes (Data Supplement). We performed recursive filtering analysis to exclude transcripts with correlation coefficients ≤ 0.4 using Pearson correlation (except three transcripts) to generate 11-20 diagnostic transcripts per PTCL subtype and 16 housekeeping genes (Data Supplement). Using these well-performing transcripts on the nCounter did not affect the classification accuracy, sensitivity, and specificity, either in FF RNA (HG-U133plus2.0) or matched FFPE RNA (nCounter, NanoString Technologies, Inc) in the training cohort (n = 105). The molecular subclassification using FFPE samples was highly comparable with the FF gold standard with an error rate of < 5% across the various PTCL subtypes.
Accuracy of PTCL Classification and Interlaboratory Reproducibility Using the Refined Signature
The reduced transcript signature retained the accuracy in classification in the HG-U133 plus2.0 platform data19-21 (Data Supplement). Therefore, the reduced diagnostic transcripts were considered the gold standard for subsequent comparisons with the nCounter assay in the training set (Fig 1B, left panel). The classification of the FFPE training cohort was recapitulated on the nCounter platform in 90% (95 of 105) of the cases (Table 1 and Data Supplement). We observed the prognostic difference between PTCL-GATA3 and PTCL-TBX21 (Data Supplement), but the number of samples was too small to reach statistical significance. Since we transitioned from a larger panel to a smaller panel of diagnostic transcripts, seven cases from different PTCL subtypes were re-evaluated with the reduced transcript set versus the original larger panel on the nCounter platform. They demonstrated concordant results, maintaining similar diagnostic accuracy (Data Supplement). When the assay was performed at two additional Clinical Laboratory Improvement Amendments sites to assess the reproducibility, we observed highly concordant results and the same classification as at the original site for all 24 PTCL cases studied (Data Supplement).
Refined Diagnostic Algorithm Across Different PTCL Entities and the Validation Cohort
To assess diagnostic performance, the molecular classification obtained with the nCounter platform was compared with the consensus pathologic diagnosis in an independent validation cohort (n = 140). This cohort had a similar clinical outcome and distribution of PTCL subtypes as the training cohort (Figs 1A and 1C). The classification obtained with the nCounter platform was highly comparable with the diagnosis rendered by expert pathologists, with an overall concordance of 91% (127 of 140, 95% CI, 0.85 to 0.95) in the validation cases, and refined the classification of challenging PTCL cases as indicated below (Table 1, Fig 1B, right panel, and Data Supplement).
AITL.
Average expression of the diagnostic signature significantly correlated with a pan T-FH gene expression signature (ie, six transcripts defined in WHO as TFH markers6; Figs 2A and 2B). The cases molecularly classified as AITL in the training and validation cohorts by nCounter showed immunomorphological features commonly associated with AITL. Consistent with previous studies, high expression of CD20 was associated with a better OS,20,21,30 which was validated using immunohistochemistry (Fig 2C). An AITL mutation spectrum (ie, TET2, DNMT3A, RHOAGI7V, and IDH2R172) was seen in 89% of the cases with available sequencing data (Fig 2D).
FIG 2.
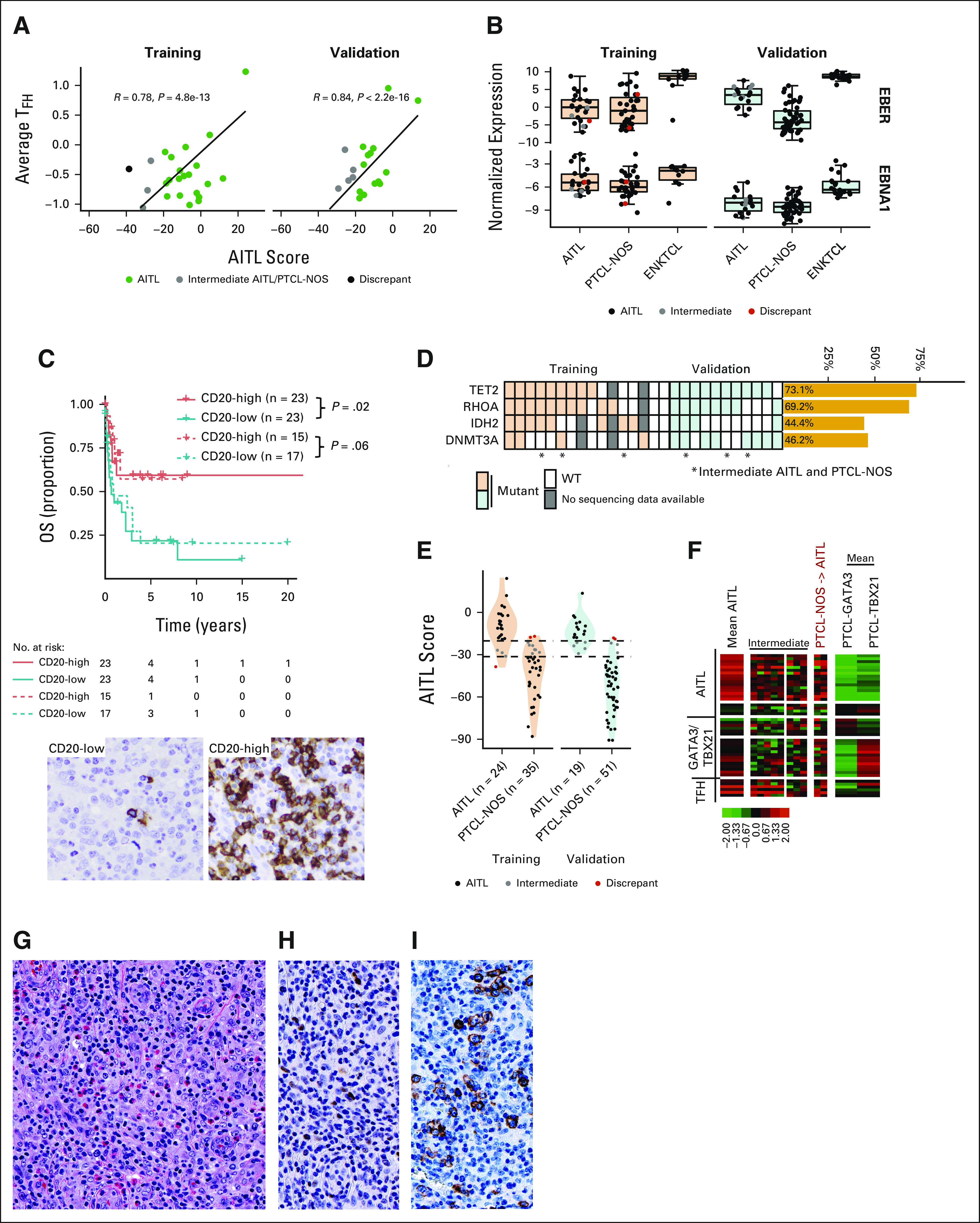
AITL classification. (A) Scatterplot of the AITL diagnostic score versus the average expression of five TFH-related genes in training and validation AITLs. (B) Boxplot of EBER and EBNA1 transcript expression in the AITLs (training/validation cohort). (C) Kaplan-Meier curve of OS of AITLs in the combined cohorts by CD20 mRNA expression. Solid lines are cases classified as AITL by molecular classification (P = .02); dotted lines are AITL by pathology (P = .06). CD20 expression in representative low- and high-expression AITL cases (400×). (D) Mutation status of cases with available sequencing data. Cases that were AITL-PTCL-NOS intermediate or did not classify as AITL on the NanoString classifier are noted with an asterisk. (E) Violin and dot plot of AITL classification diagnostic scores in AITL and PTCL-NOS cases profiled on the nCounter. Cases that were discordant between AITL and PTCL-NOS are given in red, and intermediate AITL cases in gray. (F) Heatmaps of AITL showing disagreement by NanoString classification in the validation cohort. The mean signature of the concordant cases is shown. For cases labeled intermediate, those diagnosed as AITL by consensus pathology review are on the left and intermediate cases diagnosed as PTCL-NOS are on the right. Two PTCL-NOS classified as AITL by nCounter. (G-I) Shown focal expression of BCL-6 and ICOS (400x; G, H, and I; G, H&E, H, BCL-6 and I, ICOS) seen in a PTCL-NOS case that was classified as AITL by the nCounter platform. AITL, angioimmunoblastic T-cell lymphoma; BCL-6, B-cell lymphoma 6; ENKTCL, extranodal natural killer/T-cell lymphoma; ICOS, inducible T cell co-stimulator; NOS, not otherwise specified; OS, overall survival; PTCL, peripheral T-cell lymphoma; TFH, T follicular helper; WT, wild type.
Using the nCounter platform, AITL was classified with an 83% concordance in the training cohort (20 of 24) and 74% in the validation cohort (14 of 19), whereas the remaining cases (three in training and five in validation) showed the borderline model score between AITL and PTCL-NOS (Table 1, Fig 2E, and Data Supplement). These cases missed the AITL molecular diagnosis on the basis of the threshold or cut point. Upon re-review, these cases were confirmed to have classical AITL immunomorphological features, and mutation analysis supported the diagnosis, with a classical AITL mutation spectrum including TET2, RHOAG17V, and IDH2R172 as shown in Figure 2D. These cases showed an AITL diagnostic signature expression at comparatively higher levels than other PTCLs, marginally lower than AITL, and a higher expression of the TFH signature (Figs 2A and 2F). As expected, the three follicular T-cell lymphoma cases were all classified as AITL. Two PTCL-NOS cases were molecularly classified as AITL. These cases showed focal positivity of the TFH markers B-cell lymphoma 6 (BCL-6) and inducible T cell co-stimulator (ICOS) (Figs 2G-2I), but criteria for nodal PTCL-TFH were not met. Although these two cases (Fig 2F) clearly expressed AITL signature genes and TFH mRNA signatures higher than most other PTCL-NOS cases, mutations in the genes commonly seen in AITL (TET2, IDH2R172, RHOAG17V, and DNMT3A) were not present. These two cases may represent PTCL-TFH that were not classified by the immunostains performed.
ALCL subtypes.
Initial analysis of ALCL versus other PTCLs showed an 83% (30 of 36, 95% CI, 0.67 to 0.94) concordance in the validation cohort, which was comparable with the training cohort (26 of 29; 90%, 95% CI, 0.73 to 0.98) using the refined signature (Fig 3A). The validation cohort (Table 1 and Fig 3B) showed a 90% (18 of 20) concordance in ALK+ ALCL and 75% (12 of 16) in ALK‐negative ALCL (ALK− ALCL). Interestingly, two ALK− ALCLs were classified as ALK+ ALCL because of a high ALK+ ALCL signature, but ALK mRNA expression was low (Fig 3C), and IHC did not detect ALK protein expression. As expected, ALCL-classified cases by ALK status showed that ALK+ ALCL cases had a better OS (Fig 3D). Remarkably, none of the PTCL-NOS cases, including those with strong CD30 positivity by IHC, were misclassified into ALCL. However, four ALCL (two ALK+ ALCLs and two ALK− ALCLs) showed a comparatively lower ALCL signature expression although the two ALK+ ALCLs had a high ALK signature (Fig 3C). The two ALK− ALCL cases with a low ALCL signature did not express CD30 mRNA, and the failure to classify these cases may be attributed to inadequate RNA quality or low tumor content (Fig 3E). These findings suggest that the occasional cases with a low diagnostic signature should be diagnosed with caution.
FIG 3.
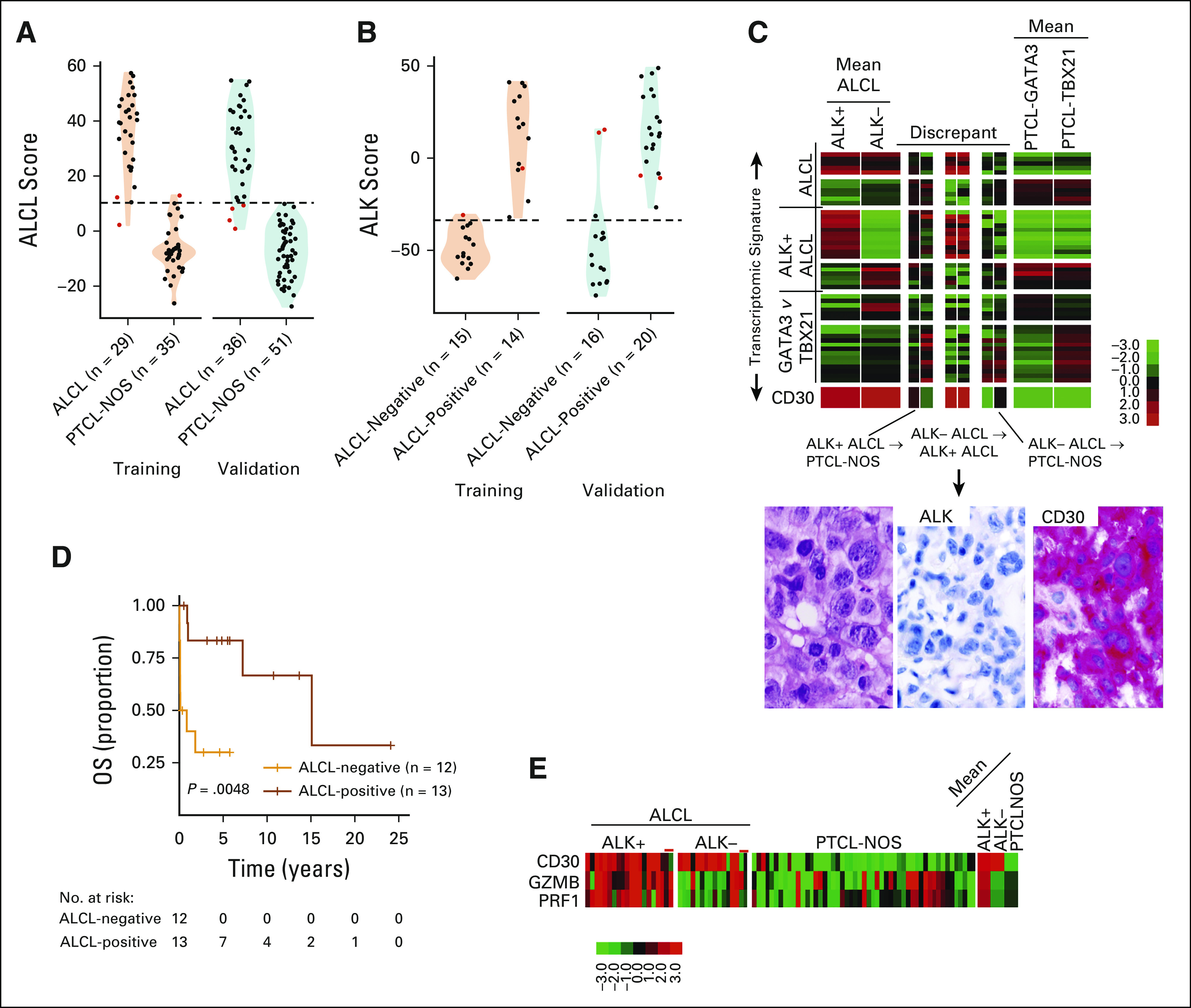
ALCL classification. (A) Violin and dot plot of ALCL classification scores versus PTCL-NOS. (B) ALK-positive ALCL versus ALK-negative ALCL on the nCounter platform. Cases that were discordant between ALCL and PTCL-NOS or ALK-negative and ALK-positive are given in red. (C) Heatmaps of ALK-negative and ALK-positive ALCL cases showing disagreement by NanoString classification in the validation cohort. H&E and CD30 and ALK immunostains of a representative ALK-negative ALCL case that was classified as ALK-positive ALCL by NanoString assay are shown (400×). (D) Kaplan-Meier curve of OS of ALCLs in the training and validation cohort by NanoString classification. (E) Heatmap of CD30 and cytotoxic transcript expression in ALCL and PTCL-NOS cases. Discrepant cases are noted with red lines. ALCL, anaplastic large cell lymphoma; H&E, hematoxylin and eosin; NOS, not otherwise specified; OS, overall survival; PTCL, peripheral T-cell lymphoma.
ATLL.
The ATLL molecular signature detected 100% of ATLL cases in the training cohort (7 of 7, 95% CI, 0.59 to 1) and 83% of the validation cohort cases (10 of 12, 95% CI, 0.52 to 0.98; Fig 4A). The two cases showing disagreement had marginal expression of the ATLL diagnostic signature, but both were confirmed to be positive for HTLV1 mRNA expression (ie, HTLV-1 bZIP factor [HBZ] by quantitative real-time polymerase chain reaction [qRT-PCR]) although in one, the expression of HBZ measured by the nCounter was lower than other ATLLs (Figs 4B and 4C). Of the two HTLV1 transcripts (HBZ and Tax-1), HBZ was consistently expressed at higher levels in ATLL cases compared with Tax-1 and showed a positive correlation with the ATLL signature (Figs 4D and 4E) and a significant correlation with HBZ expression measured by qRT-PCR (Fig 4F). Interestingly, two PTCL-NOS cases from the validation cohort were molecularly classified as ATLL (Fig 4A). Re-evaluation of these cases using HBZ-specific qRT-PCR confirmed the expression of HBZ (Fig 4E), and subsequent review of the clinical chart indicated serologic positivity for HTLV1 and a clinical presentation compatible with ATLL, unknown at the time of the initial diagnosis. Morphologically, these cases consisted of CD4-positive monomorphic T-cell lymphomas, which, in the absence of an appropriate clinical history, blood examination, and serologic testing, were misdiagnosed as PTCL-NOS7 (Fig 4B). Therefore, these two cases were reclassified as ATLL.
FIG 4.
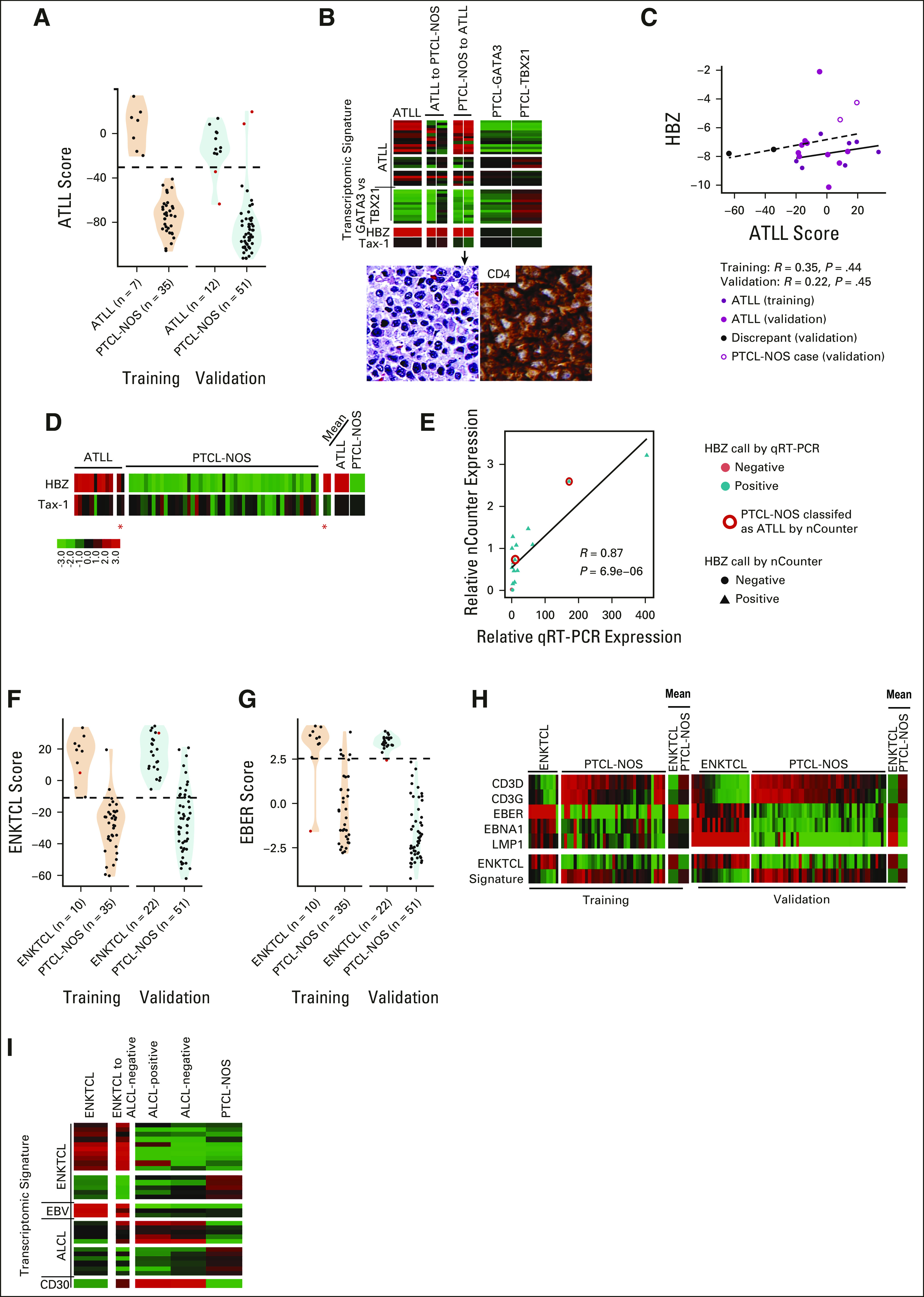
ATLL and ENKTCL classification. (A) Violin and dot plot of ATLL classification scores in ATLL and PTCL-NOS cases profiled on the nCounter. (B) Heatmaps of ATLL cases discordant by nCounter classification in the validation cohort. H&E stain and CD4 staining for a case diagnosed as PTCL-NOS but classified at ATLL by nCounter (lower panel). (C) Scatterplot of expression of the HTLV-1–specific transcripts HBZ versus ATLL score in training and validation ATLL cases. The solid fitted line represents training data, and the dashed line validation data. (D) Heatmap of HBZ and Tax-1 expression in the ATLL and PTCL-NOS validation cohorts. The discrepant cases are noted by a red asterisk. (E) Scatterplot of HBZ expression measured by qRT-PCR versus nCounter. (F and G) Violin and dot plots of (F) ENKTCL classification scores or (G) EBER scores in ENKTCLs and PTCL-NOS cases profiled in the training and validation cohorts on the nCounter. Discordant cases are given in red. (H) Heatmap of expression of CD3 gamma and delta and EBV transcripts in ENKTCL and PTCL-NOS cases in the training and validation cohorts. (I) Heatmap of expression of relevant signatures in the ENKTCL-discordant case compared with average signatures in the validation cohort. ATLL, adult T-cell leukemia/lymphoma; EBV, Epstein-Barr virus; ENKTCL, extranodal natural killer/T-cell lymphoma; H&E, hematoxylin and eosin; HBZ, HTLV-1 bZIP factor; HTLV-1, human T-lymphotropic virus; NOS, not otherwise specified; PTCL, peripheral T-cell lymphoma; qRT-PCT, quantitative real-time polymerase chain reaction.
ENKTCL.
The ENKTCL molecular classifier was able to identify 90% (9 of 10) of ENKTCL cases in the training cohort and 95% (21 of 22) in the validation cohort (Table 1 and Figs 4F and 4G). A subset of cases had elevated expression of CD3γ and CD3δ relative to other cases (Fig 4H) and may be derived from the T-lineage. Two cases were diagnosed according to current WHO classification as primary EBV-positive nodal T/NK-cell lymphoma. These cases resemble ENKTCL but with a primarily nodal presentation, and both were classified as ENKTCL by the molecular assay (Data Supplement). One molecularly classified ENTKCL case uniquely showed both ALCL and ENKTCL signatures but strong expression of EBV transcripts (Fig 4I).
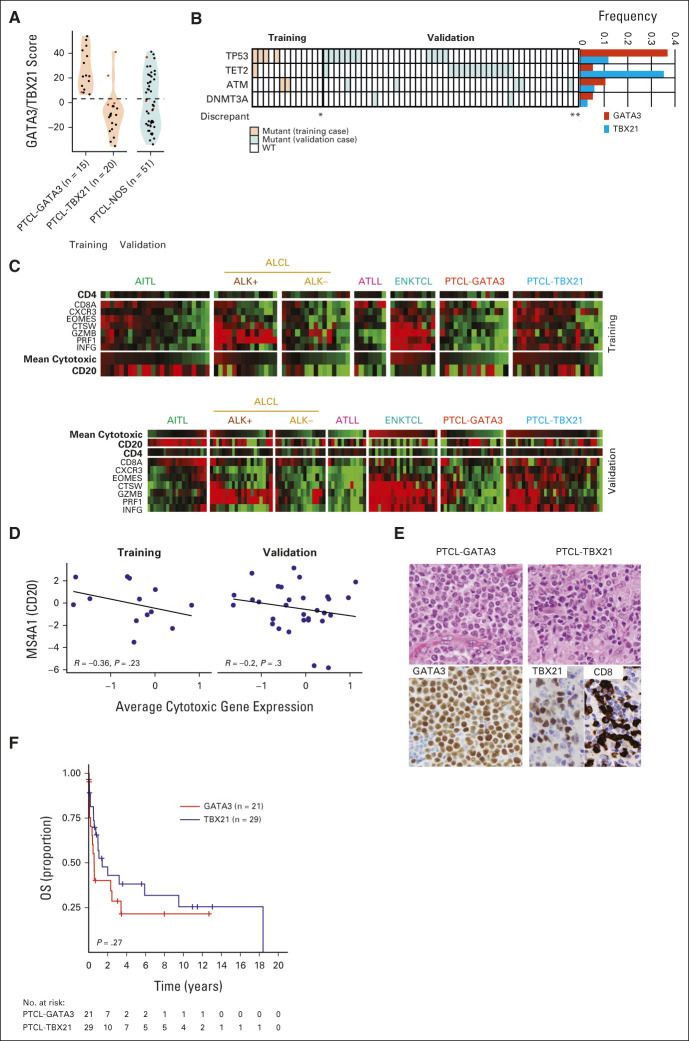
Refinement and Validation of Two Novel PTCL-NOS Subtypes (PTCL-GATA3 and PTCL-TBX21)
Of the PTCL-NOS cases in the training cohort, we separated two novel molecular subgroups with an 87% concordance (ie, PTCL-GATA3 or PTCL-TBX21) with a reduced number of transcripts (19). Of the remaining PTCL-NOS groups in the validation, 40% (21 of 52) were classified as the PTCL-GATA3 subtype and 52% (27 of 52) as the PTCL-TBX21 subtype using the nCounter platform (Table 1 and Fig 5A). This classification showed good concordance with our IHC algorithm classification (overall concordance: 80%). To further validate our transcriptomic signature classification, we compared sequencing data in these two groups. We observed that genetic alterations like TET2 mutation were frequent in the PTCL-TBX21 subtype, whereas TP53 mutations were enriched in the PTCL-GATA3 subgroup, concordant with our previous findings31 (Fig 5B).
FIG 5.
PTCL-NOS subclassification. (A) Violin and dot plots of PTCL-GATA3 classification scores in PTCL-NOS cases profiled in the training and validation cohorts on the nCounter. (B) Mutation status of the PTCL-NOS cohort with available sequencing data. (C) Heatmaps of expression of CD4, CD8, CD20, and cytotoxic genes in the training (upper) and validation (lower) cases. In CD4-positive lymphomas, the CD8 expression is likely contributed from the tumor microenvironment. (D) Scatterplot of the average expression of the cytotoxic genes versus CD20 in cases that classified as PTCL-TBX21 by NanoString. (E) H&E and IHC stains for one representative PTCL-GATA3 case showing GATA3 (left) and one PTCL-TBX21 case showing TBX21 and CD8 expression (right). (F) Kaplan-Meier curve of OS for PTCL-NOS cases with available outcome data in the combined training and validation cohorts classified as PTCL-GATA3 or PTCL-TBX21 NanoString. AITL, angioimmunoblastic T-cell lymphoma; ALCL, anaplastic large cell lymphoma; ATLL, adult T-cell lymphoma/leukemia; ENKTCL, extranodal natural killer/T-cell lymphoma; IHC, immunohistochemistry; NOS, not otherwise specified; OS, overall survival; PTCL, peripheral T-cell lymphoma; WT, wild-type.
Consistent with our previous observations,21,29 cases with the expression of cytotoxic transcripts were significantly enriched in the PTCL-TBX21 subtype (Fig 5C) and validated by a more frequent cytotoxic immunophenotype in the PTCL-TBX21 subtype than in the PTCL-GATA3 subtype (52% v 16%, P = .013; Fig 5C). An inverse correlation was noted between the average cytotoxic CD8+ T-cell signature and the pan B-cell CD20 transcripts (Fig 5D). Consistent with our previous observation,29 PTCL-TBX21 cases frequently had an enriched inflammatory background irrespective of the cytotoxic phenotype (Fig 5E). Since the clinical outcome of the PTCL-NOS was available only in a limited number of cases, the combined cohort showed a trend of inferior OS associated with cases classified as PTCL-GATA3 versus PTCL-TBX21 (median OS: 0.57 v 1.4 years; Fig 5F).
Evaluation of the PTCL-TFH Cases, Excluded from the Validation Cohort
PTCL-TFH (n = 12) cases were analyzed using the nCounter platform, and only two (17%) showed a significant association with the AITL molecular signature, whereas four cases had borderline scores between AITL and PTCL-NOS and six cases showed a clear association with the PTCL-NOS, resembling PTCL-TBX21 cases (n = 3) or PTCL-GATA3 (n = 3). When we specifically examined the TFH signature, the PTCL-TFH cases with high AITL signatures also showed higher TFH signatures than the rest (Data Supplement). We also included 10 cases of reactive hyperplasia, and none of them showed expression of the diagnostic signatures for subtypes of PTCL included in the assay.
DISCUSSION
The diagnosis of PTCL is one of the most challenging among lymphomas and more often results in an inconclusive, inconsistent, or incorrect diagnosis.19-21,32,33 Recently, novel therapeutic approaches have shown striking benefits in subgroups of PTCL, including brentuximab vedotin on CD30+ PTCL, particularly ALCL34; crizotinib in ALK+ ALCL35,36; mogamulizumab in ATLL37; HDACi and demethylating agents in AITL or TFH-PTCL38; and possibly enasidenib in IDH2-mutant AITL.39 Thus, accurate diagnosis may be important for patient treatment and in clinical trials of new drugs.40-42 We have performed extensive GEP studies on PTCL, constructed RNA-based molecular diagnostic signatures and predictors of survival, and delineated critical oncogenic mechanisms.19-21 Some of these findings have been included in the 2016 WHO classification.6 To translate this molecular information to a platform suitable for clinical application,25 we performed a systematic analysis to identify RNAs from FFPE tissues using the nCounter platform that correlated well with GEP data from FF tissues. This digital quantitation technology is more tolerant of degraded RNA typical of FFPE materials. In addition, we developed a diagnostic transcriptomic signature with a minimum number of transcripts that performed comparably in the training set with the previous GEP-derived diagnosis.13-15 In the preanalytical assessment, RNA yield and quality (ie, DV200 [% of RNA fragments above 200 nucleotides] > 50%) of older specimens were improved using the RNAstorm kit. However, in recently acquired FFPE tissues, other isolation methods may also provide good-quality RNA. In addition, this assay could be performed using limited RNA quantities (minimum required 200ng), usually obtained from a few unstained slides depending on the size of the tissue. However, we were able to have an adequate classification even using RNA extracted from core needle biopsies, which represented approximately 10% of the study samples. The interlaboratory comparison of variability and reproducibility across three Clinical Laboratory Improvement Amendments–certified laboratories also correlated very well.
As we do not have GEP data on cases for validation, we only included cases with a firm pathology diagnosis after extensive IHC studies and stringent review. The findings were very similar to those of the training set (Fig 1), maintaining high sensitivity, specificity, and accuracy. In some cases, the signature score fell just outside the cutoff points, and these were considered as borderline cases. It is worth noting that in a few cases, the molecular assay made the correct diagnosis on a retrospective analysis. However, frank discrepancies were also observed. This could be partly due to technical reasons such as tumor content and heterogeneity of the tumor, but some may be related to biology that was not known yet, such as a strong ALK signature in some ALK− ALCL cases that may represent the recently reported ALK+‐like ALCL.43 Similarly, two PTCL-NOS showed significant association with AITL and TFH transcriptomics signatures, which upon review, showed focal expression of TFH markers by immunohistochemistry (BCL-6 and ICOS). The strong TFH expression signature may indicate that these cases are more similar to PTCL-TFH but did not meet the current criteria of strong expression of at least two TFH markers. In these cases, the molecular assay revealed the complex and overlapping biology between PTCL-NOS and AITL and their poorly defined borders. We also found that the addition of EBV and HTLV1 transcripts enhanced the diagnostic performance of the molecular assay in ENTKCL and ATLL, respectively. One important contribution of this approach was the robust definition of the PTCL-GATA3 and PTCL-TBX21 cases, which have different biology and prognosis as supported by recent genetic findings.31 Although it is possible to simulate the GEP classification using an IHC panel, the stains can be challenging to optimize and interpret, which may lead to substantial variability among institutions. The assay reported here was highly reproducible among laboratories and, thus, presents a major advantage.
The GEP study that initially defined the diagnostic signatures was performed before the definition of the provisional entity of PTCL-TFH was not formally included in this study. However, the 12 PTCL-TFH cases explored with the nCounter assay showed higher mean AITL and TFH signatures, and individual signatures overlapped with the molecular signatures for AITL or PTCL subtypes, similar to the study by Dobay et al.44 These exploratory studies indicated that PTCL-TFH is unlikely to be a single entity as currently defined and needs further evaluation to determine how best it should be characterized. Similarly, our previous GEP studies20,21 indicated a cytotoxic PTCL variant within the PTCL-TBX21 subgroup. Consistent with that observation, we found 25% of the cases in PTCL-TBX21 to have a cytotoxic signature and validated an association with the CD8+ phenotype by IHC. Because of the small number of cases studied, further investigations are necessary to develop a robust signature for this group of cases.
In summary, we described an approach to translate the PTCL diagnostic signatures into a clinically applicable assay, which we envision to be a useful tool for general and academic pathologists in the diagnostically challenging field of PTCL. In addition, we believe that it can facilitate a better definition of cases for research studies and ensure more uniform cohorts for clinical trials. PTCL classification is an evolving area, and as our understanding of the underlying biology and available technology improves, modifications will be instituted to make the classification clinically relevant.
ACKNOWLEDGMENT
The authors thank the Molecular Diagnostics Laboratory at the Department of Pathology at UNMC (Dr Allison Cushman) and COH (Dr Holly Yin and Dr Raju Pallai) for facilitating nCounter and Digital Analyzer use at their facility.
George Wright
Patents, Royalties, Other Intellectual Property: Inventor on the patent Methods for Selecting And Treating Lymphoma Types, Inventor on patent Evaluation of Mantle Cell Lymphoma And Methods Related Thereto
Timothy C. Greiner
Consulting or Advisory Role: Daiichi Sankyo/UCB Japan
Lynette Smith
Consulting or Advisory Role: Therapeutic Vision Inc
Stefano Pileri
Speakers' Bureau: BeiGene
Kerry J. Savage
Honoraria: Seattle Genetics, Bristol Myers Squibb, Merck, Kyowa, Novartis Canada Pharmaceuticals Inc, Janssen Oncology
Consulting or Advisory Role: Seattle Genetics, Bristol Myers Squibb, Merck, Novartis Canada Pharmaceuticals Inc, Kyowa, Janssen Oncology
Research Funding: Roche (Inst), Bristol Myers Squibb (Inst)
Travel, Accommodations, Expenses: Seattle Genetics
Other Relationship: DSMC
Uncompensated Relationships: BeiGene
Graham Slack
Honoraria: Seattle Genetics
Consulting or Advisory Role: Seattle Genetics
James R. Cook
Patents, Royalties, Other Intellectual Property: US Provisional Patent Application No. 61/900,553, filed on November 6, 2013. Title: Methods for Selecting and Treating Lymphoma Types. Role: Coinventor
Choon Kiat Ong
Patents, Royalties, Other Intellectual Property: Publication No.: WO/2019/070204. “Methods For Treating Lymphomas” Singapore Patent Application 10201708262R (Filing date: October 6, 2017). International Application No.: PCT/SG2018/050509. International Filing Date: October 8, 2018
Philipp W. Raess
Research Funding: Scopio Labs
Piers Blombery
Honoraria: Adaptive Biotechnologies
Elaine Jaffe
Stock and Other Ownership Interests: Gilead Sciences, Merck, Teva
Lisa M. Rimsza
Consulting or Advisory Role: Roche Molecular Diagnostics
Patents, Royalties, Other Intellectual Property: Coinventor on a patent undergoing commercialization by NanoString
Julie M. Vose
Honoraria: Acerta Pharma/AstraZeneca, Roche/Genentech, MorphoSys, Conjupro Biotherapeutics, Oncternal Therapeutics, Johnson and Johnson, MEI Pharma, Lilly
Research Funding: Celgene (Inst), Incyte (Inst), Acerta Pharma (Inst), Kite, a Gilead company (Inst), Seattle Genetics (Inst), Novartis (Inst), Bristol Myers Squibb (Inst), AstraZeneca (Inst), Loxo, Epizyme
Louis Staudt
Patents, Royalties, Other Intellectual Property: Patents and patents pending regarding gene expression profiling in lymphoma that have been licensed by NanoString and for which I receive royalties
Wing C. Chan
Patents, Royalties, Other Intellectual Property: Patent on the diagnostic algorithm on GCB/ABC type DLBCL and patent on the diagnostic algorithm for PTCL
No other potential conflicts of interest were reported.
DISCLAIMER
The views expressed are the personal opinions of the authors and do not necessarily reflect the policy of the US National Cancer Institute.
SUPPORT
Supported by NIH NCI grants UH2/3CA206127-02; R41CA221466-01A1, U01CA253218A1, and P01 CA229100; the Leukemia and Lymphoma Society (TRP-6129-04); NIH NCI Eppley Cancer Center Support grant P30 CA036727 (J.I.); and National Cancer Institute Cancer center support grant P30CA033572 (W.C.C.). This study was supported by a grant from Fondazione Cassa di Risparmio di Modena, Associazione Angela Serra per la Ricerca sul Cancro, Fondazione Italiana Linfomi, Allos Therapeutics, and Spectrum Pharmaceuticals, AIRC 5×1000 (grant No. 21198 to S.P.).
C.A. and A.B. contributed equally to this work.
AUTHOR CONTRIBUTIONS
Conception and design: Catalina Amador, Alyssa Bouska, George Wright, Wing C. Chan, Javeed Iqbal
Financial support: Wing C. Chan, Javeed Iqbal
Administrative support: Wing C. Chan, Javeed Iqbal
Provision of study materials or patients: Catalina Amador, Andrew L. Feldman, Timothy C. Greiner, Stefano Pileri, German Ott, Kerry J. Savage, Won Seog Kim, Gehong Dong, Joo Song, Sarah Ondrejka, Carlos Barrionuevo, Choon Kiat Ong, Jennifer Chapman, Giorgio Inghirami, Philipp W. Raess, Clare Gould, Elaine Jaffe, Lisa M. Rimsza, Julie M Vose, Wing C. Chan
Collection and assembly of data: Catalina Amador, Alyssa Bouska, Andrew L. Feldman, Tayla Heavican, Stefano Pileri, Valentina Tabanelli, German Ott, Andreas Rosenwald, Graham Slack, Won Seog Kim, Young Hyeh, Yuping Li, Gehong Dong, Joo Song, Sarah Ondrejka, James R. Cook, Carlos Barrionuevo, Soon Thye Lim, Jennifer Chapman, Giorgio Inghirami, Philipp W. Raess, Sharathkumar Bhagavathi, Clare Gould, Piers Blombery, Elaine Jaffe, Julie M Vose, Wing C. Chan, Javeed Iqbal
Data analysis and interpretation: Catalina Amador, Alyssa Bouska, George Wright, Dennis D. Weisenburger, Andrew L. Feldman, Timothy C. Greiner, Waseem Lone, Lynette Smith, Andreas Rosenwald, Kerry J. Savage, Joo Song, Soon Thye Lim, Choon Kiat Ong, Sharathkumar Bhagavathi, Piers Blombery, Stephan W. Morris, Lisa M. Rimsza, Julie M Vose, Louis Staudt, Wing C. Chan, Javeed Iqbal
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Gene Expression Signatures for the Accurate Diagnosis of Peripheral T-Cell Lymphoma Entities in the Routine Clinical Practice
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
George Wright
Patents, Royalties, Other Intellectual Property: Inventor on the patent Methods for Selecting And Treating Lymphoma Types, Inventor on patent Evaluation of Mantle Cell Lymphoma And Methods Related Thereto
Timothy C. Greiner
Consulting or Advisory Role: Daiichi Sankyo/UCB Japan
Lynette Smith
Consulting or Advisory Role: Therapeutic Vision Inc
Stefano Pileri
Speakers' Bureau: BeiGene
Kerry J. Savage
Honoraria: Seattle Genetics, Bristol Myers Squibb, Merck, Kyowa, Novartis Canada Pharmaceuticals Inc, Janssen Oncology
Consulting or Advisory Role: Seattle Genetics, Bristol Myers Squibb, Merck, Novartis Canada Pharmaceuticals Inc, Kyowa, Janssen Oncology
Research Funding: Roche (Inst), Bristol Myers Squibb (Inst)
Travel, Accommodations, Expenses: Seattle Genetics
Other Relationship: DSMC
Uncompensated Relationships: BeiGene
Graham Slack
Honoraria: Seattle Genetics
Consulting or Advisory Role: Seattle Genetics
James R. Cook
Patents, Royalties, Other Intellectual Property: US Provisional Patent Application No. 61/900,553, filed on November 6, 2013. Title: Methods for Selecting and Treating Lymphoma Types. Role: Coinventor
Choon Kiat Ong
Patents, Royalties, Other Intellectual Property: Publication No.: WO/2019/070204. “Methods For Treating Lymphomas” Singapore Patent Application 10201708262R (Filing date: October 6, 2017). International Application No.: PCT/SG2018/050509. International Filing Date: October 8, 2018
Philipp W. Raess
Research Funding: Scopio Labs
Piers Blombery
Honoraria: Adaptive Biotechnologies
Elaine Jaffe
Stock and Other Ownership Interests: Gilead Sciences, Merck, Teva
Lisa M. Rimsza
Consulting or Advisory Role: Roche Molecular Diagnostics
Patents, Royalties, Other Intellectual Property: Coinventor on a patent undergoing commercialization by NanoString
Julie M. Vose
Honoraria: Acerta Pharma/AstraZeneca, Roche/Genentech, MorphoSys, Conjupro Biotherapeutics, Oncternal Therapeutics, Johnson and Johnson, MEI Pharma, Lilly
Research Funding: Celgene (Inst), Incyte (Inst), Acerta Pharma (Inst), Kite, a Gilead company (Inst), Seattle Genetics (Inst), Novartis (Inst), Bristol Myers Squibb (Inst), AstraZeneca (Inst), Loxo, Epizyme
Louis Staudt
Patents, Royalties, Other Intellectual Property: Patents and patents pending regarding gene expression profiling in lymphoma that have been licensed by NanoString and for which I receive royalties
Wing C. Chan
Patents, Royalties, Other Intellectual Property: Patent on the diagnostic algorithm on GCB/ABC type DLBCL and patent on the diagnostic algorithm for PTCL
No other potential conflicts of interest were reported.
REFERENCES
- 1. Bellei M, Chiattone CS, Luminari S, et al. T-cell lymphomas in South America and Europe. Rev Bras Hematol Hemoter. 2012;34:42–47. doi: 10.5581/1516-8484.20120013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vose J, Armitage J, Weisenburger D, et al. International peripheral T-cell and natural killer/T-cell lymphoma study: Pathology findings and clinical outcomes. J Clin Oncol. 2008;26:4124–4130. doi: 10.1200/JCO.2008.16.4558. [DOI] [PubMed] [Google Scholar]
- 3. Briski R, Feldman AL, Bailey NG, et al. The role of front-line anthracycline-containing chemotherapy regimens in peripheral T-cell lymphomas. Blood Cancer J. 2014;4:e214. doi: 10.1038/bcj.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rudiger T, Weisenburger DD, Anderson JR, et al. Peripheral T-cell lymphoma (excluding anaplastic large-cell lymphoma): Results from the Non-Hodgkin's Lymphoma Classification Project. Ann Oncol. 2002;13:140–149. doi: 10.1093/annonc/mdf033. [DOI] [PubMed] [Google Scholar]
- 5. Fox CP, Civallero M, Ko YH, et al. Survival outcomes of patients with extranodal natural-killer T-cell lymphoma: A prospective cohort study from the international T-cell project. Lancet Haematol. 2020;7:e284–e294. doi: 10.1016/S2352-3026(19)30283-2. [DOI] [PubMed] [Google Scholar]
- 6.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues Volume 2ed 4Lyon, France: IARC Press; 2017. https://www.iarc.who.int/news-events/who-classification-of-tumours-of-haematopoietic-and-lymphoid-tissues-2/ [Google Scholar]
- 7. Weisenburger DD, Savage KJ, Harris NL, et al. Peripheral T-cell lymphoma, not otherwise specified: A report of 340 cases from the International Peripheral T-cell Lymphoma Project. Blood. 2011;117:3402–3408. doi: 10.1182/blood-2010-09-310342. [DOI] [PubMed] [Google Scholar]
- 8. Morris SW, Kirstein MN, Valentine MB, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 9. Tsukasaki K, Tsushima H, Yamamura M, et al. Integration patterns of HTLV-I provirus in relation to the clinical course of ATL: Frequent clonal change at crisis from indolent disease. Blood. 1997;89:948–956. [PubMed] [Google Scholar]
- 10. Escalon MP, Liu NS, Yang Y, et al. Prognostic factors and treatment of patients with T-cell non-Hodgkin lymphoma: The M. D. Anderson Cancer Center experience. Cancer. 2005;103:2091–2098. doi: 10.1002/cncr.20999. [DOI] [PubMed] [Google Scholar]
- 11. Horwitz S, O'Connor OA, Pro B, et al. Brentuximab vedotin with chemotherapy for CD30-positive peripheral T-cell lymphoma (ECHELON-2): A global, double-blind, randomised, phase 3 trial. Lancet. 2019;393:229–240. doi: 10.1016/S0140-6736(18)32984-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pro B, Advani R, Brice P, et al. Five-year results of brentuximab vedotin in patients with relapsed or refractory systemic anaplastic large cell lymphoma. Blood. 2017;130:2709–2717. doi: 10.1182/blood-2017-05-780049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 14. Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 15. Dave SS, Fu K, Wright GW, et al. Molecular diagnosis of Burkitt's lymphoma. N Engl J Med. 2006;354:2431–2442. doi: 10.1056/NEJMoa055759. [DOI] [PubMed] [Google Scholar]
- 16. Monti S, Savage KJ, Kutok JL, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2005;105:1851–1861. doi: 10.1182/blood-2004-07-2947. [DOI] [PubMed] [Google Scholar]
- 17. Rosenwald A, Wright G, Wiestner A, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003b;3:185–197. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- 18. Shipp MA, Ross KN, Tamayo P, et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 19. Iqbal J, Weisenburger DD, Chowdhury A, et al. Natural killer cell lymphoma shares strikingly similar molecular features with a group of non-hepatosplenic γδ T-cell lymphoma and is highly sensitive to a novel aurora kinase A inhibitor in vitro. Leukemia. 2011;25:348–358. doi: 10.1038/leu.2010.255. [DOI] [PubMed] [Google Scholar]
- 20. Iqbal J, Weisenburger DD, Greiner TC, et al. Molecular signatures to improve diagnosis in peripheral T-cell lymphoma and prognostication in angioimmunoblastic T-cell lymphoma. Blood. 2010;115:1026–1036. doi: 10.1182/blood-2009-06-227579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iqbal J, Wright G, Wang C, et al. Gene expression signatures delineate biological and prognostic subgroups in peripheral T-cell lymphoma. Blood. 2014;123:2915–2923. doi: 10.1182/blood-2013-11-536359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lone W, Alkhiniji A, Manikkam Umakanthan J, et al. Molecular insights into pathogenesis of peripheral T cell lymphoma: A review. Curr Hematol Malig Rep. 2018;13:318–328. doi: 10.1007/s11899-018-0460-z. [DOI] [PubMed] [Google Scholar]
- 23. Iqbal J, Amador C, McKeithan TW, et al. Molecular and genomic landscape of peripheral T-cell lymphoma. Cancer Treat Res. 2019;176:31–68. doi: 10.1007/978-3-319-99716-2_2. [DOI] [PubMed] [Google Scholar]
- 24. Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161:1961–1971. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Crescenzo R, Abate F, Lasorsa E, et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer cell. 2015;27:516–532. doi: 10.1016/j.ccell.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scott DW, Wright GW, Williams PM, et al. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood. 2014;123:1214–1217. doi: 10.1182/blood-2013-11-536433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Phang KC, Akhter A, Tizen NMS, et al. Comparison of protein-based cell-of-origin classification to the Lymph2Cx RNA assay in a cohort of diffuse large B-cell lymphomas in Malaysia. J Clin Pathol. 2018;71:215–220. doi: 10.1136/jclinpath-2017-204548. [DOI] [PubMed] [Google Scholar]
- 28. Rimsza LM, Wright G, Schwartz M, et al. Accurate classification of diffuse large B-cell lymphoma into germinal center and activated B-cell subtypes using a nuclease protection assay on formalin-fixed, paraffin-embedded tissues. Clin Cancer Res. 2011;17:3727–3732. doi: 10.1158/1078-0432.CCR-10-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Amador C, Greiner TC, Heavican TB, et al. Reproducing the molecular subclassification of peripheral T-cell lymphoma-NOS by immunohistochemistry. Blood. 2019;134:2159–2170. doi: 10.1182/blood.2019000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Advani RH, Ansell SM, Lechowicz MJ, et al. A phase II study of cyclophosphamide, etoposide, vincristine and prednisone (CEOP) alternating with pralatrexate (P) as front line therapy for patients with peripheral T-cell lymphoma (PTCL): Final results from the T- cell consortium trial. Br J Haematol. 2016;172:535–544. doi: 10.1111/bjh.13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heavican TB, Bouska A, Yu J, et al. Genetic drivers of oncogenic pathways in molecular subgroups of peripheral T-cell lymphoma. Blood. 2019;133:1664–1676. doi: 10.1182/blood-2018-09-872549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maura F, Agnelli L, Leongamornlert D, et al. Integration of transcriptional and mutational data simplifies the stratification of peripheral T-cell lymphoma. Am J Hematol. 2019;94:628–634. doi: 10.1002/ajh.25450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Piccaluga PP, Fuligni F, De Leo A, et al. Molecular profiling improves classification and prognostication of nodal peripheral T-cell lymphomas: Results of a phase III diagnostic accuracy study. J Clin Oncol. 2013;31:3019–3025. doi: 10.1200/JCO.2012.42.5611. [DOI] [PubMed] [Google Scholar]
- 34. Van Der Weyden C, Dickinson M, Whisstock J, et al. Brentuximab vedotin in T-cell lymphoma. Expert Rev Hematol. 2019;12:5–19. doi: 10.1080/17474086.2019.1558399. [DOI] [PubMed] [Google Scholar]
- 35. Andraos E, Dignac J, Meggetto F, et al. NPM-ALK: A driver of lymphoma pathogenesis and a therapeutic target. Cancers (Basel) 2021;13:E144. doi: 10.3390/cancers13010144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gambacorti-Passerini C, Messa C, Pogliani EM. Crizotinib in anaplastic large-cell lymphoma. N Engl J Med. 2011;364:775–776. doi: 10.1056/NEJMc1013224. [DOI] [PubMed] [Google Scholar]
- 37. Moskowitz AJ, Lunning MA, Horwitz SM. How I treat the peripheral T-cell lymphomas. Blood. 2014;123:2636–2644. doi: 10.1182/blood-2013-12-516245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lopez AT, Bates S, Geskin L. Current status of HDAC inhibitors in cutaneous T-cell lymphoma. Am J Clin Dermatol. 2018;19:805–819. doi: 10.1007/s40257-018-0380-7. [DOI] [PubMed] [Google Scholar]
- 39.A Phase 1/2, Multicenter, Open-Label, Dose-Escalation Study of AG-221 in Subjects With Advanced Solid Tumors, Including Glioma, and With Angioimmunoblastic T-cell Lymphoma, That Harbor an IDH2 Mutation. https://clinicaltrials.gov/ct2/show/NCT02273739
- 40. Ishida T, Joh T, Uike N, et al. Defucosylated anti-CCR4 monoclonal antibody (KW-0761) for relapsed adult T-cell leukemia-lymphoma: A multicenter phase II study. J Clin Oncol. 2012;30:837–842. doi: 10.1200/JCO.2011.37.3472. [DOI] [PubMed] [Google Scholar]
- 41. O'Connor OA, Bhagat G, Ganapathi K, et al. Changing the paradigms of treatment in peripheral T-cell lymphoma: From biology to clinical practice. Clin Cancer Res. 2014;20:5240–5254. doi: 10.1158/1078-0432.CCR-14-2020. [DOI] [PubMed] [Google Scholar]
- 42. Iqbal J, Wilcox R, Naushad H, et al. Genomic signatures in T-cell lymphoma: How can these improve precision in diagnosis and inform prognosis? Blood Rev. 2016;30:89–100. doi: 10.1016/j.blre.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 43. Yeung KHRA, Russell V, Choi W, et al. ALK-negative anaplastic large cell lymphomas encompass distinct subgroups including an ALK-positive-like subgroup with favorable prognosis. Blood. 2021;138:2403. [Google Scholar]
- 44. Dobay MP, Lemonnier F, Missiaglia E, et al. Integrative clinicopathological and molecular analyses of angioimmunoblastic T-cell lymphoma and other nodal lymphomas of follicular helper T-cell origin. Haematologica. 2017;102:e148–e151. doi: 10.3324/haematol.2016.158428. [DOI] [PMC free article] [PubMed] [Google Scholar]