Abstract
SBASE (http://www.icgeb.trieste.it/sbase) is an online resource of protein domain sequences designed to facilitate detection of domain homologies based on a simple database search. The ninth release of the SBASE library of protein domain sequences contains 320 000 annotated structural, functional, ligand-binding and topogenic segments of proteins clustered into over 3481 domain groups and 483 protein families. Domain identification and functional prediction are based on a comparison of BLAST search outputs with a knowledge base of within-group (‘self’) and out-of-group (‘non-self’) similarities of the known domain groups. This is a memory-based approach wherein class-specific similarity functions are automatically learned from the database [Stanfill,C. and Waltz,D. (1986) Commun. ACM, 29, 1213–1228].
INTRODUCTION
SBASE is an online resource of protein domain sequences designed to facilitate detection of domain homologies based on simple database search (1,2). The central concept of the database is the ‘similarity group’, i.e. a group of domain sequences that have significant (e.g. P < 1) BLAST similarities to each other (3). A database versus database comparison is used to build a knowledge base of sequence similarities, and the ‘neighborhood’ of each group is represented in terms of ‘self-similarities’ (between members of the group) as well as ‘non-self similarities’ (between members and non-members; an example is shown in Fig. 1). The cumulative frequency distributions are used as a statistical description of the similarity group (3). When a sequence is compared with the domain database, the parameters shown in the inset of Figure 1 are computed and compared with the precomputed values of the similarity groups. The comparison is carried out either using a straightforward nearest neighbor approach (3), by calculating a probabilistic score (4) or by feed forward neural networks (5). In this approach, the proteins are no longer represented by the sequence, but rather by their similarities to a reference database. We termed the present method a memory-based approach because its principles are analogous to the memory-based learning paradigm described by Stanfill and Waltz (6). Clearly, the number of similarities to the group, the average of group similarities and the probabilistic score can be regarded as class-specific similarity (distance) functions that have parameters (thresholds, frequency distributions) which are automatically learned from the database. Secondly, the reference database (SBASE) and the similarity knowledge base (group statistics) can in fact be considered as the memory of the system. This is an exemplar-based description of the sequence similarity groups, which is thus different from the conventional, consensus descriptions (see 7 for a review), which are prototype-based representations.
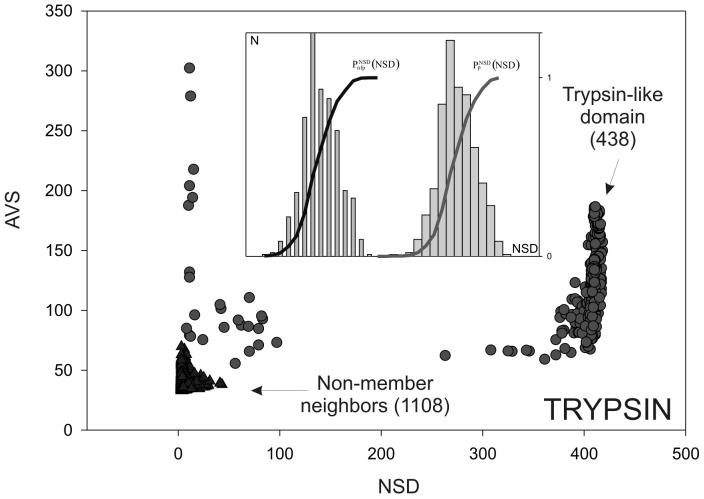
Figure 1.
An example of a domain sequence neighborhood (Trypsin-like domains). Each domain corresponds to one dot in the figure. NSD, number of similarities to the group (i.e. to trypsin domains); AVS, average of group similarities. The members of the group (‘self’) are shown as circles and domains that are not members but still have a significant (P < 1) similarity with the group (‘non-self’) are shown as triangles. (Inset) Schematic representation of the statistical parameters 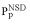 (•) and
(•) and 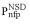 (•). Values of
(•). Values of 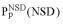 and
and 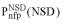 are simply read out from the precomputed empirical distributions. A similar procedure is followed for the AVS scores. The resulting six values are combined to calculate a probabilistic score (4) or used as the input parameters of artificial neural networks (5).
are simply read out from the precomputed empirical distributions. A similar procedure is followed for the AVS scores. The resulting six values are combined to calculate a probabilistic score (4) or used as the input parameters of artificial neural networks (5).
The main developments with respect to the previous release (release 8.0) can be summarized as follows:
Release 9.0 contains over 320 000 sequence entries, 11% more than release 8.0. The entries are now separated into two large groups, DOMAIN and PROTEIN FAMILY. The latter are indicated by the word FAMILY in the standard name (SN) line of the records.
The statistical description of the domain groups is now available via the web server (example shown in Fig. 1). The layout of the web server has changed.
A relational database architecture (SQL) is now used for producing and maintaining the data. This makes it possible to keep permanent accession codes and, for the servers, to process BLAST searches and statistics more rapidly.
The domain prediction system has been complemented with a new, faster boundary prediction scheme that has a graphic output.
DESCRIPTION OF THE DATA
The current release 9.0 of SBASE contains 320 000 protein segments consistently named by structure, function, biased composition, binding-specificity and/or similarity to other proteins. 1966 validated domain groups and 481 validated protein family groups are deposited in SBASE-A (157 000 records). SBASE-B contains 1520 further groups that are either (i) less well characterized than the groups of SBASE-A or (ii) are defined by composition (e.g. glycine-rich) or cellular location (e.g. transmembrane, etc.). These groups are sometimes defined in an overlapping manner, e.g. an extracellular domain (SBASE-B) may contain an EGF-module (SBASE-A).
Source and origin of data, cross-references and record structure are essentially the same as in the previous release. The boundaries of the domains are determined by homology to domains with known boundaries, such as given in the PROT-FAM (8), Pfam (9) and the INTERPRO resource (10), as well as in the original publications.
Distribution
SBASE 9.0 (October 23, 2001) is distributed by anonymous FTP file transfer from ftp://ftp.icgeb.trieste.it.
BLAST search by World Wide Web server
SBASE 9.0 can be searched by the BLAST program using the World Wide Web servers http://www.icgeb.trieste.it/sbase and http://www.abc.hu/. The services include, among others, regular expression searches and multiple alignments.
Citation
Users of SBASE and of web servers are asked to cite this article in their publications. The following citation format is suggested: ‘The sequence homologies were analyzed searching the SBASE protein domain sequence library release 9.0 via automated electronic mail (World Wide Web) server’.
Acknowledgments
ACKNOWLEDGEMENTS
The help of Suzanne Kerbavcic with the preparation of this manuscript is gratefully acknowledged. SBASE was established in 1990 and is maintained collaboratively by the International Center for Genetic Engineering and Biotechnology, Trieste, Italy and the Agricultural Biotechnology Center, Gödöllö, Hungary. This work was supported in part by EMBnet, the European Molecular Biology Network in the framework of EU grant ERBBIO4-CT96-0030.
REFERENCES
- 1.Pongor S., Skerl,V., Cserzo,M., Hatsagi,Z., Simon,G. and Bevilacqua,V. (1992) The SBASE domain library: a collection of annotated protein sequence segments. Protein Eng., 6, 391–395. [DOI] [PubMed] [Google Scholar]
- 2.Murvai J., VlahoviQek,K., Barta,E. and Pongor,S. (2001) The SBASE protein domain library, release 8.0: a collection of annotated protein sequence segments. Nucleic Acids Res., 29, 58–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murvai J., VlahoviQek,K. and Pongor,S. (2001) Towards a memory-based interpretation of proteome data. In Pifat-Mrzljak,G. (ed.), Supramolecular Structure and Function 7. Kluwer Academic Publishers, Dordrecht, pp. 155–166.
- 4.Murvai J., VlahoviQek,K. and Pongor,S. (2000) A simple probabilistic scoring method for protein domain identification. Bioinformatics, 16, 1155–1156. [DOI] [PubMed] [Google Scholar]
- 5.Murvai J., VlahoviQek,K., Szepesvári,C. and Pongor,S. (2001) Prediction of protein functional domains from sequences using artificial neural networks. Genome Res., 11, 1410–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanfill C. and Waltz,D. (1986) Toward memory-based reasoning. Commun. ACM, 29, 1213–1228. [Google Scholar]
- 7.Atwood T.K. (2000) The role of pattern databases in sequence analysis. Brief. Bioinform., 1, 45–59. [DOI] [PubMed] [Google Scholar]
- 8.Mewes H.W., Frishman,D., Gruber,C., Geier,B., Haase,D., Kaps,A., Lemcke,K., Mannhaupt,G., Pfeiffer,F., Schuller,C. et al. (2000) MIPS: a database for genomes and protein sequences. Nucleic Acids Res., 28, 37–40. Updated article in this issue: Nucleic Acids Res. (2002), 30, 31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Howe,K.L. and Sonnhammer,E.L. (2000) The Pfam protein families database. Nucleic Acids Res., 28, 263–266. Updated article in this issue: Nucleic Acids Res. (2002), 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Apweiler R., Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,L., Corpet,F., Croning,M.D. et al. (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res., 29, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]