Abstract
PALS db is a collection of Putative Alternative Splicing information from 19 936 human UniGene clusters and 16 615 mouse UniGene clusters. Alternative splicing (AS) sites were predicted by using the longest messenger RNA (mRNA) sequence in each UniGene cluster as the reference sequence. This sequence was aligned with related sequences in UniGene and dbEST to reveal the AS. This information was presented with six features: (i) literature aliases were used to improve the result of a gene name search; (ii) the quality of a prediction can be easily judged from the color-coded similarity and the scaled length of an alignment; (iii) we have clustered those EST sequences that support the same AS site together to enhance the users’ confidence on a prediction; (iv) the users can also set up the alignment criteria interactively to recover false negatives; (v) tissue distribution can be displayed by placing the mouse cursor over an alignment; (vi) gene features will be analyzed at foreign sites by submitting the selected mRNA or its encoded protein as a query. Using these features, the users cannot only discover putative AS sites in silico, but also make new observations by combining AS information with tissue distributions or with gene features. PALS db is available at http://palsdb.ym.edu.tw/.
INTRODUCTION
A single genetic locus can generate multiple gene products by combinatory strategy (1). This strategy fine-tunes gene expression at the RNA splicing level in tissue differentiation (2,3), cell cycle control (4), etc., by alternative splicing (AS). Aberrant splicing causes disorders, such as inherited diseases (5) and cancers (6). Discovering AS sites experimentally is the rate-limiting step in establishing the links between AS and cellular functions. This process can be accelerated by an in silico approach.
There are many databases or prediction tools available for providing AS information. Some databases are aimed at collecting well studied AS in organisms, such as ASDB (7). SpliceDB (8) went a step further to collect EST-confirmed AS sites. STACK (9), AsMamDB (10), TAP (11), Intronerator (12) and HASDB (13) incorporated AS information hidden in EST sequences. Among them, STACK further takes care of the problem of tissue-specific transcripts. By merging tissue-specific ESTs, STACK can provide putative tissue-specific transcripts for each gene. AsMamDB focused on genes with known AS in several mammalian species. TAP performed an EST-based gene structure prediction in genomic sequences and also collected splicing information in 1124 RefSeq genes. HASDB performed genome-wide detection of human alternative splicing and collected alternative splicing information of 6603 unique genes with mRNA sequences, which can be mapped to draft genomic sequences. Intronerator is a database of the introns of Caenorhabiditis elegans. This type of intron sequence database is very useful, because patterns residing in these sequences may govern the inclusion or exclusion of a splicing site. In addition to Intronerator, there is an intron sequence information system, ISIS (14), which contains splicesomal introns information on over 1122 human genes.
Though the number of sequences increases in an exponential manner, many AS-related databases collected only a limited fraction of known genes. In addition, the users are not allowed to judge the quality of prediction interactively. Moreover, many user interfaces are not sufficient to link AS information with functions. It appears that available databases cannot fully meet the users’ expectations. PALS db was designed to fill the gap.
RATIONALE AND STATISTICS
The more information PALS db has, the more chances a biologist can find AS information for his/her favorite genes. To collect as many known genes as possible, PALS db has taken the longest mRNA sequence in 19 936 human UniGene clusters (Build 138) and 16 615 mouse UniGene clusters (Build 93) as reference sequences to compute the putative AS sites. As shown in Table 1, AS was observed in ∼50 and 31% of the known genes in human and mouse, respectively. As expected, most of the splicing sites are supported by EST sequences, because more than half of the entries in GenBank are EST sequences. The human and mouse EST entries used in this analysis have reached 3 735 344 and 2 068 128, respectively. These two species not only have more EST sequences, but also have more known mRNA sequences than other species. This is why mRNA sequences were used as reference sequences in PALS db.
Table 1. PALS db (release 2) statistics.
| AS sites | Human (Build 138 and dbEST August 12, 2001) | Mouse (Build 93 and dbEST August 12, 2001) |
|---|---|---|
| None | 9983 (50.1%) | 11 396 (68.6%) |
| mRNA supported | 684 (3.4%) | 524 (3.1%) |
| Single EST supported | 4025 (20.2%) | 2720 (16.4%) |
| Multiple EST supported | 5244 (26.3%) | 1975 (11.9%) |
| Total | 19 936 (100%) | 16 615 (100%) |
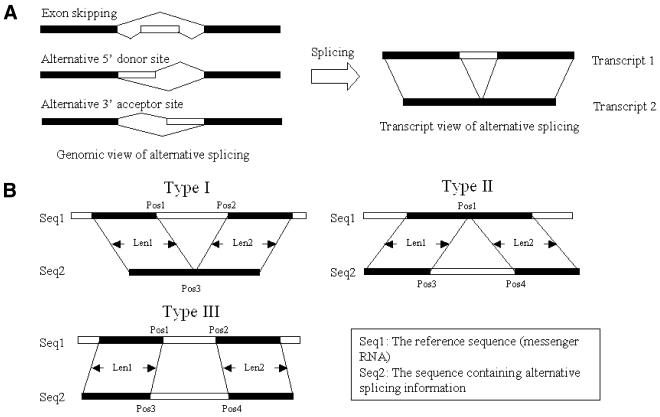
There are three possible ways to connect splicing sites in an AS event. As shown in Figure 1A (left panel), these three mechanisms, exon skipping, alternative 5′ donor sites and alternative 3′ acceptor sites, can easily be seen in a genomic view. These different mechanisms will generate different splicing products, whose relationship can best be described in the transcript view (Fig. 1A, right panel). If the reference sequence (Fig. 1B, Seq1) contains an extra sequence compared to the transcript containing AS information (Fig. 1B, Seq2), a type I AS site is saved. If the extra sequence is on Seq2, a type II AS site is saved. Combination of AS mechanisms may further complicate the difference between transcripts. In type III AS, both transcripts contain unmatched sequences that may be originated from a combination of AS mechanisms. However, low-quality EST sequences may have a similar effect on the alignment of transcripts. Thus, we discarded type three AS sites in the current release of PALS db.
Figure 1.
Major alignment types in predicting AS sites. (A) Alternative splicing may occur as exon skipping, alternative 5′ splicing donor sites and alternative 3′ acceptor sites (left panel from top to bottom). All three mechanisms may generate two alternative transcripts in which one of them (transcript 1) contains an extra fragment of sequence compared to transcript 2. By using mRNA as reference sequence, we could compare the difference between transcripts and discover putative AS sites (right panel). (B) In PALS db, the longest mRNA sequence in a UniGene cluster was used as the reference sequence to predict AS sites. The reference sequence (Seq1) was aligned with another transcript, which is in the form of either an EST sequence or an mRNA sequence (Seq2). The lengths of the matched regions on the left and right sides of an AS site were named ‘Len1’ and ‘Len2’, respectively. In type I, the reference sequence (Seq1) is like transcript 1 in (A). In type II, Seq2 (the sequence containing AS information) is like transcript 1 in (A). Pos1 and Pos2 mark locations of an AS site on the reference sequence. Pos3 and Pos4 mark locations of an AS site on the EST or mRNA sequences. In type III, combination of the three mechanisms described in (A) may cause unaligned sequences on both Seq1 and Seq2.
One EST sequence is usually sufficient to support an AS site, because EST sequences were determined experimentally. Some low-quality EST sequences may complicate the prediction. PALS db saved all the alignments that had the potential to support an AS site. We have observed that more than half of the putative human AS sites were supported by more than one EST sequence (Table 1). This redundant information cannot only enhance the confidence on prediction, but also provide a chance to discover real sites in many low-quality alignments. A friendly interface was designed to present both the AS site view and the supporting sequences view. The users can then set threshold criteria interactively to mark high-quality predictions. These unique features will be discussed in later sections.
QUERY INTERFACE
Keyword search
PALS db provided several keyword search methods, such as identification numbers, cytogenetic location, etc. The default search field is ‘gene description and gene name’. This approach is particularly useful when the full name or a complex term is used in a search, such as ‘mitogen activated protein kinase’. However, common names used in the literature may not be identical to the standard gene name suggested by the Human Genome Organisation (HUGO). For example, ‘MAPK8’ was also known as ‘JNK’ in some literature. ‘JNK’ does not retrieve anything if you search the ‘gene description and gene name’ field. We have improved this query system by using the ‘literature aliases’ collected by HUGO. This option allows the users to give common names to retrieve a gene. Nevertheless, this method is a little slower than searching the gene description and gene name. The user can look up the aliases of a gene by clicking the ‘Gene’ column in the query result.
Optional parameters
Although the default organism is human, the users can either query mouse genes or query both human and mouse genes in a keyword search. In addition to using the query interface, homologous genes in other organisms can also be linked from the graphic interface, which will be discussed in section ‘Cross reference among species’.
At default setting, PALS db marks only those AS sites that have at least 95% identity and 50 bp matches on both ends of an AS site in the EST-mRNA alignment. These stringent criteria removed false positives at the price of losing real AS sites. To recover these false negative sites, the query interface has the option of changing thresholds to filter alignments interactively. Those alignments that pass the selected threshold will be marked with an asterisk in the graphic interface. At present, PALS db provides three less stringent criteria: ‘95% 45 bases’, ‘90% 50 bases’ and ‘90% 45 bases’.
DATABASE PRESENTATION
The query results are summarized in a table to provide connections to further details. The ‘AS lists’ field lists the number of predicted AS sites for a given gene, and this field hyperlinks to a graphic ‘Summary’ page (Fig. 2) for all the predicted AS sites. The ‘Text_info’ field provides the predicted AS site position and quality information in text format. The ‘All seq info’ provides a hyperlink to the ‘Details’ page, which displays in graphics all the alignment results including those that failed to pass the preset threshold. The ‘Details’ page is linked to the ‘Summary’ page and vice versa. Both graphic pages link to the text information. The rest of the columns in the query results will display factual information about a UniGene cluster, such as the ID, gene name, description and cytogenetic location of a gene.
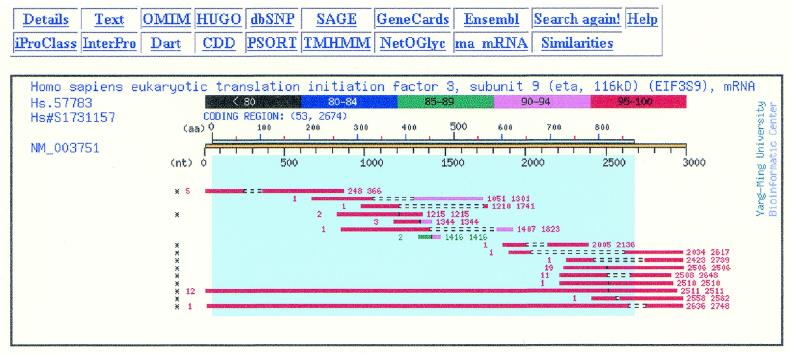
Figure 2.
Graphic summary of the alternative splicing information for the gene EIF3S9. Each horizontal bar is an EST-reference sequence alignment. The quality of alignment is color-coded. Type one and type two AS sites (Fig. 1) are characterized by two dashed lines and by a vertical line in the middle, respectively. The number of EST sequences that support a given site is shown in front of each bar. The positions of a given site are listed at the end of each bar. Those AS sites that pass the threshold are marked by asterisks. The pale blue block and the blue scale bar are the coding region and the positions on the protein sequence, respectively. The ‘Details’, ‘Text’, ‘Search again!’ and ‘Help’ buttons on the top will connect to the detailed alternative splicing information in graphics, the text information, the query interface and the user’s guide, respectively. The ‘Similarities’ button will connect to the summary page of a human gene to a paralogous gene in mouse and vice versa. The rest buttons will submit either the nucleic acid or amino acid sequence of the reference sequence to foreign web sites for further analysis.
The ‘Summary’ page lists all the possible AS sites
As mentioned in above, an AS site may be supported by multiple EST sequences. This redundant information has been clustered and merged into one AS site. The number of AS sites in a gene is counted and put into the ‘AS lists’ column in the query results. When different thresholds are used to filter the alignments, the number of AS sites will change correspondingly. This number can also be used to list all the genes that have a given number of AS sites in another query interface (http://palsdb.ym.edu.tw/cgi-bin/statistics.cgi) in order to discover interesting genes.
Furthermore, this supporting evidence can be used to increase the confidence on prediction. By clicking the numbers in the ‘AS lists’ column, all AS sites were shown on the reference sequence graphically (Fig. 2). This graphic ‘Summary’ page showed two types of AS sites. As shown in Figure 1B, the missing region of a type I AS was shown as broken lines between two aligned regions in Figure 2 (for example, the first site). For a type II AS site, the alternative transcript contained an additional fragment compared to the reference sequence (Fig. 1B, type II). The position of this site was shown as a vertical line that broke the aligned region into two parts (for example, the fourth site in Fig. 2).
The number of supporting EST sequences was summarized in front of the alignment of each AS site in Figure 2. For example, the first site has five supporting EST sequences. Those AS sites marked with an asterisk were predicted sites that passed the given threshold. By clicking the ‘Details’ button on the top-left corner, another graphic interface, the ‘Details’ page, will display all alignments that associated with an AS site regardless of the alignment quality. In the ‘Details’ page, clicking the hyperlink to NCBI UniGene or dbEST from each alignment can retrieve further sequence information. Currently, the quality of each alignment can be assessed by the color-coded display.
Quality of alignments might be helpful in recovering false negatives
Observing AS sites experimentally is much slower than verifying predicted AS sites from PALS db. An interface was thus designed to let the user examine the quality of all the alignments. The users may click the ‘All seq info’ column in the query results to get the ‘Details’ page described above. The quality of alignments is displayed by color codes and the length of matched regions can be estimated by comparing with the scale bars on the top. For example, even though the fifth alignment met the length requirement, the alignments across AS site were only 90–94% and 85–89% of the requirement, respectively. As a result, this alignment failed to pass the threshold.
Sometimes, only one EST sequence supports an AS site at a given stringency. However, other EST sequences, which fail to pass the threshold, may support the same AS site. In this case, the confidence on this predicted AS site would be enhanced. Moreover, the quality information can also be used to recall false negative site. Suppose these unqualified alignments were the only supporting evidence, this hypothetical AS site would not pass the threshold criteria. By considering all these supporting alignments together, we might be able to recall a hypothetical AS site.
Another case is the fifth AS site (position 1344) in Figure 2, which failed to pass the stringent criteria. This putative site can be supported by three alignments. Users can access the tissue distribution by switching to the detailed sequence information in graphics, the ‘Details’ page (not shown). By placing the mouse over an alignment in the ‘Details’ page, the library information will be displayed in the message line in 2 s. These different EST sequences were found in three different libraries; they are colon tumor cell line and two normal stomach libraries. Therefore, this AS site is less likely to be an artifact. As long as this site is not derived from a paralogous gene, this false negative site should be recalled. Recovering false negative sites is more efficient than discovering them experimentally.
Although the graphic information is good for presenting the relations globally, the graphic display cannot list all the details. In contrast, text can list all the details, but cannot provide a global impression. Thus, PALS db provides text information, too. The users can access text display by clicking the ‘Text’ column in the query results, or the ‘Text’ button on the two graphic interfaces. Text is formatted and can easily be imported into Microsoft™ Excel™ Advanced users may write programs to parse the text information collected from PALS db.
Position and tissue distribution of AS sites
When all the predicted AS sites are found, the priority for verification should also be determined. The significance of each AS site can be evaluated by its location on the gene and by its tissue distribution. The pale blue block in Figure 2 marks the coding region of the reference sequence. The scale bars on top of this block were designed to measure the relative position of AS sites and gene features. These features can be computed by submitting jobs to other sequence analysis web sites. They will be discussed below (‘Discovering functional differences among AS products’).
Different tissues may express different splicing forms of a gene. In addition, a given splicing form may be expressed in more than one tissue. Thus, a given AS site may be supported by EST sequences that are derived from different tissues. This tissue distribution information is implicated in the library information. For example, ∼45% of the human EST sequences are derived from the cancer cells in UniGene Build 138. It will be interesting to look for AS sites that are statistically more likely to be expressed in cancer cells.
Cross reference among species
Many human genes are highly homologous to mouse genes. Therefore, mouse is a good model animal for human. There are two ways to cross reference to similar genes in another animal. Pressing the ‘Similarities’ button (Fig. 2) will find paralogous genes in mouse. This relationship was established by the ‘Homologene’ database provided by NCBI. The ‘homologs’ button is missing in Figure 2, because the orthologous gene in mouse may not yet be confirmed. This relationship was established by the NCBI’s LocusLink database. If the EIF3S9 gene has an orthologous gene in mouse, the splicing pattern can be compared. It appears that many human and mouse genes have different splicing patterns. Further analysis is required to get a quantitative difference between the human and mouse splicing forms.
Discovering functional differences among AS products
We have included five types of tools that may yield useful predictions by comparing gene features with AS information. The first type provided protein domain or signature, such as transmembrane region, glycosylation site, etc. The available tools include InterPro, CDD, DART, TMHMM and PSORT. By comparing the relative position of AS sites and protein domains, one may discover the effect of AS on protein functions. The second type provided integrated information on human genes. The available tools are Ensembl and GeneCards. Users can use these tools to investigate the changes on gene structures produced by AS. The third type is a database that relates to gene variation or disease, such as single nucleotide polymorphism (SNP) or genetic disease. The available tools include dbSNP and OMIM. The former can reveal the effect of SNP on alternative splicing; the latter will correlate alternative splicing with disease. The fourth type provides gene expression information. The available tools include SAGE and mammalian messenger RNA (ma_mRNA). The fifth type provides protein family information, which is useful to distinguish orthologous and paralogous genes. The available tools include iProClass. Depending on the nature of remote analyses, either the sequence or a sequence ID was used to query the remote sites. By comparing the gene features with the predicted AS sites, the users may predict the functional differences among different splicing products.
Acknowledgments
ACKNOWLEDGEMENTS
Y.-H.H. and Y.-T.C. were supported by grants from National Science Council, Taiwan (NSC 89-2323-B-010-003 and NSC 89-2318-B-010-011-M51, respectively). S.-T.Y. was supported by a grant from Veterans General Hospital, Tsing-Hua University and Yang-Ming University (VTY90-P5-40). The computational resource was supported by Ministry of Education, ROC (Program for Promoting Academic Excellence of Universities, 89-B-FA22-2-4).
REFERENCES
- 1.Smith C.W. and Valcarcel,J. (2000) Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci., 25, 381–388. [DOI] [PubMed] [Google Scholar]
- 2.Grabowski P.J. (1998) Splicing regulation in neurons: tinkering with cell-specific control. Cell, 92, 709–712. [DOI] [PubMed] [Google Scholar]
- 3.Lahrtz F., Horstkorte,R., Cremer,H., Schachner,M. and Montag,D. (1997) VASE-encoded peptide modifies NCAM- and L1-mediated neurite outgrowth. J. Neurosci. Res., 50, 62–68. [DOI] [PubMed] [Google Scholar]
- 4.Jiang Z.-H. and Wu,J.Y. (1999) Alternative splicing and programmed cell death. Proc. Soc. Exp. Biol. Med., 220, 64–72. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto T., Nanba,E., Ninomiya,H., Higaki,K., Taniguchi,M., Zhang,H., Akaboshi,S., Watanabe,Y., Takeshima,T., Inui,K. et al. (1999) NPC1 gene mutations in Japanese patients with Niemann-Pick disease type C. Hum. Genet., 105, 10–16. [DOI] [PubMed] [Google Scholar]
- 6.Gessler M., Konig,A. and Bruns,G.A. (1992) The genomic organization and expression of the WT1 gene. Genomics, 12, 807–813. [DOI] [PubMed] [Google Scholar]
- 7.Dralyuk I., Brudno,M., Gelfand,M.S., Zorn,M. and Dubchak,I. (2000) ASDB: database of alternatively spliced genes. Nucleic Acids Res., 28, 296–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burset M., Seledtsov,I.A. and Solovyev,V.V. (2001) SpliceDB: database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res., 29, 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christoffels A., Gelder,A.V., Greyling,G., Miller,R., Hide,T. and Hide,W. (2001) STACK: Sequence Taq Alignment and Consensus Knowledgebase. Nucleic Acids Res., 29, 234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji H., Zhou,Q., Wen,F., Xia,H., Lu,X. and Li,Y. (2001) AsMamDB: an alternative splice database of mammals. Nucleic Acids Res., 29, 260–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kan Z., Rouchka,E.C., Gish,W.R. and States,D.J. (2001) Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res., 11, 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kent W.J. and Zahler,A.M. (2000) The Intronerator: exploring introns and alterantive splicing in Caenorhabditis elegans. Nucleic Acids Res., 28, 91–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Modrek B., Resch,A., Grasso,C. and Lee,C. (2001) Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res., 29, 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Croft L., Schandorff,S., Clark,F., Burrage,K., Arctander,P. and Mattick,J.S. (2000) ISIS, the intron information system, reveals the high frequency of alternative splicing in the human genome. Nature Genet., 24, 340–341. [DOI] [PubMed] [Google Scholar]