Abstract
yMGV (yeast Microarray Global Viewer) was designed to provide biologists with meaningful information from genome-wide yeast expression data. The database includes most of the available expression data published on yeast microarrays over the last 4 years. It provides customizable tools for the rapid visualization of expression profiles associated with a set of genes from all published experiments. It also allows users to compare the results from different publications so that they can identify genes with common expression profiles. We used yMGV to perform global analyses to find a gene expression profile specific for given biological conditions and to locate functional gene clusters on chromosomes. Other organisms will be added to this database. yMGV is accessible on the web at http://transcriptome.ens.fr/ymgv.
INTRODUCTION
The number of publications using microarrays to study gene expression in yeast has increased dramatically over the last 4 years: from five publications in 1998, to 27 in 2000. Although the authors have attempted to provide all of the corresponding data through supplemental web sites, the integration of all this information is still time consuming. Some of the yeast databases available provide data retrieval for gene expression [e.g. function junction of SGD (1), webminer (2), YPD (3), expressDB (4)]. However, these datasets are not yet complete and they can be difficult to use. For example, in the Yeast Proteome Database (YPD) changes in gene expression have to be sought experiment by experiment. As the exploitation of yeast expression data depends upon the availability of an exhaustive, user friendly, repository for yeast microarray results, we created yeast Microarray Global Viewer (yMGV), a public database containing most of the published microarray studies on yeast (1030 datasets as of August 15, 2001). The yMGV database allows biologists to search the whole dataset efficiently and rapidly. The main purpose of yMGV is to help scientists to find relevant information from published gene expression data.
DATA STORAGE IN yMGV
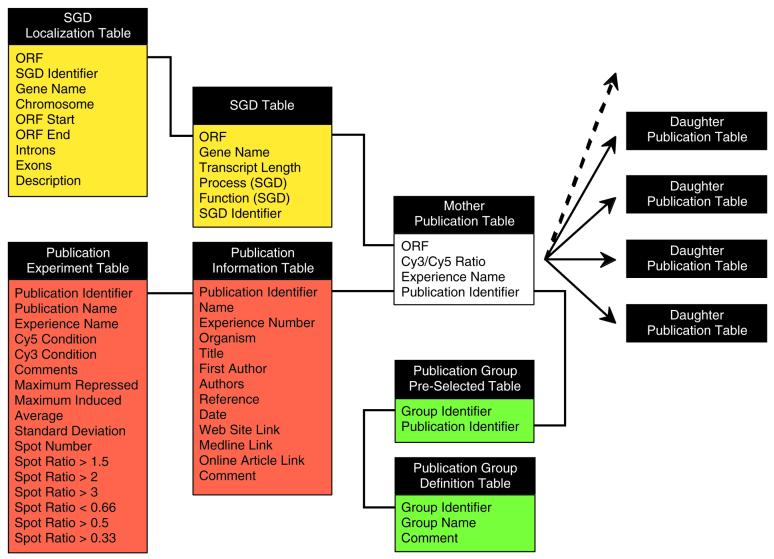
The yMGV database was constructed on the PostgreSQL software (http://www.postgresql.org/) running on the Linux system. The web page interface was made using PHP (http://www.php.net/) as a script program to query the database, to organize the results and to draw interactive online graphics. Publication data storage is organized around a mother table and each publication dataset has one inherited table (Fig. 1). Building one table per publication greatly speeds up access to a given dataset, whilst enabling all of the datasets to be queried via the mother table.
Figure 1.
Schematic representation of the structure of the yMGV database. This drawing shows the global organization of yMGV. All of the tables used are linked to the main publication table. There is one publication table for each article. These tables are inherited from a mother table, which is used as a model. The publication tables contain the results for each ORF and are linked to a complement table, which gathers information on the publication itself (Publication Information Table) and describes the corresponding experiments (Publication Experiment Table). This last table also contains statistical data for the graphical representation of experiments. Two other elements are centered around this main core. First, the data concerning pre-selected publication groups are stored together with group definition and the relationship between each article and its reference group. Secondly, part of the data from the SGD database is stored in two different tables for general information and for the graphical representation of expression maps (SGD Localization Table).
The data table contains a unique identifier for each publication, each experiment name and the filtered normalized Cy5/Cy3 ratio value for each ORF in each experiment. These data were obtained directly from the corresponding author or from the publication related web site. Another table contains information describing the publication such as the complete reference, links to PubMed and to the associated web site, the experimental methods used and statistical data (5). Some other useful information is connected to this central core, such as general data concerning yeast ORFs (using SGD definitions) or article assignment to pre-selected biological groups. We are currently working to integrate the information described in the MIAME version 1.0 (http://www.mged.org/; March 2001) in yMGV.
As of August 15, 2001, the yMGV database contained 6.4 million entries, representing the genome-wide expression data from 1030 experiments in 50 publications.
SEARCHING yMGV FOR GENE EXPRESSION PROFILES
As it is easier to browse through graphical representations than a list of numbers, yMGV displays the results as histograms representing the log2-transformed ratio values for the requested genes in each experiment. To highlight changes in gene expression, the histograms are colored red/green when expression is activated/repressed by >2-fold. To speed the search up, users can choose to only mine publications in which the expression of the selected gene changed by more than a given amount (as specified by the user) or only publications of interest. Additional information concerning the genes of interest, e.g. direct links to the corresponding SGD and YPD pages, and the experiments, e.g. complete description of the experimental protocol used (strains, growth conditions, cell preparation, dye labeling specification, etc.) is available on the results page (see http://www.biologie.ens.fr/yeast-publi.html for details).
Alternatively, genes sharing similar transcription profiles in different experimental conditions can be sought. This is done by the ‘search transcription profile’ mode, where users can select a precise transcription profile. This mode allows direct comparison of the results from different publications.
yMGV STATISTICS
One interesting feature of yMGV is that it provides a global statistical analysis of the expression results. For each gene requested, the number and percentage of variation and significant induction and repression across all experiments is available. On request, yMGV will also display a log2 distribution of the ratios for each experiment, together with the percentage of repressed and activated genes. These parameters help the users to assess the relevance of the expression changes measured for the requested genes. In addition, a list of the least and most variable genes stored in yMGV is available. Moreover, yMGV provides a graphical representation of the expression changes on the chromosomes, which allows the user to look for physical clusters of co-regulated ORFs.
OPENING NEW RESEARCH PROSPECTS USING yMGV
Looking for experimental conditions in which a gene is up- or down-regulated
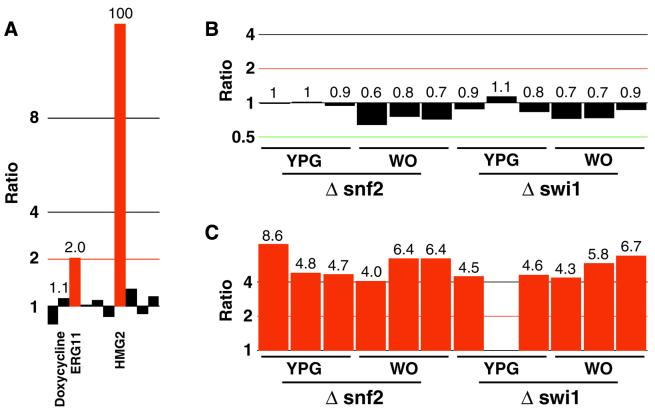
yMGV is a valuable tool that can provide an overview of the conditions in which a gene is up- or down-regulated, therefore offering clues about the function of the gene (see tutorial at http://www.biologie.ens.fr/yeast-publi.html). For example, we queried yMGV for expression profiles concerning PDR1, whose product is a zinc finger transcription factor involved in yeast multidrug resistance. To date, PDR1 has not been shown to be transcriptionally regulated. We observed that PDR1 is up-regulated in the case of a progressive inactivation of HMG2 and ERG11 (6), two genes involved in ergosterol biosynthesis (Fig. 2A). This is consistent with the role of Pdr1p in the control of the ergosterol synthesis pathway (7,8). PDR1 also seems to be activated by exposure to the DNA-damaging agent, methyl methane sulfonate (MMS) (9). It would be worth testing this drug with cells with altered Pdr1p activity. This new yMGV-derived information may stimulate pertinent new approaches to study the role of PDR1.
Figure 2.
Comparison of PDR1 and PDR3 expression profiles. The ‘alignment transcription profile for several genes’ tool was used to search yMGV for PDR1 and PDR3 genes. The main differences in their expression patterns are represented. (A) Induction of PDR1 when two components of the ergosterol pathway (ERG11 and HMG2) are inactivated (6). This graph plots the Cy3/Cy5 ratio for several conditions used in the publication. (B and C) Expression profiles of PDR1 (B) and PDR3 (C) genes when the members of the Swi/Snf system are deleted when grown in rich (YPG) or minimal (WO) glucose medium (10).
Comparing the regulation of two genes in all published experiments
We compared the transcription profiles of PDR1 with those of its functional and structural homolog PDR3 (see tutorial at http://www.biologie.ens.fr/yeast-publi.html for details). These two genes behave similarly, and only a few discriminating profiles can be observed. For example, the inactivation of the Swi1p and Snf2p transcriptional regulators (10) led to a 4-fold increase in PDR3 expression but did not significantly modify the expression of PDR1 (Fig. 2B and C). In normal conditions, Pdr1p is the most abundant regulator of the PDR network and Pdr3p is weakly expressed. PDR3 transcription is specifically activated in cells that lack the mitochondrial genome (11), but the mechanisms responsible for this activation are unknown. The above observation suggests that SWI1 and SNF2 are involved in the control of PDR3 expression.
Looking for co-regulated genes
Another useful way to mine DNA microarrays is to compare transcription profiles between a set of experiments (see tutorial at http://www.biologie.ens.fr/yeast-publi.html). This is the function of the ‘search by transcription profile’ link in the yMGV home page. We used this option to compare the effects of zinc starvation (12) with a YFH1 deletion mutant (encoding a mitochondrial protein responsible for iron binding and storage) (13). We found 17 genes that are up-regulated >3-fold in both experiments. This shows that there is a common response to zinc and iron starvation in yeast involving stress proteins (e.g. Yro2p, Hsp30p, Ygp1p and Gph1p), metal transporters (e.g. Sit1p, Ftr1p, Arn1p, Fet3p and Enb1p) and unknown genes that are thought to be specifically involved in iron uptake (e.g. Ydr534p, Yor382p, Yor383p and Tis11p). This suggests that cross-talk occurs between iron and zinc homeostasis mechanisms.
Looking for co-regulated domains on the chromosomes: the case of telomeric regulations
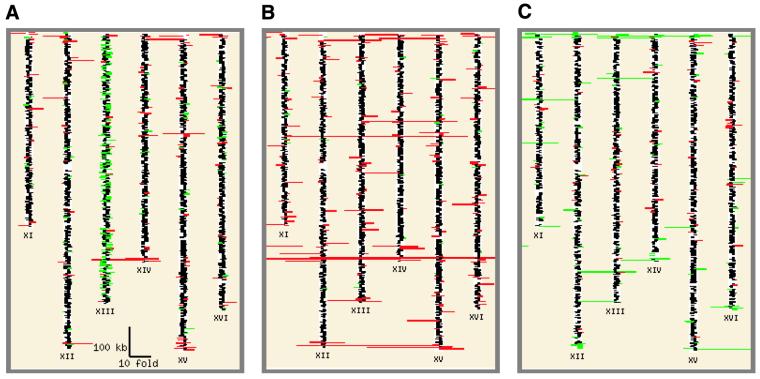
yMGV also provides a graphical representation of the gene ratio distribution along the yeast chromosome in each experiment. This feature is directly accessible by selecting the ‘expression map for one experiment’ on the yMGV home page (see tutorial at http://www.biologie.ens.fr/yeast-publi.html). This tool enables users to visualize the chromosome localization of co-expressed gene clusters easily. If we perform such a global search on the 1030 conditions available in yMGV, gene expression domains are clearly visible. For example, DeRisi et al. (14) found that all of the telomeric regions of a TUP1 deletion mutant seem to be up-regulated (Fig. 3A). This is related to the role of TUP1 in transcriptional repression in the telomeres. In addition, all of the genes on chromosome XIII seem to be down-regulated. Interestingly, a similar TUP1 deletion was studied by another team (6) and similar effects on telomeric regions were observed using yMGV, but no specific regulation was seen on chromosome XIII (Fig. 3B). The down-regulation observed by DeRisi et al. (14) could be related to an aneuploidy phenomenon (15). Similarly, a global down-regulation of 40% of the endogenous genes located within 20 kb of telomeres was shown in RPD3 deletion mutants (16). Rpd3p is involved in gene activation and silencing in yeast. This result is therefore related to the association between Tup1p and histone deacetylation (17,18). These examples highlight the importance of the yMGV chromosome display function, which can reveal fundamental, unexpected features of gene expression.
Figure 3.
Expression map for specific telomeric regulation. We used yMGV to draw an expression map of the microarray experiments. It shows that in some cases the telomeric regions are submitted to regulation. This figure represents the three main experiments in which such regulations were observed; i.e. in two publications on the inactivation of the Tup1p transcription factor [(A) 14 and (B) 6] and a publication on Rpd3p deacetylase mutants [(C) 16)]. The expression map represents the base 2 logarithm of the ratio for each gene along the entire chromosomes. Chromosomes XI–XVI are represented. The induced (red) or repressed (green) telomeric regions are clearly observable.
CONCLUSIONS AND FUTURE DEVELOPMENTS
The main feature of microarray data is to provide whole-genome views of changes in gene expression. There is far more information in microarray experiments than authors can interpret alone. This results in a large amount of ‘orphan’ information that could be of outstanding value for other scientists, provided that it can be easily accessible and compared. yMGV offers a unique visualization tool to search for gene expression data. It allows users to easily find the relevant information concerning genes or conditions of interest. The main limitation when interpreting such data is the heterogeneity of the criteria used for microarray analysis (especially normalization of ratios and filtering processes for artefactual and weak signals) in the different laboratories. International standardization of microarray results and exchange formats will soon make it possible to upgrade the database automatically and to compare results with greater confidence. In addition, one of our goals is to extend the database to other organisms. This should allow users to connect gene expression information from different organisms, which should generate a wealth of information to help us to understand the function and evolution of genomes.
REFERENCES
- 1.Ball C.A., Jin,H., Sherlock,G., Weng,S., Matese,J.C., Andrada,R., Binkley,G., Dolinski,K., Dwight,S.S., Harris,M.A. et al. (2001) Saccharomyces Genome Database provides tools to survey gene expression and functional analysis data. Nucleic Acids Res., 29, 80–81. Updated article in this issue: Nucleic Acids Res. (2002), 30, 69–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heiman M.G. and Walter,P. (2000) Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J. Cell Biol., 151, 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costanzo M.C., Crawford,M.E., Hirschman,J.E., Kranz,J.E., Olsen,P., Robertson,L.S., Skrzypek,M.S., Braun,B.R., Hopkins,K.L., Kondu,P. et al. (2001) YPD, PombePD and WormPD: model organism volumes of the BioKnowledge library, an integrated resource for protein information. Nucleic Acids Res., 29, 75–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aach J., Rindone,W. and Church,G.M. (2000) Systematic management and analysis of yeast gene expression data. Genome Res., 10, 431–445. [DOI] [PubMed] [Google Scholar]
- 5.Marc P., Devaux,F. and Jacq,C. (2001) yMGV: a database for visualization and data mining of published genome-wide yeast expression data. Nucleic Acids Res., 29, e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hughes T.R., Marton,M.J., Jones,A.R., Roberts,C.J., Stoughton,R., Armour,C.D., Bennett,H.A., Coffey,E., Dai,H., He,Y.D. et al. (2000) Functional discovery via a compendium of expression profiles. Cell, 102, 109–126. [DOI] [PubMed] [Google Scholar]
- 7.van den Hazel H.B., Pichler,H., do Valle Matta,M.A., Leitner,E., Goffeau,A. and Daum,G. (1999) PDR16 and PDR17, two homologous genes of Saccharomyces cerevisiae, affect lipid biosynthesis and resistance to multiple drugs. J. Biol. Chem., 274, 1934–1941. [DOI] [PubMed] [Google Scholar]
- 8.Kontoyiannis D.P. (2000) Efflux-mediated resistance to fluconazole could be modulated by sterol homeostasis in Saccharomyces cerevisiae. J. Antimicrob. Chemother., 46, 199–203. [DOI] [PubMed] [Google Scholar]
- 9.Jelinsky S.A., Estep,P., Church,G.M. and Samson,L.D. (2000) Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell. Biol., 20, 8157–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sudarsanam P., Iyer,V.R., Brown,P.O. and Winston,F. (2000) Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 97, 3364–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallstrom T.C. and Moye-Rowley,W.S. (2000) Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J. Biol. Chem., 275, 37347–37356. [DOI] [PubMed] [Google Scholar]
- 12.Lyons T.J., Gasch,A.P., Gaither,L.A., Botstein,D., Brown,P.O. and Eide,D.J. (2000) Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl Acad. Sci. USA, 97, 7957–7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foury F. and Talibi,D. (2001) Mitochondrial control of iron homeostasis. A genome wide analysis of gene expression in a yeast frataxin-deficient strain. J. Biol. Chem., 276, 7762–7768. [DOI] [PubMed] [Google Scholar]
- 14.DeRisi J.L., Iyer,V.R. and Brown,P.O. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science, 278, 680–686. [DOI] [PubMed] [Google Scholar]
- 15.Hughes T.R., Roberts,C.J., Dai,H., Jones,A.R., Meyer,M.R., Slade,D., Burchard,J., Dow,S., Ward,T.R., Kidd,M.J. et al. (2000) Widespread aneuploidy revealed by DNA microarray expression profiling. Nature Genet., 25, 333–337. [DOI] [PubMed] [Google Scholar]
- 16.Bernstein B.E., Tong,J.K. and Schreiber,S.L. (2000) Genomewide studies of histone deacetylase function in yeast. Proc. Natl Acad. Sci. USA, 97, 13708–13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bone J.R. and Roth,S.Y. (2001) Recruitment of the yeast Tup1p-Ssn6p repressor is associated with localized decreases in histone acetylation. J. Biol. Chem., 276, 1808–1813. [DOI] [PubMed] [Google Scholar]
- 18.Wu J., Suka,N., Carlson,M. and Grunstein,M. (2001) TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell, 7, 117–126. [DOI] [PubMed] [Google Scholar]