Abstract
Myotonic dystrophy type 1 (DM1) is one of the most common muscular dystrophies and can be potentially treated with antisense therapy decreasing mutant DMPK, targeting miRNAs or their binding sites or via a blocking mechanism for MBNL1 displacement from the repeats. Unconjugated antisense molecules are able to correct the disease phenotype in mouse models, but they show poor muscle penetration upon systemic delivery in DM1 patients. In order to overcome this challenge, research has focused on the improvement of the therapeutic window and biodistribution of antisense therapy using bioconjugation to lipids, cell penetrating peptides or antibodies. Antisense conjugates are able to induce the long-lasting correction of DM1 pathology at both molecular and functional levels and also efficiently penetrate hard-to-reach tissues such as cardiac muscle. Delivery to the CNS at clinically relevant levels remains challenging and the use of alternative administration routes may be necessary to ameliorate some of the symptoms experienced by DM1 patients. With several antisense therapies currently in clinical trials, the outlook for achieving a clinically approved treatment for patients has never looked more promising.
Keywords: antibody, bridged nucleic acids, cell-penetrating peptide, lipids, muscle, myotonic dystrophy, oligonucleotides, splicing
1. Introduction
Myotonic dystrophy type 1 (DM1) is the most common form of muscular dystrophy found in adults with a prevalence of 1 in 8000 people [1,2] although it has been reported that the genetic cause of DM1 is 3–4 times more common and mild forms of the disease are underdiagnosed [3]. In severe forms of the disease, individuals can be affected from birth [4]. The disease is characterised as a progressive, autosomal dominant, genetic disorder that affects multiple organs and systems. The clinical characteristics of DM1 vary greatly between patients but commonly include severe muscle weakness and myotonia, cardiac conduction defects, cataracts, insulin resistance and, more typically in younger patients, cognitive and neurological dysfunction. DM1 is caused by an expansion of CTG tandem repeats in the 3′-untranslated region of the myotonic dystrophy protein kinase (DMPK) gene which, upon transcription, leads to an RNA gain of function mutation [5,6]. Unaffected individuals have CTG repeat lengths of 5–37, while CTG repeats of over 50 lead to the manifestation of disease symptoms [7]. The severity of the disease is proportional to the number of CTG repeats, with increasing repeat lengths typically giving rise to an earlier onset of disease symptoms, increased symptom severity and increased somatic instability within tissues [7,8].
The muscleblind-like protein (MBNL) family has a main role in the regulation of mRNA metabolism, specifically post-transcriptional alternative splicing [9]. The family comprises three proteins with different lifetime expression patterns: MBNL1 and MBNL2 are expressed postnatally, while MBNL3 is expressed specifically during the embryonic phase and in regenerating adult tissues (e.g., liver and bone marrow) [10]. CUG repeat-expanded DMPK transcripts are retained in the nucleus as ribonucleoprotein foci and are able to sequester MBNL proteins, leading to a loss of protein function [11,12,13,14]. This alters the proteins’ splicing regulatory activity, generating predominantly foetal patterning of splicing and resulting in the identified alternative splicing of over 30 human genes [15]. Some of the most studied include CLCN1 [16], DMD [17], BIN1 [18], SCN5A [19] and INSR [20] and their missplicing can be directly correlated with classic DM1 symptoms such as muscle weakness [18], cardiac conduction defects [19] and insulin resistance [20]. Furthermore, MBNL1 is a negative regulator of the CUGBP Elav-like family member 1 (CELF1) protein, which is hyper-phosphorylated and upregulated in DM1, giving rise to further splicing misregulation [21,22,23]. Additionally, studies have shown that RNA localisation and trafficking, poly-adenylation, protein translation and micro-RNA processing are also affected by the altered behaviour of MBNL and CELF proteins in DM1 pathology [15,24,25].
There are a number of DM1 mouse models that have been used for proof-of-concept studies and the preclinical development of therapeutic approaches. These mouse models recapitulate DM1 pathology partially and they can be considered complementary. For example, the HSA-LR is a transgenic mouse expressing 250 untranslated CUGs in the human skeletal actin (HSA) that display missplicing and myotonia, but the transgene is only expressed in skeletal muscle, not in cardiac muscle or the CNS [26]. Other transgenic mice with tetracycline-inducible heart-specific expression of human mutant DMPK mRNA with 960 repeats have been used to study the cardiac pathogenesis in DM1 [27], whereas the mouse model most commonly used to study DM1 pathology in the CNS is the DMSXL mouse, with more than 1000 CTGs in the human DM1 locus (45 Kb), muscle defects and high mortality, although with mild missplicing [28]. All these mouse models have been useful in the field, but there is a need to develop large animal models that recapitulate the delivery bottleneck of new therapies in human patients.
There is currently no curative treatment for DM1, with the only treatments available to patients being aimed at managing the disease symptoms. One key reason for this is the multi-systemic nature of the disease, which requires the targeting of treatments into the cell nuclei of a range of tissues. Critically affected tissues include skeletal muscle and heart, the membranes of which are challenging to traverse, and the central nervous system (CNS), a tissue protected by the blood–brain barrier (BBB). However, advancements in gene technologies and the treatment of several other neuromuscular diseases have led to a number of therapeutic approaches being explored for DM1. These therapeutic approaches can be grouped into three categories: antisense therapy, gene therapy and small molecule drugs [29]. We and many others in this field believe that future therapeutic interventions could involve a combination of modalities, for example, an initial treatment with gene therapy followed by the repetitive administration of antisense therapy or a small molecule. Targeting RNA with antisense therapy does not elicit immune responses like the AAVs regularly used in gene therapy and provides the precision needed to avoid the off-target effects of many small molecules. This review will focus on the use and development of antisense therapy for the treatment of DM1, discussing the hurdles and current progress towards future clinical applications.
2. ASO Therapeutics
Antisense oligonucleotides (ASOs) are small (~14–30 nucleotides), single-stranded synthetic nucleic acid polymers which can be employed to modulate gene expression. ASOs can be divided into two classes, based on their mechanism of action: RNase H1-dependent ASOs or steric blocking ASOs. RNase H1-dependent ASOs form a duplex with the target RNA sequence which is recognised by RNase H1. The recruitment of RNase H1 to the duplex results in the cleavage of the target RNA and therefore a downregulation in gene expression [30,31]. Typically, ASOs acting by this mechanism are termed “gapmers” as they are comprised of a central stretch of approximately 10 nucleotides complementary to the target RNA sequence, flanked on each side by 2–5 chemically modified nucleotides [32]. The chemical modifications render the flanking regions unrecognisable to RNAse H1 but are beneficial to the overall compound due to their ability to increase the binding affinity to target RNA. Steric blocking ASOs, on the other hand, lack RNase H1 competence due to chemical modifications across the ASO and so fail to induce target degradation. Instead, they are designed to bind to target transcripts with a high affinity and interfere with RNA–RNA or RNA–protein interactions. Their clinical applications are assorted but often include modulation of alternative splicing, via exon inclusion or exclusion [33,34]. RNA gain-of-function diseases can be also treated with steric block ASOs designed to corrupt the toxic RNA function [35,36,37] while approaches to restore the translational reading frame are being employed to loss-of-function diseases [38].
Oligonucleotides have promising clinical potential due to their high target specificity, but poor biodistribution outside liver and kidney when injected systemically and they are susceptible to degradation by endonucleases and exonucleases [39]. Consequently, in order to enhance delivery and potency, a number of chemical modifications have been deployed over the years, to improve ASO pharmacological characteristics [40]. Phosphorothioation of the ASO backbone (PS), whereby a non-bridging oxygen atom is replaced with a sulphur, has been shown to improve ASO resistance to nuclease activity and increase serum half-life [41,42,43]. Meanwhile, modifications at the 2′ position of the ribose sugar have led to the development of a number of ASOs with improved safety, improved pharmokinetics and reduced immunostimulatory activity [44,45]. Bridged nucleic acid chemistries introduce a chemical bridge between the 2′ and 4′ position on the ribose ring of the ASO [46,47,48,49,50]. This constrains the pucker of the ribose sugar to the 3′ endo conformation, increasing the ASO affinity for its RNA target and enhancing stability against nucleases. Additionally, ASOs with charge-neutral backbones have been developed, including phosphorodiamidate morpholino oligonucleotide (PMOs) where the nucleotide backbone is replaced with neutral 6-membered morpholino rings [51] and peptide nucleic acids (PNAs) [52] where the ASO backbone consists of a pseudopeptide analogue. In both cases, the overall neutrality of the ASO charge increases its serum stability while facilitating the potential for bioconjugation to other chemical moieties that may aid compound delivery to target tissues [53].
3. Antisense Therapeutics for DM1
The majority of ASO therapies in development for DM1 have focused on muscle as the primary tissue target, although CNS is also an important target tissue, especially for the congenital form of DM1. Both RNAse H1-dependent and steric block-modified ASO approaches have been used as strategies in the preclinical development of therapies for DM1. RNAse H1-dependent DM1 gapmers are designed to bind to repeat expanded DMPK RNA, causing its degradation and a reduction in sequestered MBNL1 and resultant missplicing events [54,55,56,57]. Meanwhile, steric block ASOs bind with a high affinity to DMPK CUG RNA repeats, thereby preventing the binding of MBNL1 and eliciting correction of splicing [35,36,58]. For example, CAG25, a naked 25-base pair PMO that sterically blocks MBNL1 sequestration to CUG-expanded DMPK RNA, was shown to restore splicing abnormalities, reverse myotonia and reduce the number RNA foci in a DM1 mouse model upon intramuscular injection. The compound did not reduce endogenous levels of expanded CUG transcripts in wild-type (WT) mice, demonstrating a proof-of-concept for steric block DM1 ASOs selecting disease DMPK transcripts over WT. However, systemic injection failed to correct key missplicing events [59]. Similarly, several ASO gapmers have been successfully used in cellular and animal DM1 models to treat the molecular phenotype of the disease [54,55,56,57].
3.1. Bridged Nucleic Acid Therapy
Locked nucleic acids (LNAs) are a form of bridged nucleic acid modification that can be introduced into ASOs to enhance stability and target-binding affinity. A single LNA subunit substitution into RNA or DNA oligomer typically increases the Tm by 2–8 °C, greatly enhancing thermal stability [60]. Nucleic acids containing LNA subunits have been used as mixmers which contain several LNA subunits in random positions within an ASO; or as gapmers with several LNA subunits on both sides of the ASO [48,49]. Alternatively, LNA technology can also be used to synthesise all-LNA oligomers, capable of stably binding target DNA or RNA molecules without causing their endonucleolytic cleavage by RNase H [61,62]. For example, PO-LNA-CAG-10, an all-LNA ASO complementary to CUG repeats, was able to specifically correct MBNL-sensitive alternative splicing abnormalities and reduce the number and size of CUG repeat-expanded foci in vitro, although the compound was unable to enter cells without transfection agents [62]. Subsequent intramuscular injections of PO-LNA-CAG-10 in DM1 mice (HSALR) showed the promising correction of the alternative splicing patterns in skeletal muscles to near wild type levels, 48 h after treatment. More recently, a combination of 2′OMe monomers with LNA nucleotides in every third position was able to improve Mbnl1 exon 7 missplicing in primary HSALR myoblasts by directly competing with Mbnl1 for CUG binding, in order to restore protein function. Intramuscular delivery to the tibialis anterior muscle in HSALR mice resulted in the amelioration of the alternative splicing of Mbnl1-dependant exons. However, systemic subcutaneous injection of the LNA/2′-O-Methyl ASOs within the same mouse model, while exhibiting low toxicity, failed to correct missplicing of Serca2 and reduce the number of CUG-expanded foci within the skeletal muscles. It was overall concluded that the skeletal muscle uptake of their LNA/2′-O-Methyl ASOs was insufficient for intracellular efficacy without bioconjugation to a tissue-targeting moiety.
Other bridged nucleic acid chemistries such as cEt (2′,4′-constrained ethyl) have been more successful at normalising DM1 muscle phenotype after systemic administration in DM1 mice [56]. However, clinical study data suggest that the use of a systemically delivered naked ASO alone is insufficient for achieving the desired potency or efficacy in skeletal muscles in patients, even when chemically modified. The first ASO compound to be tested for DM1 in clinical trials was a naked cEt-modified gapmer developed by IONIS. The compound had previously shown high efficacy in reversing the DM1 skeletal muscle phenotype and low toxicity in preclinical animal models [63]. However, the clinical trial (NCT02312011) was discontinued due to a failure to achieve skeletal muscle concentrations required for functional and biological efficacy [64]. Therefore, further enhancements such as bioconjugation or alternative ASO strategies and delivery methods are being developed in order to improve the stability and biodistribution of ASOs for the treatment of DM1.
3.2. Bioconjugation: Lipid Conjugates
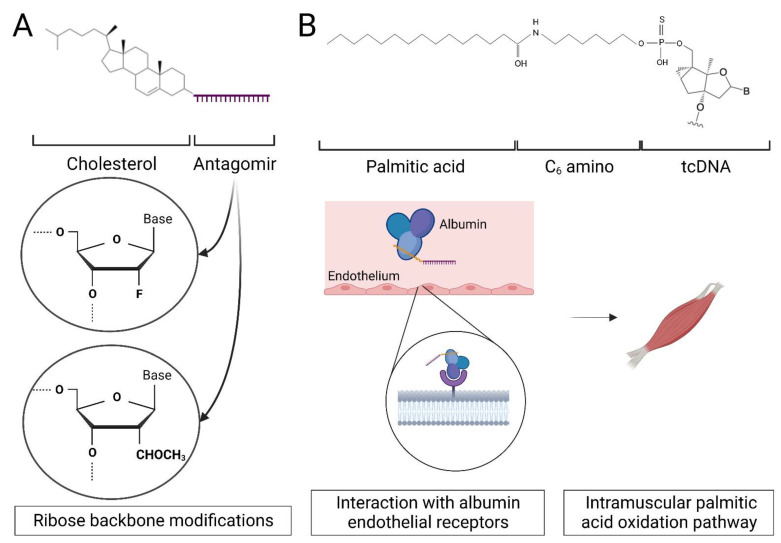
Bioconjugation involves the direct covalent linkage of various chemical moieties to ASOs and is now widely used to enhance intracellular uptake, tissue specificity and reduce renal clearance from systemic circulation [40]. ASO conjugation with hydrophobic lipid moieties improves distribution to skeletal and cardiac muscle in both mice and non-human primates, enhancing association with lipoproteins such as HDL and albumin [65] (Figure 1). Albumin is the most abundant plasma protein in human blood, with circulating levels of 35–50 g/L [66]. As skeletal and cardiac muscle cells rely on the oxidation of long-chain fatty acids for contractile work, fatty acids are transported through blood to these cells and cross the tissue endothelium via albumin interactions with endothelial surface receptors [67,68]. Conjugation of PS ASOs with fatty acids, such as palmitic acid, improves compound affinity for serum albumin decreasing renal clearance and therefore potency in muscle tissues [69]. Moreover, lipid conjugation enhances the endosomal release of ASOs in cells [70].
Figure 1.
Antisense molecules can be conjugated to lipids to improve their pharmacokinetic properties. (A) Single-stranded RNA analogues complementary to miRNAs (AntagomiR) conjugated to cholesterol are delivered to non-CNS tissues at clinically relevant concentrations. Crossing the BBB into the CNS can be improved by conjugating the lipid to a DNA/RNA heteroduplex oligonucleotide [71]. (B) Conjugation of tricyclo-DNA (tcDNA) [72] ASOs with fatty acids such as palmitic acid has also been shown to improve ASO distribution by binding to serum albumin and indirectly with endothelial surface receptors facilitating transport across the continuous capillary endothelium in muscle.
In the context of DM1, the group of Ruben Artero (University of Valencia) developed DM1 therapeutic antisense compounds conjugated to lipids that silence microRNAs (miRNAs) associated with the disease pathology [73]. These compounds, called antagomiRs or anti-miRs [74], are steric blocking ASOs that are complementary to miRNAs and also chemically modified at different positions in the backbone and on the sugar molecule [75]. AntagomiR-23b silences miRNA-23b-3p leading to an increase in MBNL protein levels which can ameliorate DM1 pathology [73]. The effect of antagomiR-23b was studied via subcutaneous and intravenous administrations in the DM1 HSALR mice and demonstrated an improvement in skeletal muscle phenotype after single injections at 12.5 mg/kg [76]. Further analysis showed MBLN1 splicing correction and increased expression, with restored normal MBNL1/2 distribution at the nucleus and cytoplasm level in muscle fibres. The presence of RNA foci remained unchanged [73]. The silencing of has-miR-218-5p, an endogenous MBNL1/2 repressor in human myotubes, overexpressed in DM1 animal disease models and patient muscle biopsies also led to MBNL1/2 upregulation in myotubes [73]. The systemic administration of antagomiR-218 to HSALR mice induced similar improvements in molecular and physical phenotypes as observed for antagomiR-23b [77].
3.3. Bioconjugation: Antibody Conjugates
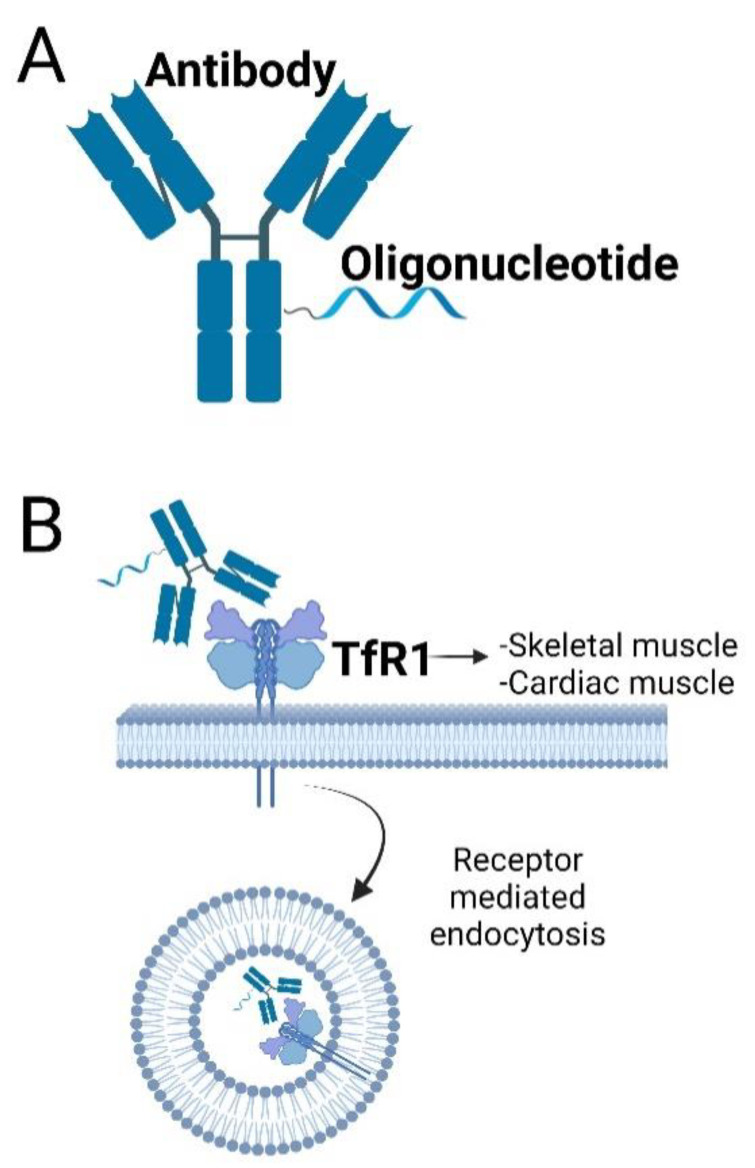
Antibody–drug conjugates have long been tested in oncology for targeting chemotherapeutic agents to specific tissues or cell types [78]. The conjugation of ASOs to antibodies shows promise for enhancing ASO delivery to DM1-affected tissues due the ability of antibodies to increase serum stability, membrane permeability and tissue selectivity [79]. The transferrin receptor, TfR1, is one of the most common cell-surface receptors to be exploited for antibody-facilitated delivery or oligonucleotides [80,81,82,83]. Antibody targeting of the TfR1 allows for the delivery of conjugated cargo into cells via receptor-mediated endocytosis and it is highly expressed in both skeletal and cardiac muscles (Figure 2). Two pharmaceutical companies (Avidity and Dyne Therapeutics) are exploiting this high TfR1 expression through the conjugation of DM1 small interfering RNA (siRNA) [84] targeting DMPK transcripts to TfR1 targeting antibodies or antibody fragments. The conjugation of TfR1-targeting antibodies or antibody fragments to siRNA-targeting DM1 repeat-expanded DMPK RNA allows for increased skeletal muscle penetration and therefore an overall enhancement in treatment efficacy. AOC1001 (Avidity), comprises of a monoclonal antibody against the TfR1 conjugated to a DMPK siRNA, and is currently being tested in a P1/2 clinical trial (MARINA: NCT05027269) for tolerability and safety at single and multiple doses [84]. In preclinical testing (data currently unpublished), the compound was reported to be successfully delivered to skeletal, cardiac and smooth muscles in both mouse and cynomolgus monkeys, producing a reduction in DMPK RNA expression and an amelioration of the disease phenotype in DM1 mice. Meanwhile, Dyne Therapeutics is recruiting patients for the clinical testing of Dyne-101, a DMPK ASO conjugated to an anti-TfR1 antibody fragment. Treatment of a DM1 mouse model with Dyne-101 caused a 40–50% and a 49% splicing correction in the skeletal muscle and heart biomarkers, respectively, while studies in cynomolgus monkeys did not reveal any toxicity after 13 weeks of treatment (ACHIEVE: NCT05481879).
Figure 2.
Antisense Oligonucleotides can be conjugated to antibodies targeting the Transferrin Receptor 1 (TfR1) and penetrate cells by receptor-mediated endocytosis. (A) Schematic design of an antibody–ASO conjugate. The ASO is conjugated directly to the antibody using covalent unions. (B) Antibodies targeting TfR1, a highly expressed cell-surface receptor of DM1 in target tissues such as skeletal and cardiac muscles, have been used to deliver antisense cargo. The AOC enters the cell by receptor-mediated endocytosis resulting on an enhanced ASO delivery. The affinity of TfR1antibodies can be fine-tuned to maximise the interaction with receptors in the BBB allowing for the compound to be delivered to the CNS (in mice) after systemic administration [85].
3.4. Bioconjugation: Peptide Conjugates
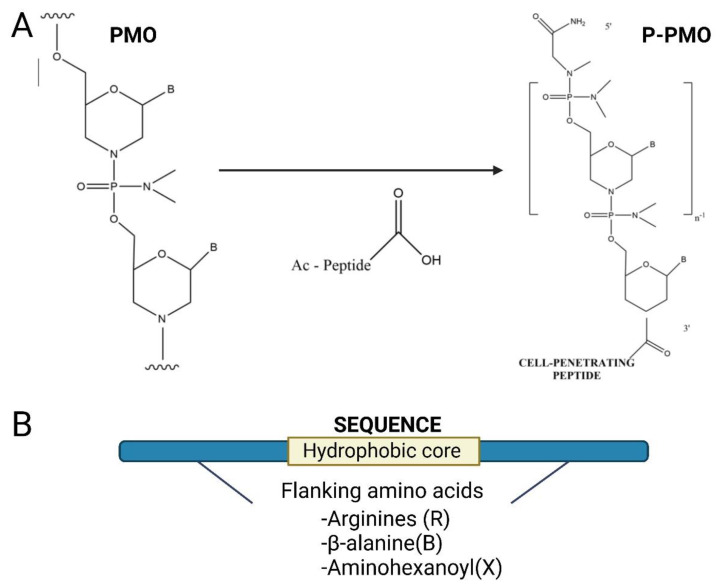
Peptides can confer cell-penetrating, or tissue/cell targeting properties onto therapeutic ASOs. Cell-penetrating peptides (CPPs) can be directly conjugated to charge-neutral ASO chemistries, such as PMO and PNA, and this approach has been taken in several different splice-switching diseases [86,87,88,89,90,91]. The development of peptide PMOs (PPMOs) for the treatment of neuromuscular diseases in the groups of Matthew Wood (University of Oxford) and Mike Gait (MRC Laboratory of Molecular Biology, Cambridge) demonstrated the efficacy of this approach treating DM1, DMD and SMA with PMOs conjugated to different CPPs such as Pip6a [35,86,87] (Figure 3). Pip6a is a member of a series of PMO/PNA internalisation peptides (Pips) comprising arginines (R), β-alanine (B) and aminohexanoyl (X) amino acids flanking an internal core containing hydrophobic residues. The overall arginine-rich (RXRRBR)2XB-PMO structure is able to enhance ASO delivery to cardiac and skeletal muscle [89].
Figure 3.
Antisense oligonucleotides can be conjugated to cell-penetrating peptides (CPPs) to increase cellular uptake. (A) CPPs can be directly conjugated to charge-neutral oligonucleotides, such as PMOs or PNAs using direct covalent unions [35]. (B) The peptide sequence is usually positively charged and rich in arginine. Here, a cell-penetrating peptide comprising a hydrophobic core and two flanking regions enriched with cationic amino acids such as arginine or B-alanine is shown.
The conjugation of CPPs to PMOs was shown to improve the DM1 phenotype in HSALR mice whether targeting toxic DM1 CUG repeats via a blocking mechanism for Mbnl1 displacement [35] or targeting a miR-23b binding site on the Mbnl1 3′UTR increasing Mbln1 protein levels using ASOs called blockmiRs [92]. In the case of PPMOs targeting the repetitive sequence, a single tail vein injection of Pip6a-PMO at 12.5 mg/kg induced a significant correction of Mbnl1-dependent splicing defects 2 weeks after treatment, with a 50% reduction in the number of RNA foci and a 60% decrease in CUGexp-RNA [35]. This is in contrast to three injections at 200 mg/kg of naked PMO which did not have any effect in the molecular phenotype. Repeated systemic injections of Pip6a-PMO at 12.5 mg/kg led to the complete correction of Cncl1 and Mbnl1 DM1 alternative splicing while heatmap results demonstrate a global gene expression correction in treated mice [35]. Moreover, hind-limb myotonia was completely reversed after treatment and the quantification of corrected splicing changes revealed long-lasting activity of Pip6a-PMO 26 weeks after treatment [35]. The Wood laboratory continued to develop cell-penetrating peptides with a wider therapeutic window than Pip6a, forming the basis of an MRC-University of Oxford spin-off company (PepGen) to take the peptide PMO chemistry to clinical trials. PepGen has recently started a phase 1 clinical trial of a peptide-conjugated oligonucleotide (PGN-EDO51) for the treatment of DMD and is intending to start a clinical trial with PGN-EDODM1 in DM1 patients [36].
4. Alternative Routes of Delivery
While advances in ASO chemical modification and conjugation have rendered the delivery of DM1 ASOs to skeletal and cardiac muscle more effective, delivering enough compound to the CNS to induce a clinically relevant effect remains a challenge. The majority of treatments in development for neuromuscular diseases are unable to cross the BBB upon systemic administration. Therefore, in order to circumvent this biological barrier, routes of administration that deliver ASOs directly into the CNS have been utilised for a number of diseases such as SMA [93], Parkinson’s disease [94] and Huntington disease (HD) [95].
The most advanced CNS treatments with ASOs in patients have been deployed via intrathecal administration, with Nusinersen, a 2′-MOE ASO for SMA, being approved by the FDA in 2016 [96]. More recently, in 2019, Tabrizi et al. [95] tested the 2′-MOE RG6042, also known as HTTRX, designed to reduce concentrations of Huntingtin (HTT) messenger RNA in HD [95]. Data from the phase 1/2a clinical trial (NCT02519036) in 46 adult patients with early HD showed an ASO-mediated HTT reduction of up to 30% 24 h after intrathecal administration. The compound demonstrated a good safety profile, with no evidence of adverse events after a dosing regimen of four repeated monthly intrathecal administrations; however, the phase 3 clinical trial (NCT03761849) was halted based on the benefit–risk profile.
Intracerebroventricular (ICV) administration has been attempted since the first stages of ASO development [97,98,99]. ICV delivery of ASOs is now being explored for DM1 in the DMSXL mouse model, a DM1 transgenic mouse model harbouring a human DMPK gene with over 1000 CTG repeats and displaying a CNS phenotype [28]. In the case of IONIS 486178 (a 16-nucleotide ASO gapmer containing BNA (cEt) modifications) [100], a single 75 µg ICV bolus administration of the ASO-induced DMPK mRNA knockdown, nuclear redistribution of MBNL1/2 and a reduction in RNA foci in the CNS of the DMSXL mouse model for at least 3 weeks [28]. Additionally, there was a correction in the phenotypic behavioural abnormalities. IONIS 486178 ASO showed distribution throughout the brain, with the greatest compound accumulations in the hypothalamus, cortex and cerebellum. Furthermore, no CNS inflammation or peripheral toxicity was detected post treatment, suggesting that the compound is well tolerated in adult mice [28,100]. These results are particularly promising for congenital or childhood DM1 patients that typically experience more severe CNS symptoms [101,102].
5. Future Challenges and Concluding Remarks
Nowadays, with the efficacy of antisense therapy being repeatedly demonstrated in DM1 models, the largest challenge remaining for this class of compounds is delivery to critically affected tissues in humans. Advancements in chemical modifications and bioconjugations have rendered overcoming this barrier more achievable, although safety and tolerability studies have shown that approaches to improve delivery and potency need to be carefully balanced with maintaining low toxicity. The challenge of delivery to the CNS at clinically relevant levels also remains and direct CNS administration routes may be necessary to ameliorate some of the symptoms experienced by patients. Nevertheless, with several ASO therapies currently in clinical trials, the outlook for achieving a clinically approved treatment for patients has never looked more promising.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing is not applicable to this article.
Conflicts of Interest
MJAW and MAV are co-inventors of patents licensed to PepGen, a company dedicated to the peptide-based enhancement of delivery of therapeutic oligonucleotide analogs.
Funding Statement
This work was supported by Medical Research Council (MRC) grants (MRW0147421 and MRP01741X1) and by John Fell Fund (0007895).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Norwood F.L.M., Harling C., Chinnery P.F., Eagle M., Bushby K., Straub V. Prevalence of genetic muscle disease in Northern England: In-depth analysis of a muscle clinic population. Brain. 2009;132:3175–3186. doi: 10.1093/brain/awp236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siciliano G., Manca M., Gennarelli M., Rocchi A., Iudice A., Miorin M., Mostacciuolo M. Epidemiology of myotonic dystrophy in Italy: Re-apprisal after genetic diagnosis. Clin. Genet. 2001;59:344–349. doi: 10.1034/j.1399-0004.2001.590508.x. [DOI] [PubMed] [Google Scholar]
- 3.Johnson N.E., Butterfield R.J., Mayne K., Newcomb T., Imburgia C., Dunn D., Duval B., Feldkamp M.L., Weiss R.B. Population-Based Prevalence of Myotonic Dystrophy Type 1 Using Genetic Analysis of Statewide Blood Screening Program. Neurology. 2021;96:e1045–e1053. doi: 10.1212/WNL.0000000000011425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Echenne B., Bassez G. Congenital and infantile myotonic dystrophy. Handb. Clin. Neurol. 2013;113:1387–1393. doi: 10.1016/B978-0-444-59565-2.00009-5. [DOI] [PubMed] [Google Scholar]
- 5.Mahadevan M., Tsilfidis C., Sabourin L., Shutler G., Amemiya C., Jansen G., Neville C., Narang M., Barceló J., O’Hoy K. Myotonic dystrophy mutation: An unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 6.Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.P., Hudson T. Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 7.Tsilfidis C., MacKenzie A.E., Mettler G., Barceló J., Korneluk R.G. Correlation between CTG trinucleotide repeat length and frequency of severe congenital myotonic dystrophy. Nat. Genet. 1992;1:192–195. doi: 10.1038/ng0692-192. [DOI] [PubMed] [Google Scholar]
- 8.Fortune M.T., Vassilopoulos C., Coolbaugh M.I., Siciliano M.J., Monckton D.G. Dramatic, expansion-biased, age-dependent, tissue-specific somatic mosaicism in a transgenic mouse model of triplet repeat instability. Hum. Mol. Genet. 2000;9:439–445. doi: 10.1093/hmg/9.3.439. [DOI] [PubMed] [Google Scholar]
- 9.Konieczny P., Stepniak-Konieczna E., Sobczak K. MBNL proteins and their target RNAs, interaction and splicing regulation. Nucleic Acids Res. 2014;42:10873–10887. doi: 10.1093/nar/gku767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanadia R.N., Urbinati C.R., Crusselle V.J., Luo D., Lee Y.J., Harrison J.K., Oh S.P., Swanson M.S. Developmental expression of mouse muscleblind genes Mbnl1, Mbnl2 and Mbnl3. Gene Expr. Patterns. 2003;3:459–462. doi: 10.1016/S1567-133X(03)00064-4. [DOI] [PubMed] [Google Scholar]
- 11.Taneja K.L., McCurrach M., Schalling M., Housman D., Singer R.H. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis B.M., McCurrach M.E., Taneja K.L., Singer R.H., Housman D.E. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl. Acad. Sci. USA. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee K.Y., Li M., Manchanda M., Batra R., Charizanis K., Mohan A., Warren S.A., Chamberlain C.M., Finn D., Hong H., et al. Compound loss of muscleblind-like function in myotonic dystrophy. EMBO Mol. Med. 2013;5:1887–1900. doi: 10.1002/emmm.201303275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller J.W., Urbinati C.R., Teng-Umnuay P., Stenberg M.G., Byrne B.J., Thornton C.A., Swanson M.S. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.López-Martínez A., Soblechero-Martín P., de-la-Puente-Ovejero L., Nogales-Gadea G., Arechavala-Gomeza V. An Overview of Alternative Splicing Defects Implicated in Myotonic Dystrophy Type I. Genes. 2020;11:1109. doi: 10.3390/genes11091109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lueck J.D., Mankodi A., Swanson M.S., Thornton C.A., Dirksen R.T. Muscle chloride channel dysfunction in two mouse models of myotonic dystrophy. J. Gen. Physiol. 2007;129:79–94. doi: 10.1085/jgp.200609635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rau F., Lainé J., Ramanoudjame L., Ferry A., Arandel L., Delalande O., Jollet A., Dingli F., Lee K.Y., Peccate C., et al. Abnormal splicing switch of DMD’s penultimate exon compromises muscle fibre maintenance in myotonic dystrophy. Nat. Commun. 2015;6:7205. doi: 10.1038/ncomms8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fugier C., Klein A.F., Hammer C., Vassilopoulos S., Ivarsson Y., Toussaint A., Tosch V., Vignaud A., Ferry A., Messaddeq N., et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat. Med. 2011;17:720–725. doi: 10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- 19.Freyermuth F., Rau F., Kokunai Y., Linke T., Sellier C., Nakamori M., Kino Y., Arandel L., Jollet A., Thibault C., et al. Splicing misregulation of SCN5A contributes to cardiac-conduction delay and heart arrhythmia in myotonic dystrophy. Nat. Commun. 2016;7:11067. doi: 10.1038/ncomms11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savkur R.S., Philips A.V., Cooper T.A. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 21.Kuyumcu-Martinez N.M., Wang G.S., Cooper T.A. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward A.J., Rimer M., Killian J.M., Dowling J.J., Cooper T.A. CUGBP1 overexpression in mouse skeletal muscle reproduces features of myotonic dystrophy type 1. Hum. Mol. Genet. 2010;19:3614–3622. doi: 10.1093/hmg/ddq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ladd A.N. CUG-BP, Elav-like family (CELF)-mediated alternative splicing regulation in the brain during health and disease. Mol. Cell. Neurosci. 2013;56:456–464. doi: 10.1016/j.mcn.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Batra R., Charizanis K., Manchanda M., Mohan A., Li M., Finn D.J., Goodwin M., Zhang C., Sobczak K., Thornton C.A., et al. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol. Cell. 2014;56:311–322. doi: 10.1016/j.molcel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang E.T., Cody N.A., Jog S., Biancolella M., Wang T.T., Treacy D.J., Luo S., Schroth G.P., Housman D.E., Reddy S., et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mankodi A., Logigian E., Callahan L., McClain C., White R., Henderson D., Krym M., Thornton C.A. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 27.Rao A.N., Campbell H.M., Guan X., Word T.A., Wehrens X.H., Xia Z., Cooper T.A. Reversible cardiac disease features in an inducible CUG repeat RNA-expressing mouse model of myotonic dystrophy. JCI Insight. 2021;6:e143465. doi: 10.1172/jci.insight.143465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huguet A., Medja F., Nicole A., Vignaud A., Guiraud-Dogan C., Ferry A., Decostre V., Hogrel J.Y., Metzger F., Hoeflich A., et al. Molecular, physiological, and motor performance defects in DMSXL mice carrying >1,000 CTG repeats from the human DM1 locus. PLoS Genet. 2012;8:e1003043. doi: 10.1371/journal.pgen.1003043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pascual-Gilabert M., López-Castel A., Artero R. Myotonic dystrophy type 1 drug development: A pipeline toward the market. Drug Discov. Today. 2021;26:1765–1772. doi: 10.1016/j.drudis.2021.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu H., Lima W.F., Zhang H., Fan A., Sun H., Crooke S.T. Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J. Biol. Chem. 2004;279:17181–17189. doi: 10.1074/jbc.M311683200. [DOI] [PubMed] [Google Scholar]
- 31.Crooke S.T. Molecular Mechanisms of Antisense Oligonucleotides. Nucleic Acid Ther. 2017;27:70–77. doi: 10.1089/nat.2016.0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monia B.P., Lesnik E.A., Gonzalez C., Lima W.F., McGee D., Guinosso C.J., Kawasaki A.M., Cook P.D., Freier S.M. Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem. 1993;268:14514–14522. doi: 10.1016/S0021-9258(19)85268-7. [DOI] [PubMed] [Google Scholar]
- 33.Singh R.N., Singh N.N. Mechanism of Splicing Regulation of Spinal Muscular Atrophy Genes. Adv. Neurobiol. 2018;20:31–61. doi: 10.1007/978-3-319-89689-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dominski Z., Kole R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc. Natl. Acad. Sci. USA. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein A.F., Varela M.A., Arandel L., Holland A., Naouar N., Arzumanov A., Seoane D., Revillod L., Bassez G., Ferry A., et al. Peptide-conjugated oligonucleotides evoke long-lasting myotonic dystrophy correction in patient-derived cells and mice. J. Clin. Investig. 2019;129:4739–4744. doi: 10.1172/JCI128205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulders S.A., van den Broek W.J., Wheeler T.M., Croes H.J., van Kuik-Romeijn P., de Kimpe S.J., Furling D., Platenburg G.J., Gourdon G., Thornton C.A., et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc. Natl. Acad. Sci. USA. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanadia R.N., Shin J., Yuan Y., Beattie S.G., Wheeler T.M., Thornton C.A., Swanson M.S. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc. Natl. Acad. Sci. USA. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aartsma-Rus A., Straub V., Hemmings R., Haas M., Schlosser-Weber G., Stoyanova-Beninska V., Mercuri E., Muntoni F., Sepodes B., Vroom E., et al. Development of Exon Skipping Therapies for Duchenne Muscular Dystrophy: A Critical Review and a Perspective on the Outstanding Issues. Nucleic Acid Ther. 2017;27:251–259. doi: 10.1089/nat.2017.0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eder P.S., DeVine R.J., Dagle J.M., Walder J.A. Substrate specificity and kinetics of degradation of antisense oligonucleotides by a 3′ exonuclease in plasma. Antisense Res. Dev. 1991;1:141–151. doi: 10.1089/ard.1991.1.141. [DOI] [PubMed] [Google Scholar]
- 40.Roberts T.C., Langer R., Wood M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020;19:673–694. doi: 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rifai A., Brysch W., Fadden K., Clark J., Schlingensiepen K.H. Clearance kinetics, biodistribution, and organ saturability of phosphorothioate oligodeoxynucleotides in mice. Am. J. Pathol. 1996;149:717–725. [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe T.A., Geary R.S., Levin A.A. Plasma protein binding of an antisense oligonucleotide targeting human ICAM-1 (ISIS 2302) Oligonucleotides. 2006;16:169–180. doi: 10.1089/oli.2006.16.169. [DOI] [PubMed] [Google Scholar]
- 43.Eckstein F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 2014;24:374–387. doi: 10.1089/nat.2014.0506. [DOI] [PubMed] [Google Scholar]
- 44.Hamm S., Latz E., Hangel D., Müller T., Yu P., Golenbock D., Sparwasser T., Wagner H., Bauer S. Alternating 2′-O-ribose methylation is a universal approach for generating non-stimulatory siRNA by acting as TLR7 antagonist. Immunobiology. 2010;215:559–569. doi: 10.1016/j.imbio.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Manoharan M. 2′-carbohydrate modifications in antisense oligonucleotide therapy: Importance of conformation, configuration and conjugation. Biochim. Biophys. Acta. 1999;1489:117–130. doi: 10.1016/S0167-4781(99)00138-4. [DOI] [PubMed] [Google Scholar]
- 46.Obika S., Onoda M., Morita K., Andoh J., Koizumi M., Imanishi T. 3′-amino-2′,4′-BNA: Novel bridged nucleic acids having an N3′-->P5′ phosphoramidate linkage. Chem. Commun. 2001;19:1992–1993. doi: 10.1039/b105640a. [DOI] [PubMed] [Google Scholar]
- 47.Obika S., Uneda T., Sugimoto T., Nanbu D., Minami T., Doi T., Imanishi T. 2′-O,4′-C-Methylene bridged nucleic acid (2′,4′-BNA): Synthesis and triplex-forming properties. Bioorg. Med. Chem. 2001;9:1001–1011. doi: 10.1016/S0968-0896(00)00325-4. [DOI] [PubMed] [Google Scholar]
- 48.Veedu R.N., Wengel J. Locked nucleic acid as a novel class of therapeutic agents. RNA Biol. 2009;6:321–323. doi: 10.4161/rna.6.3.8807. [DOI] [PubMed] [Google Scholar]
- 49.Vester B., Wengel J. LNA (locked nucleic acid): High-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 50.Morita K., Hasegawa C., Kaneko M., Tsutsumi S., Sone J., Ishikawa T., Imanishi T., Koizumi M. 2′-O,4′-C-ethylene-bridged nucleic acids (ENA): Highly nuclease-resistant and thermodynamically stable oligonucleotides for antisense drug. Bioorg. Med. Chem. Lett. 2002;12:73–76. doi: 10.1016/S0960-894X(01)00683-7. [DOI] [PubMed] [Google Scholar]
- 51.Summerton J., Weller D. Morpholino antisense oligomers: Design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 1997;7:187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- 52.Saarbach J., Sabale P.M., Winssinger N. Peptide nucleic acid (PNA) and its applications in chemical biology, diagnostics, and therapeutics. Curr. Opin. Chem. Biol. 2019;52:112–124. doi: 10.1016/j.cbpa.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 53.Larsen H.J., Bentin T., Nielsen P.E. Antisense properties of peptide nucleic acid. Biochim. Biophys. Acta. 1999;1489:159–166. doi: 10.1016/S0167-4781(99)00145-1. [DOI] [PubMed] [Google Scholar]
- 54.Wheeler T.M., Leger A.J., Pandey S.K., MacLeod A.R., Nakamori M., Cheng S.H., Wentworth B.M., Bennett C.F., Thornton C.A. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee J.E., Bennett C.F., Cooper T.A. RNase H-mediated degradation of toxic RNA in myotonic dystrophy type 1. Proc. Natl. Acad. Sci. USA. 2012;109:4221–4226. doi: 10.1073/pnas.1117019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jauvin D., Chrétien J., Pandey S.K., Martineau L., Revillod L., Bassez G., Lachon A., MacLeod A.R., Gourdon G., Wheeler T.M., et al. Targeting DMPK with Antisense Oligonucleotide Improves Muscle Strength in Myotonic Dystrophy Type 1 Mice. Mol. Ther. Nucleic Acids. 2017;7:465–474. doi: 10.1016/j.omtn.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yadava R.S., Yu Q., Mandal M., Rigo F., Bennett C.F., Mahadevan M.S. Systemic therapy in an RNA toxicity mouse model with an antisense oligonucleotide therapy targeting a non-CUG sequence within the DMPK 3′UTR RNA. Hum. Mol. Genet. 2020;29:1440–1453. doi: 10.1093/hmg/ddaa060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wheeler T.M., Sobczak K., Lueck J.D., Osborne R.J., Lin X., Dirksen R.T., Thornton C.A. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leger A.J., Mosquea L.M., Clayton N.P., Wu I.H., Weeden T., Nelson C.A., Phillips L., Roberts E., Piepenhagen P.A., Cheng S.H., et al. Systemic delivery of a Peptide-linked morpholino oligonucleotide neutralizes mutant RNA toxicity in a mouse model of myotonic dystrophy. Nucleic Acid Ther. 2013;23:109–117. doi: 10.1089/nat.2012.0404. [DOI] [PubMed] [Google Scholar]
- 60.Koshkin A.A., Singh S.K., Nielsen P., Rajwanshi V.K., Kumar R., Meldgaard M., Olsen C.E., Wengel J. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. doi: 10.1016/S0040-4020(98)00094-5. [DOI] [Google Scholar]
- 61.Jeong W., Rapisarda A., Park S.R., Kinders R.J., Chen A., Melillo G., Turkbey B., Steinberg S.M., Choyke P., Doroshow J.H., et al. Pilot trial of EZN-2968, an antisense oligonucleotide inhibitor of hypoxia-inducible factor-1 alpha (HIF-1α), in patients with refractory solid tumors. Cancer Chemother. Pharmacol. 2014;73:343–348. doi: 10.1007/s00280-013-2362-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wojtkowiak-Szlachcic A., Taylor K., Stepniak-Konieczna E., Sznajder L.J., Mykowska A., Sroka J., Thornton C.A., Sobczak K. Short antisense-locked nucleic acids (all-LNAs) correct alternative splicing abnormalities in myotonic dystrophy. Nucleic Acids Res. 2015;43:3318–3331. doi: 10.1093/nar/gkv163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pandey S.K., Wheeler T.M., Justice S.L., Kim A., Younis H.S., Gattis D., Jauvin D., Puymirat J., Swayze E.E., Freier S.M., et al. Identification and characterization of modified antisense oligonucleotides targeting DMPK in mice and nonhuman primates for the treatment of myotonic dystrophy type 1. J. Pharmacol. Exp. Ther. 2015;355:329–340. doi: 10.1124/jpet.115.226969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ionis Pharmaceuticals Reports DMPKRx Phase 1/2 Trial Results. [(accessed on 12 September 2022)]. Available online: https://us8.campaign-archive.com/?u=8f5969cac3271759ce78c8354&id=8cc67ae9b8&e=cd1f4d18fe.
- 65.Østergaard M.E., Jackson M., Low A., Chappell A.E., Lee R.G., Peralta R.Q., Yu J., Kinberger G.A., Dan A., Carty R., et al. Conjugation of hydrophobic moieties enhances potency of antisense oligonucleotides in the muscle of rodents and non-human primates. Nucleic Acids Res. 2019;47:6045–6058. doi: 10.1093/nar/gkz360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weaving G., Batstone G.F., Jones R.G. Age and sex variation in serum albumin concentration: An observational study. Ann. Clin. Biochem. 2016;53:106–111. doi: 10.1177/0004563215593561. [DOI] [PubMed] [Google Scholar]
- 67.Seth P.P., Tanowitz M., Bennett C.F. Selective tissue targeting of synthetic nucleic acid drugs. J. Clin. Investig. 2019;129:915–925. doi: 10.1172/JCI125228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larsen M.T., Kuhlmann M., Hvam M.L., Howard K.A. Albumin-based drug delivery: Harnessing nature to cure disease. Mol. Cell. Ther. 2016;4:3. doi: 10.1186/s40591-016-0048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prakash T.P., Mullick A.E., Lee R.G., Yu J., Yeh S.T., Low A., Chappell A.E., Østergaard M.E., Murray S., Gaus H.J., et al. Fatty acid conjugation enhances potency of antisense oligonucleotides in muscle. Nucleic Acids Res. 2019;47:6029–6044. doi: 10.1093/nar/gkz354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang S., Allen N., Prakash T.P., Liang X.H., Crooke S.T. Lipid Conjugates Enhance Endosomal Release of Antisense Oligonucleotides into Cells. Nucleic Acid Ther. 2019;29:245–255. doi: 10.1089/nat.2019.0794. [DOI] [PubMed] [Google Scholar]
- 71.Nagata T., Dwyer C.A., Yoshida-Tanaka K., Ihara K., Ohyagi M., Kaburagi H., Miyata H., Ebihara S., Yoshioka K., Ishii T., et al. Cholesterol-functionalized DNA/RNA heteroduplexes cross the blood-brain barrier and knock down genes in the rodent CNS. Nat. Biotechnol. 2021;39:1529–1536. doi: 10.1038/s41587-021-00972-x. [DOI] [PubMed] [Google Scholar]
- 72.Relizani K., Echevarría L., Zarrouki F., Gastaldi C., Dambrune C., Aupy P., Haeberli A., Komisarski M., Tensorer T., Larcher T., et al. Palmitic acid conjugation enhances potency of tricyclo-DNA splice switching oligonucleotides. Nucleic Acids Res. 2022;50:17–34. doi: 10.1093/nar/gkab1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cerro-Herreros E., Sabater-Arcis M., Fernandez-Costa J.M., Moreno N., Perez-Alonso M., Llamusi B., Artero R. miR-23b and miR-218 silencing increase Muscleblind-like expression and alleviate myotonic dystrophy phenotypes in mammalian models. Nat. Commun. 2018;9:2482. doi: 10.1038/s41467-018-04892-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khan T., Weber H., DiMuzio J., Matter A., Dogdas B., Shah T., Thankappan A., Disa J., Jadhav V., Lubbers L., et al. Silencing Myostatin Using Cholesterol-conjugated siRNAs Induces Muscle Growth. Mol. Ther. Nucleic Acids. 2016;5:e342. doi: 10.1038/mtna.2016.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Krützfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 76.Cerro-Herreros E., González-Martínez I., Moreno-Cervera N., Overby S., Pérez-Alonso M., Llamusí B., Artero R. Therapeutic Potential of AntagomiR-23b for Treating Myotonic Dystrophy. Mol. Ther. Nucleic Acids. 2020;21:837–849. doi: 10.1016/j.omtn.2020.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cerro-Herreros E., González-Martínez I., Moreno N., Espinosa-Espinosa J., Fernández-Costa J.M., Colom-Rodrigo A., Overby S.J., Seoane-Miraz D., Poyatos-García J., Vilchez J.J., et al. Preclinical characterization of antagomiR-218 as a potential treatment for myotonic dystrophy. Mol. Ther. Nucleic Acids. 2021;26:174–191. doi: 10.1016/j.omtn.2021.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sievers E.L., Senter P.D. Antibody-drug conjugates in cancer therapy. Annu. Rev. Med. 2013;64:15–29. doi: 10.1146/annurev-med-050311-201823. [DOI] [PubMed] [Google Scholar]
- 79.Dugal-Tessier J., Thirumalairajan S., Jain N. Antibody-Oligonucleotide Conjugates: A Twist to Antibody-Drug Conjugates. J. Clin. Med. 2021;10:838. doi: 10.3390/jcm10040838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnsen K.B., Burkhart A., Thomsen L.B., Andresen T.L., Moos T. Targeting the transferrin receptor for brain drug delivery. Prog. Neurobiol. 2019;181:101665. doi: 10.1016/j.pneurobio.2019.101665. [DOI] [PubMed] [Google Scholar]
- 81.Paterson J., Webster C.I. Exploiting transferrin receptor for delivering drugs across the blood-brain barrier. Drug Discov. Today Technol. 2016;20:49–52. doi: 10.1016/j.ddtec.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 82.Sonoda H., Morimoto H., Yoden E., Koshimura Y., Kinoshita M., Golovina G., Takagi H., Yamamoto R., Minami K., Mizoguchi A., et al. A Blood-Brain-Barrier-Penetrating Anti-human Transferrin Receptor Antibody Fusion Protein for Neuronopathic Mucopolysaccharidosis II. Mol. Ther. 2018;26:1366–1374. doi: 10.1016/j.ymthe.2018.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Webster C.I., Hatcher J., Burrell M., Thom G., Thornton P., Gurrell I., Chessell I. Enhanced delivery of IL-1 receptor antagonist to the central nervous system as a novel anti-transferrin receptor-IL-1RA fusion reverses neuropathic mechanical hypersensitivity. Pain. 2017;158:660–668. doi: 10.1097/j.pain.0000000000000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Johnson N., Day J., Hamel J., Statland J., Subramony S.H., Arnold W.D., Thornton C., Wicklund M., Soltanzadeh P., Knisley B., et al. Study Design of AOC 1001-CS1, a Phase 1/2 Clinical Trial Evaluating the Safety, Tolerability, Pharmacokinetics and Pharmacodynamics of AOC 1001 Administered Intravenously to Adult Patients with Myotonic Dystrophy Type 1 (DM1) (MARINA) (S23.006) Neurology. 2022;98:2711. [Google Scholar]
- 85.Hammond S.M., Abendroth F., Goli L., Burrell M., Thom G., Gurrell I., Stoodley J., Ahlskog N., Gait M.J., Wood M.J.A., et al. Systemic antibody-oligonucleotide delivery to the central nervous system ameliorates mouse models of spinal muscular atrophy. bioRxiv. 2021 doi: 10.1101/2021.07.29.454272. [DOI] [Google Scholar]
- 86.Betts C.A., Saleh A.F., Carr C.A., Hammond S.M., Coenen-Stass A.M., Godfrey C., McClorey G., Varela M.A., Roberts T.C., Clarke K., et al. Prevention of exercised induced cardiomyopathy following Pip-PMO treatment in dystrophic mdx mice. Sci. Rep. 2015;5:8986. doi: 10.1038/srep08986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hammond S.M., Hazell G., Shabanpoor F., Saleh A.F., Bowerman M., Sleigh J.N., Meijboom K.E., Zhou H., Muntoni F., Talbot K., et al. Systemic peptide-mediated oligonucleotide therapy improves long-term survival in spinal muscular atrophy. Proc. Natl. Acad. Sci. USA. 2016;113:10962–10967. doi: 10.1073/pnas.1605731113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gao X., Zhao J., Han G., Zhang Y., Dong X., Cao L., Wang Q., Moulton H.M., Yin H. Effective dystrophin restoration by a novel muscle-homing peptide-morpholino conjugate in dystrophin-deficient mdx mice. Mol. Ther. 2014;22:1333–1341. doi: 10.1038/mt.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yin H., Moulton H.M., Seow Y., Boyd C., Boutilier J., Iverson P., Wood M.J. Cell-penetrating peptide-conjugated antisense oligonucleotides restore systemic muscle and cardiac dystrophin expression and function. Hum. Mol. Genet. 2008;17:3909–3918. doi: 10.1093/hmg/ddn293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ivanova G.D., Arzumanov A., Abes R., Yin H., Wood M.J.A., Lebleu B., Gait M.J. Improved cell-penetrating peptide–PNA conjugates for splicing redirection in HeLa cells and exon skipping in mdx mouse muscle. Nucleic Acids Res. 2008;36:6418–6428. doi: 10.1093/nar/gkn671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hammond S.M., Aartsma-Rus A., Alves S., Borgos S.E., Buijsen R.A.M., Collin R.W.J., Covello G., Denti M.A., Desviat L.R., Echevarría L., et al. Delivery of oligonucleotide-based therapeutics: Challenges and opportunities. EMBO Mol. Med. 2021;13:e13243. doi: 10.15252/emmm.202013243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Overby S.J., Cerro-Herreros E., González-Martínez I., Varela M.A., Seoane-Miraz D., Jad Y., Raz R., Møller T., Pérez-Alonso M., Wood M.J., et al. Proof of concept of peptide-linked blockmiR-induced MBNL functional rescue in myotonic dystrophy type 1 mouse model. Mol. Ther. Nucleic Acids. 2022;27:1146–1155. doi: 10.1016/j.omtn.2022.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Goodkey K., Aslesh T., Maruyama R., Yokota T. Nusinersen in the Treatment of Spinal Muscular Atrophy. In: Yokota T., Maruyama R., editors. Exon Skipping and Inclusion Therapies. 1st ed. Volume 1828. Humana Press; New York, NY, USA: 2018. pp. 69–76. [DOI] [PubMed] [Google Scholar]
- 94.Yang J., Luo S., Zhang J., Yu T., Fu Z., Zheng Y., Xu X., Liu C., Fan M., Zhang Z. Exosome-mediated delivery of antisense oligonucleotides targeting α-synuclein ameliorates the pathology in a mouse model of Parkinson’s disease. Neurobiol. Dis. 2021;148:105218. doi: 10.1016/j.nbd.2020.105218. [DOI] [PubMed] [Google Scholar]
- 95.Tabrizi S.J., Leavitt B.R., Landwehrmeyer G.B., Wild E.J., Saft C., Barker R.A., Blair N.F., Craufurd D., Priller J., Rickards H., et al. Targeting Huntingtin Expression in Patients with Huntington’s Disease. N. Engl. J. Med. 2019;380:2307–2316. doi: 10.1056/NEJMoa1900907. [DOI] [PubMed] [Google Scholar]
- 96.Corey D. Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy. Nat. Neurosci. 2017;20:497–499. doi: 10.1038/nn.4508. [DOI] [PubMed] [Google Scholar]
- 97.King M., Pan Y.X., Mei J., Chang A., Xu J., Pasternak G.W. Enhanced kappa-opioid receptor-mediated analgesia by antisense targeting the sigma1 receptor. Eur. J. Pharmacol. 1997;331:R5–R6. doi: 10.1016/S0014-2999(97)01064-9. [DOI] [PubMed] [Google Scholar]
- 98.Rossi G., Pan Y.X., Cheng J., Pasternak G.W. Blockade of morphine analgesia by an antisense oligodeoxynucleotide against the mu receptor. Life Sci. 1994;54:PL375–PL379. doi: 10.1016/0024-3205(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 99.Leventhal L., Cole J.L., Rossi G.C., Pan Y.X., Pasternak G.W., Bodnar R.J. Antisense oligodeoxynucleotides against the MOR-1 clone alter weight and ingestive responses in rats. Brain Res. 1996;719:78–84. doi: 10.1016/0006-8993(96)00089-3. [DOI] [PubMed] [Google Scholar]
- 100.Ait Benichou S., Jauvin D., De Serres-Bérard T., Pierre M., Ling K.K., Bennett C.F., Rigo F., Gourdon G., Chahine M., Puymirat J. Antisense oligonucleotides as a potential treatment for brain deficits observed in myotonic dystrophy type 1. Gene Ther. :2022. doi: 10.1038/s41434-022-00316-7. in press . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Douniol M., Jacquette A., Cohen D., Bodeau N., Rachidi L., Angeard N., Cuisset J.M., Vallée L., Eymard B., Plaza M., et al. Psychiatric and cognitive phenotype of childhood myotonic dystrophy type 1. Dev. Med. Child Neurol. 2012;54:905–911. doi: 10.1111/j.1469-8749.2012.04379.x. [DOI] [PubMed] [Google Scholar]
- 102.Weijs R., Okkersen K., van Engelen B., Küsters B., Lammens M., Aronica E., Raaphorst J., van Cappellen van Walsum A.M. Human brain pathology in myotonic dystrophy type 1: A systematic review. Neuropathology. 2021;41:3–20. doi: 10.1111/neup.12721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article.