Abstract
Maize seedlings contain high amounts of 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOA), and the effect of DIMBOA is directly associated with multiple insect-resistance against insect pests such as Asian corn borer and corn leaf aphids. Although numerous genetic loci for multiple insect-resistant traits have been identified, little is known about genetic controls regarding DIMBOA content. In this study, the best linear unbiased prediction (BLUP) values of DIMBOA content in two ecological environments across 310 maize inbred lines were calculated; and their phenotypic data and BLUP values were used for marker-trait association analysis. We identified nine SSRs that were significantly associated with DIMBOA content, which explained 4.30–20.04% of the phenotypic variation. Combined with 47 original genetic loci from previous studies, we detected 19 hot loci and approximately 11 hot loci (in Bin 1.04, Bin 2.00–2.01, Bin 2.03–2.04, Bin 4.00–4.03, Bin 5.03, Bin 5.05–5.07, Bin 8.01–8.03, Bin 8.04–8.05, Bin 8.06, Bin 9.01, and Bin 10.04 regions) supported pleiotropy for their association with two or more insect-resistant traits. Within the 19 hot loci, we identified 49 candidate genes, including 12 controlling DIMBOA biosynthesis, 6 involved in sugar metabolism/homeostasis, 2 regulating peroxidases activity, 21 associated with growth and development [(auxin-upregulated RNAs (SAUR) family member and v-myb avian myeloblastosis viral oncogene homolog (MYB)], and 7 involved in several key enzyme activities (lipoxygenase, cysteine protease, restriction endonuclease, and ubiquitin-conjugating enzyme). The synergy and antagonism interactions among these genes formed the complex defense mechanisms induced by multiple insect pests. Moreover, sufficient genetic variation was reported for DIMBOA performance and SSR markers in the 310 tested maize inbred lines, and 3 highly (DIMBOA content was 402.74–528.88 μg g−1 FW) and 15 moderate (DIMBOA content was 312.92–426.56 μg g−1 FW) insect-resistant genotypes were major enriched in the Reid group. These insect-resistant inbred lines can be used as parents in maize breeding programs to develop new varieties.
Keywords: maize, DIMBOA accumulation, Asian corn borer/corn leaf aphid, genetic diversity, association mapping, general linear model, mixed linear model, pleiotropy, candidate genes
1. Introduction
Maize (Zea mays), is an important agro-economical crop that is utilized globally as food, animal feed, and biofuel products with production of more than 1.14 billion tons in 2018 [1]. Despite the large production area for maize, extensive losses are common in China due to a range of biotic stressors, such as multiple insect injury [1]. Over 350 insect pests have been identified for maize. Of these, Asian corn borer (ACB) (Ostrinia furnacalis; Lepidoptera, Pyralidae) is one of the most destructive insect pests in maize. Newly hatched ACB larvae primarily feed on leaves during the whorl stage; subsequently, the 3rd or 4th instars bore into the stalk. This may cause yield losses of 10 to 30% in the outbreaks recorded in China [2]. In addition, the aboveground parts of maize are susceptible to corn leaf aphid (CLA) (Rhopalosiphum maidis; Homoptera, Aphididae), especially in tropical and warmer temperature regions [3,4]. CLA infestation in maize seedlings slows down plant development, reduces plant height, and decreases grain yield [5]. CLA damage can even occur through the maize tassel in which the accumulation of sticky honeydew can prevent the shedding of pollen, with yield losses of up to 90% [4,6]. Several aphid species can also transmit maize dwarf mosaic virus in a non-persistent manner [7], causing yield losses as high as 70% [8].
Chemical insecticides are widely used to reduce yield losses in maize incurred by insect pests, including ACB and CLA. However, residues from these insecticides can pose a significant health hazard to maize consumers and also cause harm to the environment [9,10]. Furthermore, the overuse of pesticides may also result in the development of chemical resistance to insects and the emergence of secondary pests [9,11]. Fortunately, maize is a genetically diverse crop that exhibits a wide variation in its resistance to multiple insects [9,12,13]; the varieties of maize that show high resistance to insect pests can be used to thwart multiple insect attacks via their various defense mechanisms [9,14]. In maize, the chemical defense metabolite 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOA), a cyclic hydroxamic acid derivative (benzoxazinoids, bxs), is known to confer resistance to leaf-feeding by both ACB [2,15] and CLA [4,16] via inhibitory and toxicity mechanisms. Previous studies reported that the DIMBOA could constitute > 1% of the dry weight of maize seedlings [17] and could be more abundant in adult maize plants after the induction of defense mechanisms [18]. The advances in molecular biology and statistical methods over recent years have led to the identification of multiple quantitative trait loci (QTLs), associated markers, and candidate genes for maize resistance to leaf-feeding by different corn borers and aphids [2,4,12,19,20]. However, only one study reported the involvement of QTLs responsible for DIMBOA content [15], and the low resolution of QTL linkage mapping methodology did not provide a convincing conclusion that genetic variation in maize resistance to multiple insect pests was associated with DIMBOA content and bxs alleles.
Association mapping based on linkage disequilibrium (LD) provides a fine-mapping method that enables researchers to identify functional variation in a broader germplasm background [21]. Both QTL linkage mapping and association mapping methods could be complementary strategies for investigating genetic variation in maize related to DIMBOA content. At present, only a few maize genotypes (Mc37, Mp708, Mp704, D06, CML103, A637, and EP39) are known to be ACB-resistant germplasms [20]; little is known with regard to multiple insect-resistance breeding in maize. Therefore, there is an urgent need to screen genetic resources and characterize the mechanisms associated with multiple insect resistance. The testing of inbred maize lines in terms of their DIMBOA levels in response to ACB and CLA feeding may serve as a potent approach to evaluate and develop new varieties with multiple insect-resistance. Gansu Province in China is one of the core bases for maize seed production and breeding, and a large number of elite inbred lines have been cultivated over the last decade. However, the development of breeding programs for multiple insect-resistance has been slow in this region. For this reason, we collected and investigated 310 elite inbred maize lines with broad genetic backgrounds from different ecological environments in Gansu Province, China. Our objectives were to (i) evaluate the accumulation of DIMBOA in the V6 stage of these different genotypes in different ecological environments and to identify elite multiple insect-resistant parents for use in breeding programs; (ii) explore the genetic diversity and population structure of these maize lines by 186 polymorphic simple repeated sequences (SSRs) in the 10 maize chromosomes, and (iii) identify SSRs that were significantly associated with DIMBOA content and the best linear unbiased prediction (BLUP) values of DIMBOA content in two ecological environments via a general linear model (GLM) and a mixed linear model (MLM). We also detected hot genetic loci and candidate genes associated with multiple insect-resistance by combining our research findings with those of previous studies. These findings have laid a foundation for further multiple insect-resistance marker-assisted selection (MAS) breeding in maize.
2. Results
2.1. DIMBOA Content Variation
The maize-feeding insects begin to damage maize leaves at the V5-V6 seedlings stage in the field [15]. At the V6 stage, sufficient variation was observed for DIMBOA content in 310 maize inbred lines in both Zhangye (E1) and Longxi (E2) ecological environments (Figure 1A). The average DIMBOA content of 310 inbred lines seedlings in the E1 environment ranged from 8.66 (T58) to 528.88 μg g−1 FW (ZY19-Jiu1101) with an overall mean of 94.21μg g−1 FW; similarly, the average DIMBOA content of these seedlings in the E2 environment varied from 8.84 (T58) to 493.40 μg g−1 FW (RX20-1006) with an overall mean of 93.26 μg g−1 FW (Figure 1B). Moreover, factorial analysis of variance also proved significant for the tested variables (genotypes and environments) and their interaction with the DIMBOA content in the seedlings of these maize genotypes (Figure 1C). These data indicated that the DIMBOA formation and accumulation were controlled by a number of factors, including maize’s genetic constitutions, external environments, and their interaction. Therefore, after maize-feeding insects, such as ACB and CLA feed on leaves, DIMBOA can be induced rapidly in maize to confer resistance to these insects, along with other adverse responses, such as anorexia and toxic reactions in these insects.
Figure 1.
Statistical analysis of DIMBOA content at V6 stage in Zhangye (E1) and Longxi (E2) ecological environments among 310 maize inbred lines seedlings in Gansu Province, China in 2020 and their best linear unbiased prediction (BLUP) values. (A) Violin plot shows the DIMBOA content in E1 and E2 environments, and their BLUP values among all inbred lines. (B) The statistics including mean ± standard deviation (SD), minimum, maximum, confidence interval (CI), skewness, kurtosis, and genetic variation coefficient (CVg) of DIMBOA content and their BLUP values among all inbred lines in E1 and E2 environments. (C) Frequency of the genotypic variance (σg2), environmental variance (σe2), genotype × environment interaction variance (σge2), and error variance (σε2) for DIMBOA content; *** indicated a significant difference with p < 0.01 (ANOVA). (D) The broad-sense heritability () and genotype × environment interaction heritability () of DIMBOA content.
In addition, the DIMBOA content of these maize materials in the two ecological environments followed a typical partial distribution, as the skewness and kurtosis values of the DIMBOA content were > 1.0 (Figure 1B). We speculated that this phenomenon may be related to the unequal enrichment of DIMBOA content in the selected maize inbred lines. Further analysis showed that the estimated broad-sense heritability () and genotype × environment interaction heritability () of DIMBOA content in all inbred lines across both ecological environments were 74.64 and 37.32%, respectively (Figure 1D). The data demonstrated that DIMBOA content was less affected by the environment when compared to other insect-resistant traits, especially the leaf feeding rating of corn borer (LFR) and the number of holes of corn borer (HO), and their were 37.5 and 46.4% [2], respectively. Meanwhile, the genetic variation coefficient (CVg) of DIMBOA content among 310 maize inbred lines in both E1 and E2 environments were 108.47 and 106.22%, respectively (Figure 1B). Thereby, it is necessary to detect the genetic loci responsible for DIMBOA content in maize.
2.2. Genetic Diversity and Population Structure Analysis
Genetic diversity and population structure analysis are important tools for germplasm characterization and subsequent utilization in multiple insect-resistance improvements. A total of 186 polymorphic SSRs were used to analyze the distribution of 748 alleles in 310 maize inbred lines. The number of alleles varied from two to eight at a locus. The umc1917 (Bin 1.04, ctg14), umc2314 (Bin 6.01, ctg268) and umc2031 (Bin 8.06, ctg361) exhibited the maximum number of alleles (8 alleles) (Table S1). It is well known that the marker attributes, i.e., the polymorphism information content (PIC) value and Shannon-Wiener’s index (I) value are routinely used to evaluate the informativeness of the primers. In this study, the PIC value of each SSR ranged between 0.294 (umc2297, Bin 5.03, ctg220) and 0.826 (umc2314, Bin 6.01, ctg268). Out of 186 SSRs, 69 markers were highly polymorphic, with a PIC ≥ 0.600 (Table S1). Similarly, the average I value was 1.359 and ranged between 0.585 (umc2112, Bin 1.04, ctg21; umc2043, Bin 10.05, ctg415) and 2.410 (umc2314, Bin 6.01, ctg268) (Table S1). These data showed that a PIC value ≥ 0.600 was observed in approximately 40% of the markers, suggesting that these SSRs were very informative and useful in the assessment of genetic diversity, population structure, and marker-DIMBOA content association analysis.
In addition, the population structure of these inbred lines was analyzed by STRUCTURE software. When various groups (K value) ranging from 1 to 12 were compared, the ΔK reached the maximum value when K = 5, thus the 310 maize inbred lines were divided into five optimal groups (Figure 2A). The inbred lines with a membership probability of ≥ 0.500 were assigned to the same groups, and if the inbred lines had a membership probability of less than this value, they were assigned to a mixed group (not assigned to any of the five groups) [22,23]. Of the 310 inbred lines, 294 (accounting for 94.84%) were assigned to either one of the five groups, including the Lüda red cob (LRC; 44 lines) group, Tang si ping tou (TSPT; 81 lines) group, Lancaster (Lan; 52 lines) group, P (47 lines) group, and Reid (70 lines) group, respectively, and the remaining 16 inbred lines (accounting for 5.16%) were categorized as a mixed group (Figure 2B and Figure 3A; Table S2).
Figure 2.
Population structure of 310 maize inbred lines revealed by 186 polymorphic SSR markers. (A) Change curve in the log probability data of ΔK value against K (group number) value. (B) Population structure of 310 inbred lines based on 186 SSR markers at K = 5. Each inbred line was represented by a vertical line, which indicated the membership coefficient for that individual, and the five groups were the Lüda red cob (LRC), Tang si ping tou (TSPT), Lancaster (Lan), P, and Reid groups, respectively.
Figure 3.
Evaluation of multiple insect-resistance among 310 maize inbred lines in both Zhangye (E1) and Longxi (E2) ecological environments by unweighted pair-group method with arithmetic means (UPGMA) cluster analysis, and their attributive group dissection. (A) Statistics for group number and frequency of each optimal group. (B) Attributive groups and their frequency distribution of three high insect-resistant and 15 moderate insect-resistant inbred lines; average of DIMBOA content for each attributive group (including Lüda red cob (LRC), Lancaster (Lan), P, and Reid group, respectively) in E1 and E2, respectively. (C) Evaluation of multiple insect-resistance among 310 inbred lines in two environments were performed using R package (http://www.R-project.org/; accessed on 20 April 2022) with UPGMA (the K value sets to 5), including type I (high insect-resistant lines), type II (moderate insect-resistant lines), type III (insect-resistant lines), type IV (moderate insect-susceptible lines), and type V (insect-susceptible lines), respectively.
2.3. Evaluation of Multiple Insect-Resistance of Inbred Lines and Dissection of Their Attributive Groups
According to the performance of DIMBOA content among all inbred lines in the two ecological environments, these germplasms were divided into five types (type I to type V). Type I (DIMBOA content in E1/E2: 467.11–528.88/402.74–493.40 μg g−1 FW; accounted for 0.97%) had three genotypes, which were 19LX-1230, RX20-1006, and ZY19-Jiu1101; they were defined as high insect-resistant germplasms. Type II (DIMBOA content in E1/E2: 312.92–426.56/321.41–424.15 μg g−1 FW; accounted for 4.84%) had 15 inbred lines including F1227, M1005, F0501, LongF1008, 1201, M1009, PH1CRW, 1512, F3202, F1220, F3208, F2260, F1233, F2303, and F3210, and they were defined as moderate insect-resistant germplasms. Type III (DIMBOA content in E1/E2: 184.88–292.42/176.52–296.96 μg g−1 FW; accounted for 8.71%) had 27 genotypes, including ShanM3304, M1001, MeizaS2–3, Jizaoyu, M0803, F0306, M10202, ly6305, F2211, M1202, F0311, M1409Ying, F1501, M0124, M0822, M0105, F2502, M0505, PHHJC, 8723-2, M0306, F2222, F2213, M0824, M0125, 747, and F3226, and they were defined as insect-resistant germplasms. Type IV (DIMBOA content in E1/E2: 103.88–184.62/99.76–187.68 μg g−1 FW; accounted for 16.77%) included 52 moderate insect-susceptible germplasms. Type V (DIMBOA content in E1/E2: 8.66–101.14/8.84–99.40 μg g−1 FW; accounted for 68.71%) included 213 insect-susceptible germplasms (Figure 3C). Furthermore, the three high insect-resistant inbred lines and 15 moderate insect-resistant inbred lines belonged to the Reid (accounted for 50.00%), LRC (accounted for 22.22%), P (accounted for 16.67%), and Lan (accounted for 11.11%) groups, respectively (Figure 3B). The data suggested that Reid was an important insect-resistant group, and more attention should be given to the improvement of new insect-resistant varieties using Reid genotypes.
2.4. Association Analysis of DIMBOA Content and BLUP Values
In this study, we retrieved the genetic distance (centimorgan, cM) of 186 SSRs on an IBM2 2008 Neighbors map frame (https://www.maizegdb.org/data_center/map (accessed on 18 September 2022)) (Table S1) to build a genetic linkage map, which spanned a total length of 6684.4 cM (Figure 4). Then, the associated SSR loci of DIMBOA content in both ecological environments and the BLUP values were assessed using Tassel 3.0 software with GLM (Q) and MLM (Q + K) approach. We detected nine and three significant (p < 0.01) SSR loci associated with DIMBOA content in both environments (E1 and E2) by GLM and MLM, respectively; these SSR loci were in Bin 1.04, Bin 1.11, Bin 2.01, Bin 4.00, Bin 4.01, Bin 6.02, Bin 8.04, and Bin 10.04. The phenotypic variation explained by these SSR loci ranged from 4.30% (umc2363, Bin2.01, ctg69) in the E1 environment to 20.04% (umc1008, Bin 4.00, ctg154) in the E2 environment via GLM, and ranged from 4.74% (umc1858, Bin 8.04, ctg349) in the E2 environment to 10.57% (umc1017, Bin 4.01, ctg155) in the E1 environment via MLM (Figure 4; Table 1). Likewise, a total of six and two SSR loci in Bin 1.04, Bin 1.11, Bin 4.00, Bin 4.01, and Bin 10.04 were identified to be significantly (p < 0.01) associated with BLUP values by GLM and MLM, respectively, which explained 5.41% (umc1054, Bin 10.04, ctg412)–19.42% (umc1017, Bin 4.01, ctg155) by GLM, and explained 9.29% (umc1917, Bin 1.04, ctg14)–13.70% (umc1008, Bin 4.00, ctg154) by MLM (Figure 4; Table 1).
Figure 4.
Genetic linkage map construction according to the genetic distance (centimorgan, cM) of 186 polymorphic SSRs on IBM2 2008 Neighbors map frame. The identified SSRs were significantly (p < 0.01) associated with DIMBOA content in Zhangye (E1; pink circle) and Longxi (E2; blue circle) ecological environments, Gansu Province, China in 2020. The best linear unbiased prediction (BLUP; green circle) values for these SSRs were assessed using a general linear model (GLM) and mixed linear model (MLM), respectively.
Table 1.
SSR markers were found significantly (p < 0.01) associated with DIMBOA content in both ecological environments, and their best linear unbiased prediction (BLUP) values in marker-trait association analysis were assessed using a general linear model (GLM) and mixed linear model (MLM), respectively.
| Code | Associated SSR Marker | Bin Location |
Contig | Physical Location (bp) |
GLM | MLM | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| R2 in E1 | R2 in E2 | R2 in BLUP | R2 in E1 | R2 in E2 | R2 in BLUP | |||||
| 1 | umc1917 | 1.04 | ctg14 | 62,677,623 to 62,680,874 | 6.14 | 11.44 | 9.29 | |||
| 2 | umc1009 | 1.11 | ctg64 | 292,965,682 to 292,971,925 | 13.52 | 10.16 | 10.97 | |||
| 3 | umc2363 | 2.01 | ctg69 | 4,164,953 to .4,170,212 | 4.30 | |||||
| 4 | umc1008 | 4.00 | ctg154 | 1,078,106 to 1,080,829 | 15.39 | 20.04 | 12.61 | 9.48 | 13.70 | |
| 5 | umc1017 | 4.01 | ctg155 | 3,002,162 to 3,006,932 | 19.11 | 10.83 | 19.42 | 10.57 | 9.94 | |
| 6 | umc1758 | 4.01 | ctg156 | 5,004,917 to 5,009,075 | 10.53 | 16.74 | 9.90 | |||
| 7 | umc1178 | 6.02 | ctg281 | 89,065,425 to 89,072,192 | 7.97 | 9.19 | ||||
| 8 | umc1858 | 8.04 | ctg349 | 112,055,118 to ..112,059,831 | 5.07 | 4.92 | 4.74 | |||
| 9 | umc1054 | 10.04 | ctg412 | 114,307,530 to 114,311,922 | 7.59 | 5.41 | ||||
SSR, simple sequence repeat; GLM, general linear model; MLM, mixed linear model; BLUP, best linear unbiased prediction; R2 in E1/E2, the phenotypic variation explained by associated SSR loci for DIMBOA content in Zhangye/Longxi, Gansu Province, China in 2020; R2 in BLUP, the phenotypic variation explained by associated SSR loci for BLUP values.
2.5. Identification of Hot Genetic Loci and Candidate Genes Associated with Multiple Insect-Resistant Traits
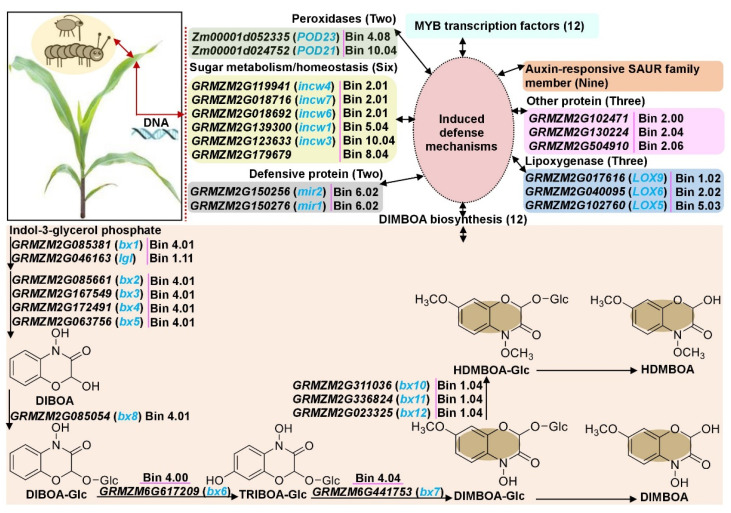
Next, we attempted to obtain hot genetic loci for multiple insect-resistant traits of ACB and CLA to lay a foundation for fine mapping and candidate gene prediction, verification, and breeding application. First, we collected 47 original genetic loci for ACB-/CLA-resistant traits including DIMBOA content, tunnel length of corn borer (TL), aphid incidence rate (AIR), average aphid incidence grade (AIG), LFR, HO, tunnel length/number of holes of corn borer (TL/HO), and aphid resistance (AR) from previous studies (Table 2), and combined our results (Table 1) to construct a physical map (Figure 5). Further, 19 hot genetic loci (Loci 1-Loci 19) for multiple insect resistance were identified on 8 chromosomes except chromosomes 3 and 7, resulting in the identification of 49 candidate genes, including 12 controlling DIMBOA biosynthesis, 6 involved in sugar metabolism/homeostasis, 2 participating in peroxidases (POD) activity, 9/12 associated with auxin-upregulated RNAs (SAUR) family member/ v-myb avian myeloblastosis viral oncogene homolog (MYB), and 3/2/1/1 responsible for the lipoxygenase/cysteine protease/restriction endonuclease/ubiquitin conjugating-enzyme (Figure 5; Table 3; Table S3); these candidate genes may be utilized to determine the multiple insect-resistance to ACB and CLA through their complex interaction network.
Table 2.
Summary of original genetic loci for multiple maize insect-resistant traits of Asian corn borer (ACB) and corn leaf aphid (CLA) from previous studies.
| Trait | Marker Interval |
Chr. (Bin Location) | Marker Type | Marker Physical Location (bp) |
Contig | R2 (%) | Population (Size) | Method | Reference |
|---|---|---|---|---|---|---|---|---|---|
| TL | rs747464 | 4 (4.01) | SNP | 4,759,749 | Inbred lines (301) | GWAS | Gao 2018 [19] | ||
| rs665864 | 2 (2.04) | SNP | 48,777,177 | ||||||
| rs624256 | 2 (2.00) | SNP | 2,806,677 to 2,812,518 | ||||||
| rs653464 | 2 (2.04) | SNP | 28,768,661 to 28,772,516 | ||||||
| rs650025 | 2 (2.03) | SNP | 23,620,411 to 23,625,163 | ||||||
| rs658849 | 2 (2.04) | SNP | 37,078,026 to .37,083,625 | ||||||
| AIR | phi072–umc1164 | 4 (4.00–4.01) |
SSR | 1,078,106 to 1,080,829 3,264,213 to 3,268,368 |
ctg154 ctg155 |
9.26/ 11.55 |
BT-1 × N6 RILs (250) | QTL | Li 2016 [24] |
| umc1666–bnlg1200 | 7 (7.01–7.02) |
SSR | 47,948,200 to 47,950,815 16,678,518 to 16,683,850 |
ctg301 ctg297 |
6.54 | ||||
| AIG | phi072–umc1164 | 4 (4.00–4.01) |
SSR | 1,078,106 to 1,080,829 3,264,213 to 3,268,368 |
ctg154 ctg155 |
8.60/ 11.53 |
|||
| bnlg1700–umc1935 | 5 (5.03–5.03) |
SSR | 33,303,797 to 33,305,797 52,145,125 to 52,169,752 |
ctg217 ctg219 |
4.73 | ||||
| bnlg1792–umc1666 | 7 (7.02–7.02) |
SSR | 85,162,973 to 85,165,402 47,948,200 to 47,950,815 |
ctg305 ctg301 |
6.59 | ||||
| AR | PZA03561.1 | 1 (1.04) | SNP | 60,091,612 to 60,097,741 | B73 × Ky21 RILs (122) | QTL | Tzin et al. 2015 [12] | ||
| PZA01426.1 | 7 (7.00) | SNP | 774,537 to 776,537 | ||||||
| AR | AC213878–AC204415 | 4 (4.01–4.01) |
SNP | 3,021,364 to 3,062,132 4,379,100 to 4,395,483 |
ctg155 ctg156 |
B73 × Mo17 RILs (142) | QTL | Betsiashvili et al. 2015 [4] | |
| DIMBOA | PZA03189 | 1 (1.04) | SNP | 64,242,765 to 64,246,594 | ctg14 | 2.80 | Genetically diverse inbred lines (281) | GLM | Butrón et al. 2010 [15] |
| PZA00635 | 2 (2.04) | SNP | 60,393,632 to 60,395,632 | ctg80 | 3.41 | ||||
| PHM1184 | 4 (4.01) | SNP | 3,050,215 to 3,055,036 | ctg155 | 15.74 | ||||
| PZA02002 | 4 (4.04) | SNP | 29,007,117 to 29,016,300 | ctg163 | 2.44 | ||||
| PZA00980 | 5 (5.06) | SNP | 204,965,179 to 204,967,179 | 3.34 | |||||
| PZA01527 | 6 (6.01) | SNP | 58,422,170 to 58,431,307 | 2.40 | |||||
| PZA00473 | 6 (6.05) | SNP | 124,327,526 to 124,335,707 | 1.61 | |||||
| PZA02746 | 8 (8.06) | SNP | 163,286,695 to 163,294,127 | ctg362 | 2.26 | ||||
| LFR | umc1991 | 1 (1.08) | SSR | 245,317,331 to 245,319,777 | ctg50 | 7.24 | Mc37 × Zi330 F2:3 (162) | QTL | Li et al. 2010 [2] |
| umc2079 | 2 (2.04) | SSR | 57,593,899 to 57,596,593 | ctg79 | 7.01 | ||||
| mmc0401 | 2 (2.05) | SSR | 145,615,676 to 145,621,417 | ctg90 | 19.28 | ||||
| umc1759 | 4 (4.01) | SSR | 5,004,917 to 5,009,075 | ctg156 | 12.18 | ||||
| phi084 | 10 (10.04) | SSR | 87,282,919 to 87,285,844 | ctg406 | 15.27 | ||||
| HO | umc1635 | 2 (2.05) | SSR | 84,120,011 to 84,124,101 | ctg85 | 12.63 | |||
| phi033 | 9 (9.01) | SSR | 11,499,426 to 11,509,187 | ctg371 | 6.03 | ||||
| TL/HO | phi427913 | 1 (1.01) | SSR | 8,441,182 to 8,447,730 | ctg4 | 13.01 | |||
| umc1958 | 9 (9.01) | SSR | 11,778,648 to 11,782,406 | ctg371 | 6.54 | ||||
| LFR | bnlg1429–bnlg1016 | 1 (1.02–1.04) |
SSR | 16,117,043 to 16,123,450 56,158,913 to 56,164,047 |
ctg7 ctg14 |
12.80 | H21 × Mo17 F2:3 (120) | QTL | Yu 2003 [20] |
| phi008–phi085 | 5 (5.03–5.06) |
SSR | 14,083,193 to 14,086,595 205,292,914 to 205,298,428 |
ctg209 ctg251 |
12.80 | ||||
| umc1858–bnlg1176 | 8 (8.04–8.05) |
SSR | 112,055,118 to 112,059,831 121,984,323 to 121,999,950 |
ctg349 ctg4 |
35.10 | ||||
| HO | umc1466 | 4 (4.08) | SSR | 182,908,212 to 182,910,212 | ctg184 | 50.80 | |||
| umc1336 | 10 (10.04) | SSR | 86,430,405 to 86,433,004 | ctg406 | 51.80 | ||||
| TL | bnlg1434–bnlg1126 | 4 (4.01–4.03) |
SSR | 1,093,958 to 1,099,174 11,086,040 to 11,089,955 |
ctg154 ctg184 |
7.70 | |||
| bnlg1352–bnlg1031 | 8 (8.02–8.06) |
SSR | 11,570,168 to 11,575,230 164,821,348 to 164,827,624 |
ctg326 ctg363 |
11.00 | ||||
| TL/HO | umc1022–nc004 | 4 (4.01–4.03) |
SSR | 3,259,762 to 3,264,006 13,408,584 to 13,413,986 |
ctg155 ctg158 |
12.70 | |||
| LFR | umc1509–phi079 | 4 (4.02–4.05) |
SSR | 5,459,368 to 5,463,263 36,911,994 to 36,918,779 |
ctg156 ctg164 |
15.80 | Zi330 × K36 F2:3 (114) | QTL | Yu 2003 [20] |
| HO | bnlg147–umc2236 | 1 (1.02–1.06) |
SSR | 39,062,881 to 39,072,502 198,164,070 to 198,166,779 |
ctg11 ctg41 |
9.60 | |||
| mmc0081–phi128 | 5 (5.05–5.07) |
SSR | 172,138,316 to 172,143,312 210,153,373 to 210,156,706 |
ctg238 ctg253 |
12.10 | ||||
| umc1139–umc1627 | 8 (8.01–8.03) |
SSR | 2,989,632 to 3,048,953 78,519,415 to 78,523,602 |
ctg326 ctg344 |
15.90 | ||||
| TL | phi96100 | 2 (2.01) | SSR | 2,835,084 to 2,837,748 | ctg68 | 49.60 | |||
| umc1139–umc1627 | 8 (8.01–8.03) |
SSR | 2,989,632 to 3,048,953 78,519,415 to 78,523,602 |
ctg326 ctg344 |
15.80 | ||||
| phi033–umc1958 | 9 (9.01–9.01) |
SSR | 11,237,266 to 11,239,266 11,778,648 to 11,782,406 |
ctg371 ctg371 |
12.80 | ||||
| TL/HO | phi049–phi453121 | 3 (3.00–3.01) |
SSR | 1,728,270 to 1,730,270 1,627,384 to 1,635,027 |
ctg111 ctg114 |
8.80 |
TL, tunnel length of corn borer; AIR, aphid incidence rate; AIG, average aphid incidence grade; AR, aphid resistance; DIMBOA, DIMBOA content; LFR, leaf feeding rating of corn borer; HO, number of holes of corn borer; TL/HO, tunnel length/number of holes of corn borer; Chr., chromosome; SNP, single nucleotide polymorphisms; SSR, simple sequence repeat; R2, the phenotypic variation explained by identified genetic loci; RILs, recombinant inbred lines; GWAS, genome-wide association studies; QTL, quantitative trait loci; GLM, general linear model.
Figure 5.
Physical map construction, and the distribution of hot genetic loci (Loci, red dashed box), candidate genes, and original genetic loci for multiple insect-resistant traits including tunnel length of corn borer (TL), aphid incidence rate (AIR), average aphid incidence grade (AIG), aphid resistance (AR), DIMBOA content, leaf feeding rating of corn borer (LFR), number of holes of corn borer (HO), and tunnel length/number of holes of corn borer (TL/HO) in the physical map. The light green rectangles represent original genetic loci from previous studies. The blue rectangles represent our results. The blue markers represent the associated markers in our results.
Table 3.
The hot genetic loci of multiple insect-resistance and corresponding candidate genes in these hot genetic loci.
| Hot Loci | Bin Location | Candidate Gene ID | Encoded Protein | Gene Location (bp) | Orthologs |
|---|---|---|---|---|---|
| Loci 1 | 1.01–1.02 | GRMZM2G017616 (LOX9) | Lipoxygenase 9 | Bin 1.02 16,572,327 to 16,582,222 |
LOC_Os03g08220 (Oryza sativa) |
| Loci 2 | 1.04 | GRMZM2G147698 (MYB156) | MYB-transcription factor 156 | Bin 1.04 64,242,265 to 64,247,094 |
LOC_Os03g25550 (Oryza sativa) |
| GRMZM2G311036 (bx10) | DIMBOA-glucoside O-methyltransferase | Bin 1.04 66,309,137 to 66,314,243 |
Sb01g033880 (Sorghum bicolor) |
||
| GRMZM2G336824 (bx11) | DIMBOA-glucoside O-methyltransferase | Bin 1.04 66,391,909 to 66,397,349 |
Sb01g033880 (Sorghum bicolor) |
||
| GRMZM2G023325 (bx12) | DIMBOA-glucoside O-methyltransferase | Bin 1.04 66,504,957 to 66,508,971 |
Sb01g033880 (Sorghum bicolor) |
||
| Loci 3 | 1.11 | GRMZM2G046163 (lgl) | Indole-3-glycerol phosphate lyase | Bin 1.11 288,336,957 to 288,342,082 |
LOC_Os03g58290 (Oryza sativa) |
| Loci 4 | 2.00–2.01 | GRMZM2G102471 (uce4) | Ubiquitin-conjugating enzyme 4 | Bin 2.00 2,806,677 to 2,812,518 |
LOC_Os04g57220 (Oryza sativa) |
| GRMZM2G119941 (incw4) | Invertase cell wall 4 | Bin 2.01 3,188,791 to 3,195,146 |
LOC_Os04g56920 (Oryza sativa) | ||
| GRMZM2G018716 (incw7) | Invertase cell wall 7 | Bin 2.01 3,227,856 to 3,233,490 |
LOC_Os04g56930 (Oryza sativa) |
||
| GRMZM2G018692 (incw6) | Invertase cell wall 6 | Bin 2.01 3,232,233 to 3,237,644 |
LOC_Os04g56920 (Oryza sativa) |
||
| GRMZM2G365166 (SAUR14) | Auxin-responsive protein SAUR36 | Bin 2.01 3,590,872 to 3,594,536 |
LOC_Os04g56690 (Oryza sativa) | ||
| GRMZM2G365162 (SAUR15) | Indole-3-acetic acid-induced protein ARG7 | Bin 2.01 3,600,982 to 3,605,199 |
LOC_Os04g56680 (Oryza sativa) |
||
| GRMZM2G040095 (LOX6) | Lipoxygenase 6 | Bin 2.02 4,190,652 to 4,197,763 |
Sb06g031350 (Sorghum bicolor) |
||
| Loci 5 | 2.03–2.04 | GRMZM2G123202 (MYB54) | MYB-transcription factor 54 | Bin 2.03 23,620,411 to 23,625,163 |
LOC_Os04g45060 (Oryza sativa) |
| GRMZM2G089806 (SAUR20) | Auxin-induced protein X10A | Bin 2.04 28,768,661 to 28,772,516 |
LOC_Os04g43740 (Oryza sativa) |
||
| Loci 6 | 2.04 | GRMZM2G130224 | Restriction endonuclease type II-like superfamily protein | Bin 2.04 37,078,026 to 37,083,625 |
LOC_Os04g40900 (Oryza sativa) |
| Loci 7 | 2.05 | GRMZM2G504910 | Tetratricopeptide repeat protein 27 homolog | Bin 2.06 168,638,340 to 168,652,992 |
LOC_Os07g27180 (Oryza sativa) |
| Loci 8 | 4.00 –4.03 | GRMZM6G617209 (bx6) | 2-oxoglutarate-dependent dioxygenase | Bin 4.00 1,251,138 to 1,255,544 |
LOC_Os03g48430.1 (Oryza sativa) |
| GRMZM2G167549 (bx3) | Indolin-2-one monooxygenase | Bin 4.01 3,001,662 to 3,007,432 |
Sb05g026080 (Sorghum bicolor) |
||
| GRMZM2G172491 (bx4) | 3-hydroxy-indolin-2-one monoxygenase (P450) | Bin 4.01 3,049,715 to 3,055,536 |
Sb05g026080 (Sorghum bicolor) |
||
| GRMZM2G063756 (bx5) | BHBOA monoxgenase (P450) | Bin 4.01 3,111,425 to 3,117,001 |
LOC_Os08g01510.1 (Oryza sativa) |
||
| GRMZM2G085054 (bx8) | 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3 (4H)-one 2-D-glucosyltransferase | Bin 4.01 3,213,147 to 3,218,058 |
LOC_Os11g25454.1 (Oryza sativa) |
||
| GRMZM2G085381 (bx1) | Indole-3-glycerol phosphate lyase | Bin 4.01 3,259,262 to 3,264,506 |
LOC_Os03g58300.1 (Oryza sativa) |
||
| GRMZM2G085661 (bx2) | Indole-2-monooxygenase | Bin 4.01 3,263,713 to 3,268,868 |
Sb05g026080 (Sorghum bicolor) |
||
| Loci 9 | 4.04 | GRMZM2G441753 (bx7) | TRIBOA-glc O methyl transferase | Bin 4.04 18,243,663 to 18,246,553 |
LOC_Os12g25450.1 (Oryza sativa) |
| Loci 10 | 4.05 | GRMZM2G157306 (MYB92) | MYB-related-transcription factor 92 | Bin 4.05 34,497,413 to 34,505,954 |
LOC_Os08g05510 (Oryza sativa) |
| Loci 11 | 4.08 | Zm00001d052335 (POD23) | Peroxidase 23 | Bin 4.08 187,346,362 to 187,351,257 |
|
| Loci 12 | 5.03 | GRMZM2G102760 (LOX5) | Lipoxygenase 5 | Bin 5.03 12,284,156 to 12,292,064 |
LOC_Os03g49380 (Oryza sativa) |
| GRMZM2G332390 (SAUR48) | Auxin-responsive SAUR family member | Bin 5.03 15,029,401 to 15,033,175 |
LOC_Os03g45850 (Oryza sativa) |
||
| GRMZM2G113135 (SAUR49) |
Auxin-responsive SAUR family member | Bin 5.03 60,953,699 to 60,958,116 |
LOC_Os10g36703 (Oryza sativa) |
||
| GRMZM2G330012 (SAUR50) | Auxin-responsive protein SAUR19 | Bin 5.03 56,965,982 to 56,969,995 |
LOC_Os06g48860 (Oryza sativa) |
||
| GRMZM2G361993 (SAUR51) | Auxin-responsive protein SAUR32 | Bin 5.03 60,953,699 to 60,958,116 |
LOC_Os06g50040 (Oryza sativa) |
||
| Loci 13 | 5.05–5.07 | GRMZM2G139300 (incw1) | Invertase cell wall 1 | Bin 5.04 169,496,097 to 169,503,589 |
LOC_Os02g33110 (Oryza sativa) |
| GRMZM2G159547 (MYB48) | MYB-transcription factor 48 | Bin 5.07 208,396,628 to 208,401,248 |
LOC_Os02g51799 (Oryza sativa) |
||
| Loci 14 | 6.02 | GRMZM2G150256 (mir2) | Maize insect resistance 2-cysteine protease | Bin 6.02 89,064,767 to 89,072,691 |
Sb10g028000 (Sorghum bicolor) |
| GRMZM2G150276 (mir1) | Maize insect resistance 1-cysteine protease | Bin 6.02 89,070,747 to 89,075,683 |
Sb10g028010 (Sorghum bicolor) |
||
| GRMZM2G423833 (MYB115) | MYB-transcription factor 115 | Bin 6.02 89,299,116 to 89,305,100 |
LOC_Os06g40330.1 (Oryza sativa) |
||
| GRMZM2G093789 (MYB59) | MYB-transcription factor 59 | Bin 6.02 89,401,621 to 89,417,741 |
LOC_Os06g40330.1 (Oryza sativa) |
||
| Loci 15 | 8.01–8.03 | GRMZM2G312419 (MYB60) | MYB-transcription factor 60 | Bin 8.01 2,760,493 to 2,764,284 |
LOC_Os01g16810 (Oryza sativa) |
| GRMZM2G096358 (MYB22) | MYB-transcription factor 22 | Bin 8.02 11,485,302 to 11,489,135 |
LOC_Os01g03720 (Oryza sativa) |
||
| GRMZM2G143274 (MYB76) | MYB-transcription factor 76 | Bin 8.02 16,771,384 to 16,774,583 |
LOC_Os01g07430 (Oryza sativa) |
||
| GRMZM2G119693 (MYB91) | MYB-transcription factor 91 | Bin 8.03 63,067,162 to 63,070,647 |
LOC_Os05g46610 (Oryza sativa) |
||
| Loci 16 | 8.04–8.05 | GRMZM2G179679 | Sugars will eventually be exported transporter 3a | Bin 8.04 112,055,118 to 112,059,831 |
LOC_Os05g12320 (Oryza sativa) |
| GRMZM2G171781 (MYB30) | MYB-transcription factor 30 | Bin 8.05 128,604,416 to 128,609,501 |
LOC_Os05g04820 (Oryza sativa) |
||
| Loci 17 | 8.06 | GRMZM2G431066 (SAUR70) | Auxin-responsive protein SAUR50 | Bin 8.06 164,748,431 to 164,752,132 |
|
| Loci 18 | 9.01 | GRMZM2G451533 (SAUR71) | SAUR-like auxin-responsive protein family | Bin 9.01 11,435,786 to 11,439,677 |
LOC_Os06g09550 (Oryza sativa) |
| Loci 19 | 10.04 | Zm00001d024752 (POD21) | Peroxidase 21 | Bin 10.04 86,134,891 to 86,138,913 |
|
| GRMZM2G127490 (MYB149) | MYB-transcription factor 149 | Bin 10.04 87,799,992 to 87,806,148 |
LOC_Os05g04820 (Oryza sativa) |
||
| GRMZM2G123633 (incw3) | Invertase cell wall 3 | Bin 10.04 114,307,030 to 114,312,422 |
LOC_Os04g33720 (Oryza sativa) |
2.6. Expression Levels of Five Candidate Genes Responsible for DIMBOA Biosynthesis
We randomly selected 5 of the 12 candidate genes that control DIMBOA biosynthesis, including GRMZM2G085381 (bx1), GRMZM6G617209 (bx6), GRMZM2G441753 (bx7), GRMZM2G311036 (bx10), and GRMZM2G336824 (bx11) to examine their relative expression levels in maize seedlings of RX20-1006 (high insect-resistant line) and T58 (insect-susceptible line) in the E1 environment at the V6 stage using real-time PCR (RT-qPCR). The results showed that the relative expression levels of the five genes were significantly correlated with the DIMBOA content in the two maize genotypes (Figure 6A–C).
Figure 6.
The relative expression level of five candidate genes involved in DIMBOA biosynthesis, DIMBOA content observation and their correlation in maize seedlings of RX20-1006 (high insect-resistant line) and T58 (insect-susceptible line) in Zhangye (E1) environment, Gansu Province, China. (A) The relative expression level (mean ± standard deviation) of the five candidate genes; different lowercase letters indicate a significant difference (p < 0.05) in gene expression. (B) DIMBOA content (mean ± standard deviation) in high insect-resistant (RX20-1006) and insect-susceptible line (T58); different lowercase letters indicate a significance difference (p < 0.05) in DIMBOA content. (C) The correlational relationships between the relative expression level of five candidate genes and DIMBOA content; lines represent significant correlations (p < 0.05).
3. Discussion
3.1. Defense Strategies of Maize against ACB-/CLA-Feeding
It is well known that the plethora of ACB and CLA that either simultaneously or concurrently attack multiple maize parts, such as newly hatched ACB feeds on whorl leaves and later instars, tunnel into the stalk or the ear to feed on pith tissues or fresh kernels [25], resulting in a reduction of photosynthetic property, disruption of nutrient and water transport, an increase of stalk lodging and bacterial/fungal infections [25,26], and ultimately complicates harvesting practices and reduces grain yield and quality [27,28]. In addition, CLA is a phloem sap-sucking pest [29], and it can absorb nutrients from phloem sap and alter source-sink patterns [30]; its digestive waste products, i.e., honeydew, can also deposit on the leaf surface of maize and promote mold growth [6]. CLA is also a vector for some plant viral diseases that facilitate pathogen entry and cause maize leaves to curl, discolor, and wilt after serious CLA infestations [6].
In response to insect attack, maize has evolved an extensive array of defense strategies to prevent ACB or CLA feeding and colonization. Increasing evidence has verified that maize-derived compounds bxs, e.g., 2,4-dihydroxy-7-1,4-benzoxazin-3-one glucoside (DIMBOA-Glc), DIMBOA, and 6-methoxy-3h-1,3-benzoxazol-ne (MBOA) are multifunctional defense metabolites that can protect maize against insect pests feeding and pathogens [31,32,33]. DIMBOA acts as a feeding deterrent in maize that can decrease in vivo endoproteinase activity in the larval midgut of the European corn borer, thus limiting the availability of amino acids, reducing larval growth [34], influencing some nervous system and detoxification, and inactivating some hydrolysis enzymes of ACB larvae [35]. Infiltration of DIMBOA into maize leaves stimulated callose accumulation and elevated maize CLA resistance [29]. At 60 h after the 1st instar larvae of Sesamia nonagrwides infestation, the leaves of infested maize were injured, with a significant increase in leaf DIMBOA content of 42–96% [36]. Indeed, these studies indicated that bxs metabolites, especially DIMBOA were involved in maize resistance of corn borer and aphid, which can be a good indicator for screening multiple insect-resistant maize genotypes. Results presented here, together with previous findings [29,34,35], showed that both accumulation and catabolism of DIMBOA varied greatly among maize inbred lines, which may contribute to the resistance of borer and aphid in maize (Figure 1A–C and Figure 3C). Meihls et al. [16] also reported that DIMBOA content was highly correlated with its precursor DIMBOA-Glc abundance, and these endogenous compounds potentially built up maize’s resistance to insect pests. Dafoe et al. [37] further found that jasmonic acid and ethylene were produced rapidly in response to corn borer feeding, and their induction differentially regulated bxs in maize stems; even other phytohormones, i.e., salicylic acid and indole-3-acetic acid, generally considered antagonists of jasmonic acid signaling, were also involved in regulating defense responses [38].
3.2. Genetic Loci Comparison between DIMBOA Content and Multiple Insect-Resistant Traits, and Their Hot Loci Identification
Understanding the genetic basis of the multiple insect resistance of maize is critical to the control of combinatorial attacks of ACB and CLA in the field. In this study, we observed a key trait for multiple insect resistance, i.e., the DIMBOA content in two ecological environments and their BLUP values, to detect nine significant associated SSR markers using association mapping via GLM and MLM across 310 diverse maize inbred lines from Gansu Province, China; these nine SSR markers were located in Bin 1.04, Bin 1.11, Bin 2.01, Bin 4.00, Bin 4.01, Bin 6.02, Bin 8.04, and Bin 10.04, respectively (Figure 4; Table 1). Using the same method, Butrón et al. [15] also identified eight linked single nucleotide polymorphisms (SNP) markers related to DIMBOA content across 281 genetically diverse inbred lines located in Bin 1.04, Bin 2.04, Bin 4.01, Bin 4.04, Bin 5.06, Bin 6.01, Bin 6.05, and Bin 8.06, respectively (Table 2). They also mapped 12 and 7 QTLs associated with DIMBOA/DIMBOA-Glc/DIMBOA-T content in both B73 × CML322 recombinant inbred lines (RILs) and B73 × IL14H RILs populations, respectively; these QTLs were distributed on chromosomes 1, 3, 4, 6, 7, and 8, respectively [15]. These findings demonstrate that there are multiple genetic loci involved in DIMBOA biosynthesis and decomposition on nine chromosomes except for Chromosome 9. Because of their clear differences in genetic effects and phenotypic variance in these loci, there is a great potential to obtain different multiple insect-resistant lines (materials) based on MAS application. In addition, we further combined the genetic loci from previous foliar studies and our current results of DIMBOA content as well as seven other multiple insect-resistant traits (Table 1 and Table 2) to gain a better understanding of hot loci in maize resistance to multiple insect pests and to explore avenues for multiple insect-resistance breeding.
Interestingly, we identified 19 hot loci (Loci 1-Loci 19) involved in maize multiple insect resistance in the present study (Figure 5; Table 3). Of these, Loci 2 in Bin 1.04 (bnlg147-umc1917 interval) is involved in LFR, HO, AR, and DIMBOA content; Loci 4 in Bin 2.00–2.01 (rs624256-umc2363 interval) controls TL and DIMBOA content; Loci 5 in Bin 2.03–2.04 (rs650025-rs65 interval) is responsible for TL and LFR; Loci 8 in Bin 4.00–4.03 (phi072-umc1509 interval) is related to TL, LFR, AR, AIR, AIG, TL/HO, and DIMBOA content; Loci 12 in Bin 5.03 (phi008-umc1935 interval) is associated with AIG and LFR; Loci 13 in Bin 5.05–5.07 (mmc0081-phi128 interval) is associated with LFR, HO, and DIMBOA content; Loci 15 in Bin 8.01–8.03 (umc1139-umc1627 interval) regulates HO and TL; Loci 16 in Bin 8.04–8.05 (umc1858-bnlg1176 interval) is responsible for TL, LFR, and DIMBOA content; Loci 17 in Bin 8.06 (PZA02746-bnlg1031 interval) is related to TL and DIMBOA content; Loci 18 in Bin 9.01 (phi033-umc1958 interval) is involved in TL, HO, and TL/HO; and Loci 19 in Bin 10.04 (umc1336-umc1054 interval) is associated with LFR, HO, and DIMBOA content. Thus, these 11 hot loci have a pleiotropic effect on 2 to 7 multiple insect-resistant traits, and Bin 1.04, Bin 2.00–2.01, Bin 4.00–4.03, Bin 5.05–5.07, Bin 8.04–8.05, Bin 8.06, and Bin 10.04 regions play important roles in conferring DIMBOA accumulation and other aspects of maize multiple-insect resistance to ACB and CLA. Consistent with previous findings, LFR was significantly correlated with HO (phenotypic correlation coefficient (r); r = 0.252) and TL/HO (r = 0.229) in 162 F3 maize population to ACB resistance [2]. Meanwhile, 3 of 19 hot loci, i.e., Loci 2, Loci 8, and Loci 12 are co-involved in multiple insect resistance to both ACE and CLA; thus, we speculate that the resistance to ACB and CLA for leaf feeding damage is partially controlled by the same mechanisms in these three hot loci. For future research, the contribution of Loci 2, Loci 8, and Loci 12, as well as their functional genes must be examined when developing elite maize varieties with multiple insect resistance to ACE and CLA.
3.3. Validation of Candidate Genes in Hot Loci
According to the physical interval of the above 19 hot loci controlling 8 insect-resistant traits and the GO annotations of corresponding genes in these hot intervals, a total of 49 candidate genes were identified (Table 3 and Table S3); they may play important roles in maize multiple insect-resistance.
The 12 candidate genes were identified within three hot loci; namely, GRMZM2G311036 (bx10), GRMZM2G336824 (bx11), and GRMZM2G023325 (bx12) were detected in Loci 1 (Bin 1.04), and they encoded DIMBOA-glucoside O-methyltransferase; GRMZM2G046163 (lgl; encoded indole-3-glycerol phosphate lyase) was mapped in Loci 3 (Bin 1.11); GRMZM2G167549 (bx3; encoded indolin-2-one monooxygenase), GRMZM2G172491 (bx4; encoded 3-hydroxy-indolin-2-one monoxygenase (P450)), GRMZM2G063756 (bx5; encoded BHBOA monoxgenase (P450)), GRMZM6G617209 (bx6; encoded 2-oxoglutarate-dependent dioxygenase), GRMZM2G085054 (bx8; encoded 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3 (4H)-one 2-D-glucosyltransferase), GRMZM2G085381 (bx1; encoded indole-3-glycerol phosphate lyase), and GRMZM2G085661 (bx2; encoded indole-2-monooxygenase) were identified in Loci 8 (Bin 4.00–4.03); and GRMZM2G441753 (bx7; encoded 2,4,7-trihydroxy-2H-1,4-benzoxazin-3(4H)-one (TRIBOA-Glc) O methyl transferase) was located in Loci 9 (Bin 4.04) (Table 3). The lgl and bx1 also displayed similar enzyme functions; lgl catalyzed the formation of free indole and was selectively activated by volicitin in the saliva of lepidopterous larvae [15,39,40]. Then, the enzyme actions of four maize cytochrome P450-dependent monooxygenases (bx2, bx3, bx4, and bx5) converted free indole to 2,4-dihydroxy-1,4-benoxazin-3-one (DIBOA) [15]. The bx1 was a modified form of the tryptophan synthase alpha subunit, and it was expressed constitutively in young seedlings, while igl was induced in more advanced stages of plant development and contributed to the blend of odors that attracted beneficial parasitoids [15,39,40]. The next step in the bxs biosynthesis pathway was the conversion of DIBOA to DIBOA-Glc by the action of specific gluocosyltransferases. The bx8 and GRMZM2G161335 (bx9) were involved in the glucosylation of DIBOA [41], and the conversion of DIBOA to DIMBOA-Glc required hydroxylation and methylation. The bx6 was responsible for the hydroxylation step that converted DIBOA-Glc to TRIBOA-Glc, and this conversion likely took place in the cytosol [42,43]. Methylation was catalyzed by bx7, making DIMBOA-Glc [15]. In addition, the bx10, bx11, and bx12 were likely candidates for catalyzing the conversion of DIMBOA-Glc to HDMBOA-Glc [16]. These findings also supported the results of the relative expression levels of five candidate genes that controlled DIMBOA biosynthesis; their expression patterns were positively and significantly correlated with the DIMBOA accumulation (Figure 6A–C). Therefore, these candidate genes in Bin 1.04, Bin 1.11, and Bin 4.00–4.03 regions were involved in DIMBOA biosynthesis, and the four associated SSR markers (umc1917, umc1008, umc1017, and umc1758) can be used to distinguish DIMBOA accumulation in maize.
The two candidate genes, GRMZM2G150256 (mir2) and GRMZM2G150276 (mir1) were mapped within Loci 14 and encoded a maize insect resistance-cysteine protease (key defensive protein) against chewing insect pests in maize (Table 3). The synthetic diet aphid feeding trial bioassays with recombinant mir1-cysteine protease demonstrated that mir1-Cysteine protease triggered direct toxicity to CLA [44], and ethylene acted as a central node in regulating mir1 expression to different feeding guilds of insect herbivores [44].
Other phytohormones may also be responsive to multiple insect-resistance in maize. A total of nine auxin-responsive SAUR family member genes were detected within Loci 4 (Bin 2.00–2.01), Loci 5 (Bin 2.03–2.04), Loci 12 (Bin 5.03), Loci 17 (Bin 8.06), and Loci 18 (Bin 9.01), respectively (Table 3). Similarly, two auxin-responsive SAUR family member genes that closely related to ACB-resistance were detected on chromosome 2 (near rs653464 and rs649775 marker, respectively) based on genome-wide association study (GWAS) [19]. Recent characterization of European corn borer attack on maize stems showed a rapid and sustained accumulation of indole-3-acetic acid and jasmonic acid in damaged tissues [38].
Lipoxygenase (LOX) plays critical roles in plant defense against multiple insect pests and pathogens [1,45,46]; and, consistent with LOX’s function, we also found three LOX genes, i.e., GRMZM2G017616 (LOX9) within Loci 1 (Bin 1.01–1.02), GRMZM2G040095 (LOX6) within Loci 4 (Bin 2.00–2.01), and GRMZM2G102760 (LOX5) within Loci 12 (Bin 5.03) (Table 3). LOX6 was strongly induced by jasmonic acid and the fungal pathogen Cochliobolus carbonum in maize [47].
The MYB transcription factor can interact with mRNA/proteins to form a fine regulatory network to activate the expression of downstream defense genes and induce insect-resistance defense response. Interestingly, the previous findings [19] supported our results, i.e., 12 MYB genes were validated within Loci 2 (Bin 1.04), Loci 5 (Bin 2.03–2.04), Loci 10 (Bin 4.05), Loci 13 (Bin 5.05–5.07), Loci 14 (Bin 6.02), Loci 15 (Bin 8.01–8.03), Loci 16 (Bin 8.04–8.05), and Loci 19 (Bin 10.04) in this study, respectively (Table 3). Thus, these MYBs may be involved in the defense response to ACB and CLA, and further studies are needed to explore the downstream target genes of MYB in herbivore-induced resistance.
Cell wall invertases (incw) catalyze the irreversible hydrolysis of sucrose into glucose and fructose and play important roles in sucrose partitioning, plant development, and defense responses to biotic stresses [48]. The hydrolysis of cell wall polysaccharides during pathogen infection generates sugar signaling to stimulate intracellular defense response [49]. The five incw genes, i.e., GRMZM2G119941 (incw4), GRMZM2G018716 (incw7), and GRMZM2G018692 (incw6) within Loci 4 (Bin 2.00–2.01), GRMZM2G139300 (incw1) within Loci 13 (Bin 5.05–5.07), and GRMZM2G123633 (incw3) within Loci 19 (Bin 10.04) were identified in this study (Table 3). Essmann et al. [50] reported that RNA interference-mediated repression of an incw gene resulted in reduced defense of transgenic tobacco plants. Moreover, we also detected GRMZM2G179679 encoding SWEET 3a (Sugars Will Eventually be Exported Transporter 3a) within Loci 16 (Bin 8.04–8.05) (Table 3). Thereby, sugar metabolism/homeostasis may play a positive role in maize defense response to ACB and CLA.
Plant POD participates in multiple physiological processes, such as auxin metabolism [51], lignin biosynthesis [52,53], and tolerance against osmotic stress [54]. Additionally, reactive oxygen species (ROS), especially H2O2 induces POD activity, which then oxidizes and polymerizes p-coumaryl/coniferyl-/sinapyl-alcohol into lignin monomers on the cell wall [52]; thereby, POD is involved in the loosening and stiffening of the cell wall during plant development. Moreover, López-Castillo et al. [55] reported that ZmPrx35 as the prevailing POD was involved in defense against pathogens and insects. Similarly, we also identified Zm00001d024752 (POD21) and Zm00001d052335 (POD23) within Loci 19 (Bin 10.04) and Loci 11 (Bin 4.08), respectively (Table 3). Therefore, we speculate that host maize can limit food supplies to multiple insect pests and against larval boring via a key physical barrier, such as cell wall rigidity in the pre-ingestion phase.
In addition, three other candidate genes, i.e., GRMZM2G102471 (uce4) responsible for ubiquitin-conjugating enzyme 4, RMZM2G130224 responsible for restriction endonuclease type II-like superfamily protein, and GRMZM2G504910 responsible for tetratricopeptide repeat protein 27 homolog, were identified within Loci 4 (Bin 2.00–2.01), Loci 6 (Bin 2.04), and Loci 7 (Bin 2.05) in the present study, respectively (Table 3). Interestingly, the GRMZM2G130225 (near rs658849) and GRMZM2G102471 (near rs624256) were also identified to be involved in TL of ACB using GWAS analysis [19].
In summary, according to the above studies, a possible molecular network underlying maize multiple insect-resistance to ACB and CLA was constructed (Figure 7), which could benefit the development of new maize varieties with multiple insect resistance.
Figure 7.
Molecular defense mechanisms underlying resistance to multiple insect pests of Asian corn borer (ACB) and corn leaf aphid (CLA) in maize. The synergy and antagonism of 49 candidate genes within 19 hot genetic loci formed the complex maize multiple insect pest-induced defense mechanisms, as well as DIMBOA biosynthesis pathway network.
3.4. Germplasms Diversity and Multiple Insect-Resistance Evaluation
When assessing the genetic diversity in maize genotypes, SSR remains the preferred choice due to their co-dominant and multi-allelic nature, abundance, and loci specificity [23]. In the current study, 748 alleles with a range of 2 to 8 per locus were identified among 310 maize inbred lines using 186 polymorphic SSRs (Table S1). The finding showed a wide range of diversity among genotypes [23], which will benefit broadening the genetic base in any breeding program. The average PIC value of all SSR markers was 0.543 (range from 0.294 to 0.826) in our result (Table S1). The data demonstrates the presence of many informative allelic variations in this maize population [22,23]. Moreover, considering the membership probability of ≥ 0.500, the population structure further indicated that the 294 inbred lines were divided into 5 optimal groups (Table S2); this result was consistent with the findings of Liu et al. [22]. Further, we comprehensively evaluated the multiple insect resistance of 310 inbred lines in 2 ecological environments, and screened 3 high and 15 moderate insect-resistant germplasms, which were mainly in the Reid group (accounting for 50.00%) (Figure 3B–C). These findings have laid a foundation for the improvement of insect-resistant maize varieties in the future.
4. Materials and Methods
4.1. Plant Materials
In this study, the collected 310 elite maize inbred lines from different ecological environments (Zhangye, 216 lines; Longxi, 64 lines, Jingtai, 21 lines; Pingliang, nine lines) in Gansu Province, China (Table S2). A total of 20 seeds of these germplasms per row were randomly planted in 4.0 m rows, 0.2 m aisles, and 0.5 m between rows on April 16 in Zhangye (E1; 38.83° N, 106.93° E, 1536 m altitude) and in Longxi (E2; 34.97° N, 104.40° E, 2074 m altitude), Gansu Province, China in 2020. Before sowing, the soil surface was covered with plastic film (0.08 mm thick, 1.2 m wide). At the V6 stage, the 6th leaf of each inbred line in the E1 and E2 environments was collected, frozen in liquid nitrogen, and stored at –70 °C for subsequent DNA extraction and DIMBOA content assay.
4.2. DIMBOA Content Assay
The DIMBOA content was determined using high-performance liquid chromatography (HPLC; Shimadzu LCMS8040 system, Beijing, China). Namely, freeze-dried leaves (0.2 g per sample) were homogenized and weighted into screw-capped 10 mL polypropylene centrifuge tubes, and 5 mL of HPLC grade methanol-methanoic acid solution (0.01%, v/v) was added to each tube. The tubes were rotated and placed in the dark for 12 h and then centrifuged at 12,000 rpm (Centrifuge 5425/5425 R; Eppendorf, Hamburg, Germany) for 20 min at 4 °C. Supernatants (600 μL) were slowly passed the corresponding Millex® needle filter and transferred into auto-sample vials for analysis by HPLC. Standard DIMBOA (CAS No.: 15893-52-4) was purchased (Sigma-Aldrich, MA, USA) and was used to optimize the mass spectrometric parameters and fragment spectra.
4.3. Genetic Diversity Analysis and Marker-DIMBOA Content Association Mapping
Genomic DNA was extracted from the 6th leaf of 310 inbred lines using the cetyltrimethyl ammonium bromide (CTAB) method [56]. Then, a total of 186 polymorphic SSR markers covering the entire maize genome from the MaizeGDB (http://www.maizegdb.org/ (accessed on 10 January 2020)) were used to perform SSR analysis. Fragments were separated using polyacrylamide gel electrophoresis. The polymorphism information content (PIC) value and Shannon-Wiener’s index (I) value were determined as follows [23]:
| (1) |
| (2) |
where was the i allele frequency. Allele identity was used for an unweighted pair-group method with arithmetic means (UPGMA) cluster analysis. Population structure [57] was analyzed using STRUCTURE v. 2.3.3 software (http://web.stanford.edu/group/pritchardlab/structure_software/release_versions/v2.3.4/html/structure.html (accessed on 10 August 2022)) for the assessment of groups and genetic relationships among the 310 maize inbred lines. The project was run with the set parameters of the population admixture model, and the allele frequency correlated. The optimum group number was determined by ΔK value [58]. A linkage map was developed according to the genetic distance (centimorgan, cM) of corresponding SSR markers on an IBM2 2008 Neighbors map frame (https://www.maizegdb.org/data_center/map (accessed on 18 September 2022)) using BioMercator v. 4.2 software (http://www.bioinformatics.org/mqtl/wiki/ (accessed on 18 September 2022)). The linkage disequilibrium (LD) values for r2 [59] and D′ [57] between SSR loci on chromosomes were calculated using Tassel 3.0 software (https://tassel.bitbucket.io/ (accessed on 10 August 2022)), following a permutation test of 10,000. The K matrix and marker-DIMBOA content association mapping was completed in Tassel 3.0 software using the genotypic data of 186 polymorphic SSRs and phenotypic data on DIMBOA content for a set of 310 inbred lines. The association analysis was conducted by a GLM with Q matrix (individuals’ probability of membership in the population) [60] and an MLM with a K + Q matrix [61]. The associated SSRs for DIMBOA content were filtered out based on the phenotypic variance (R2) of the marker at p < 0.01 level and with the lowest false discovery rate (FDR). Using MaizeGDB (http://www.maizegdb.org/ (accessed on 20 September 2022)) and nucleotide and primer blast tools, the physical locations of associated SSR markers for DIMBOA content were determined on chromosomes.
4.4. Hot Genetic Loci and Candidate Genes Detection
Using public databases, i.e., MaizeGDB (http://www.maizegdb.org/ (accessed on 23 September 2022)), NCBI (http://www.ncbi.nlm.nih.gov (accessed on 23 September 2022)), and CNKI (http://www.cnki.net (accessed on 23 September 2022)), we collected information on corresponding QTLs and associated markers for multiple insect-resistant traits from our results and previous studies via QTL analysis, GWAS, and association mapping. The hot genetic loci were the overlapping regions combining multiple genetic loci responsible for multiple insect-resistant traits or single genetic loci that explained the large R2 (10%) in 10 Mb physical intervals. Further, the physical map of all hot genetic loci and candidate genes involving multiple insect-resistant traits in these hot genetic loci regions was developed by BioMercator v. 4.2 software (http://www.bioinformatics.org/mqtl/wiki/ (accessed on 28 September 2022)) [62]. The functional annotation of corresponding candidate genes was performed using the tool AgBase v2.00 (https://agbase.arizona.edu/ (accessed on 6 October2022)) [63].
4.5. RT-qPCR Analysis
Among 310 maize inbred lines, we selected the two inbred lines with the largest difference in multiple insect resistance, i.e., RX20-1006 with the highest DIMBOA content and T58 with the lowest DIMBOA content. Their total RNAs were extracted from the 6th leaf with TRIZOL reagent (Invitrogen, USA), and cDNA was made using a kit (Starscript II First-stand cDNA Synthesis Mix With gDNA Remover, GenStar, Beijing, China), according to the manufacturer’s instructions. RT-PCR was conducted using TransStart Tip Green qPCR SuperMix (Tran, Beijing, China). The primers for five candidate genes [12,64] were designed using Primer Premier 5.0 software (http://www.premierbiosoft.com/ (accessed on 9 October 2022)) (Table S4). Relative gene expression levels were assessed by the 2−∆∆Ct method, with GRMZM2G126010 as an internal reference gene [63].
4.6. Statistical Analysis
The average DIMBOA content in 310 inbred lines from five biological replicates in each ecological environment were analyzed, respectively. A mixed linear model was fitted using the lmer function in lme4 package of R (http://www.R-project.org/ (accessed on 16 December 2021)) to calculate the BLUP of DIMBOA content values [65]. These data were then compared statistically using IBM-SPSS Statistics v. 19.0 (SPSS Inc., Chicago, IL, USA; http://www.ibm.com/products/spss-statistics (accessed on 16 December 2021)). The significance of the total and residual variances of DIMBOA content in 310 inbred lines under both ecological environments was estimated by a GLM for univariate data and by one-way analysis of variance (ANOVA). The broad-sense heritability () and genotype × environment interaction heritability () values for DIMBOA content under both environments were estimated as follows [53,66,67]:
| HB2 = σg2/(σg2 + σge2/n + σε2/nr), | (3) |
| HGE2 = (σg2/n)/(σg2 + σge2/n + σε2/nr) | (4) |
where σg2 was the genotypic variance, σe2 was the environmental variance, σε2 was the error variance, σge2 was the variance of genotype × environment interaction, n was the number of ecological environments (n = 2), and r was the number of replications (r = 5). The CVg [63] of DIMBOA content among all inbred lines under each environment was calculated as follows:
| (5) |
where was the average value of DIMBOA content among all inbred lines in each environment, and δ was the standard deviation.
5. Conclusions
In summary, the DIMBOA confers significant resistance to ACB and CLA. In this study, SSR analysis revealed a wide genetic diversity in the 310 tested maize inbred lines from four type regions of China’s Gansu Province, which is the largest maize seed production and breeding area in China. Population structure indicated that 294 inbred lines were successfully assigned to one or another group at a membership probability of ≥ 0.500. DIMBOA performance evaluation screened out 3 high and 15 moderate insect-resistant inbred lines, which can be used as parents in breeding programs to develop new maize varieties with multiple insect resistance. Using linkage mapping, we detected nine significant SSRs associated with DIMBOA content in both environments. We then combined the 47 original genetic loci for 8 multiple insect-resistant traits from previous studies to detect 19 hot loci. Among them, 11 hot loci were located in Bin 1.04, Bin 2.00–2.01, Bin 2.03–2.04, Bin 4.00–4.03, Bin 5.03, Bin 5.05–5.07, Bin 8.01–8.03, Bin 8.04–8.05, Bin 8.06, Bin 9.01, and Bin 10.04 regions, and they supported pleiotropy for their association with two or more insect-resistant traits. Further, the 49 candidate genes involved in DIMBOA biosynthesis, sugar metabolism/homeostasis, and other multiple insect-resistant defense mechanisms in maize were identified in all 19 hot loci, and their highly interconnected network may form complex maize, multiple insect, pest-induced defense mechanisms.
Acknowledgments
This work was supported in part by the U.S. Department of Agriculture, Agricultural Research Service through project 3060-21220-033-00D. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Abbreviations
| ACB | Asian corn borer |
| AIG | Average aphid incidence grade |
| AIR | Aphid incidence rate |
| AG | Aphid resistance |
| BLUP | The best linear unbiased prediction |
| bxs | Benzoxazinoids |
| CLA | Corn leaf aphid |
| DIMBOA | 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one |
| GLM | General linear model |
| GWAS | Genome-wide association studies |
| HO | Number of holes of corn borer |
| I | Shannon-Wiener’s index |
| LD | Linkage disequilibrium |
| LFR | Leaf feeding rating of corn borer |
| MLM | Mixed linear model |
| MAS | Marker-assisted selection |
| PIC | Polymorphism information content |
| QTL | Quantitative trait locus |
| TL | Tunnel length of corn borer |
| TL/HO | Tunnel length/number of holes of corn borer |
| SNP | Single-nucleotide polymorphism |
| SSRs | Simple sequence repeats |
Supplementary Materials
The following supporting information can be downloaded from https://www.mdpi.com/article/10.3390/ijms24032138/s1. References [12,65,66] are cited in the supplementary materials.
Author Contributions
Conceptualization, X.Z. and Y.N.; methodology, X.Z. and Y.N.; resources, Y.N., X.Z., X.B. and T.M.; software, X.Z. and P.L.; writing—review and editing, Y.N., X.Z., and W.C.; project administration, X.Z. and Y.N. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The research was funded by the Major Scientific and Technological Special Project of Gansu, China (22ZD6NA010), the Gansu Province Science Foundation for Youths, China (21JR7RA840), the National Natural Science Foundation of China (32060486; 32160526), and the Research Program Sponsored by the State Key Laboratory of Aridland Crop Science, Gansu Agricultural University, China (GSCS-2022-1; GSCS-2020-Z02).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Wang W.X., Wang X.Y., Liao H.M., Feng Y.J., Guo Y.S., Shu Y.H., Wang J.W. Effects of nitrogen supply on induced defense in maize (Zea mays) against fall armyworm (Spodoptera frugiperda) Int. J. Mol. Sci. 2022;23:10457. doi: 10.3390/ijms231810457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li X., He K.L., Wang Z.Y., Bai S.X. Quantitative trait loci for Asian corn borer resistance in maize population. Agric. Sci. China. 2010;9:77–84. doi: 10.1016/S1671-2927(09)60070-5. [DOI] [Google Scholar]
- 3.Chen W.B., Shakir S., Bigham M., Richter A., Fei Z.J., Jander G. Genome sequence of the corn leaf aphid (Rhopalosiphum maidis Fitch) Gigascience. 2019;8:giz003. doi: 10.1093/gigascience/giz033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betsiashvili M., Ahern K.R., Jander G. Additive effects of two quantitative trait loci that confer Rhopalosiphum maidis (corn leaf aphid) resistance in maize inbred line Mo17. J. Exp. Bot. 2015;66:571–578. doi: 10.1093/jxb/eru379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bing J.W., Guthrie W.D. Generation mean analysis for resistance in maize to the corn leaf aphid (Homoptera, Aphididae) J. Econ. Entomol. 1991;84:1080–1082. doi: 10.1093/jee/84.3.1080. [DOI] [Google Scholar]
- 6.Carena M.J., Glogoza P. Resistance of maize to the corn leaf aphid: A review. Maydica. 2004;49:241–254. [Google Scholar]
- 7.Kannan M., Ismail I., Bunawan H. Maize dwarf mosaic virus: From genome to disease management. Viruses. 2018;10:492. doi: 10.3390/v10090492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mikel M.A., D’Arcy C.J., Rhodes A.M., Ford R.E. Yield loss in sweet corn correlated with time of inoculation with maize dwarf mosaic virus. Plant Dis. 1981;65:902–904. doi: 10.1094/PD-65-902. [DOI] [Google Scholar]
- 9.Badji A., Kwemoi D.B., Machida L., Okii D., Mwila N., Agbahoungba S., Kumi F., Ibanda A., Bararyenya A., Solemanegy M., et al. Genetic basis of maize resistance to multiple insect pests: Integrated genome-wide comparative mapping and candidate gene prioritization. Genes. 2020;11:689. doi: 10.3390/genes11060689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kebede M., Shimalis T. Out-break, distribution and management of fall armyworm, spodoptera frugiperda J.E. Smith in African: The status and prospects. Acad. Agric. J. 2018;3:551–568. [Google Scholar]
- 11.Devi S. Fall armyworm threatens food security in southern Africa. Lancet. 2018;391:727. doi: 10.1016/S0140-6736(18)30431-8. [DOI] [PubMed] [Google Scholar]
- 12.Tzin V., Lindsay P.L., Christensen S.A., Meihls L.N., Blue L.B., Jander G. Genetic mapping shows intraspecific variation and transgressive segregation for caterpillar-induced aphid resistance in maize. Mol. Ecol. 2015;24:5739–5750. doi: 10.1111/mec.13418. [DOI] [PubMed] [Google Scholar]
- 13.Meihls L.N., Kaur H., Jander G. Natural variation in maize defence against insect herbivores. Cold Spring Harb. Sym. Quant. Biol. 2012;77:269–283. doi: 10.1101/sqb.2012.77.014662. [DOI] [PubMed] [Google Scholar]
- 14.Murenga M., Derera J., Mugo S., Tongoona P. A review of genetic analysis and response to selection for resistance to Busseola fusca and Chilo partellus, stem borers in tropical maize germplasm: A Kenyan perspective. Maydica. 2016;61:M4. [Google Scholar]
- 15.Butrón A., Chen Y.C., Rottinghaus G.E., McMullen M.D. Genetic variation at bx1 controls DIMBOA content in maize. Theor. Appl. Genet. 2010;120:721–734. doi: 10.1007/s00122-009-1192-1. [DOI] [PubMed] [Google Scholar]
- 16.Meihls L.N., Handrick V., Glauser G., Barbier H., Kaur H., Haribal M.M., Lipka A.E., Gershenzon J., Buckler E.S., Erb M., et al. Natural variation in maize aphid resistance is associated with 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside methyltransferase activity. Plant Cell. 2013;25:2341–2355. doi: 10.1105/tpc.113.112409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cambier V., Hance T., de Hoffmann E. Variation of DIMBOA and related compounds content in relation to the age and plant organ in maize. Phytochemistry. 2000;53:223–229. doi: 10.1016/S0031-9422(99)00498-7. [DOI] [PubMed] [Google Scholar]
- 18.Huffaker A., Pearce G., Veyrat N., Erb M., Turlings T.C.J., Sartor R., Shen Z.X., Briggs S.P., Vaughan M.M., Alborn H.T., et al. Plant elicitor peptides are conserved signals regulating direct and indirect antiherbivore defense. Proc. Natl. Acad. Sci. USA. 2012;11:5707–5712. doi: 10.1073/pnas.1214668110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao H.J. Genome-Wide Association Study with Resistance to the Asian Corn Borer in Maize. Shenyang Agricultural University; Shenyang, China: 2018. [Google Scholar]
- 20.Yu Y.T. QTL Analysis of Resistance to Asian Corn Borer in Maize. Agricultural University of Hebei; Baoding, China: 2003. [Google Scholar]
- 21.Flint-Garcia S.A., Thuillet A.C., Yu J.M., Pressoir G., Romero S.M., Mitchell S.E., Doebley J., Kresovich S., Goodman M.M., Buckler E.S. Maize association population: A high-resolution platform for quantitative trait locus dissection. Plant J. 2005;44:1054–1064. doi: 10.1111/j.1365-313X.2005.02591.x. [DOI] [PubMed] [Google Scholar]
- 22.Liu Z.Z., Wu X., Liu H.L., Li Y.X., Li Q.C., Wang F.G., Shi Y.S., Song Y.C., Song W.B., Zhao J.R., et al. Genetic diversity and population structure of important Chinese maize inbred lines revealed by 40 core simple sequence repeats (SSRs) Sci. Agric. Sin. 2012;45:2107–2138. [Google Scholar]
- 23.Kumar B., Choudhary M., Kumar P., Kumar K., Kumar S., Singh B.K., Lahkar C., Meenakshi, Kumar P., Dar Z.A., et al. Population structure analysis and association mapping for turcicum leaf blight resistance in tropical maize using SSR markers. Genes. 2022;13:618. doi: 10.3390/genes13040618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X.P. Inheritance of Resistance to Rhopalosiphum maidis in Maize. Henan Agricultural University; Zhengzhou, China: 2016. [Google Scholar]
- 25.Guo J.F., He K.L., Meng Y.J., Hellmich R.L., Chen S.J., Lopez M.D., Lauter N., Wang Z.Y. Asian corn borer damage is affected by rind penetration strength of corn stalks in a spatiotemporally dependent manner. Plant Direct. 2022;6:e381. doi: 10.1002/pld3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin S.A., Darrah L.L., Hibbard B.E. Divergent selection for rind penetrometer resistance and its effects on European corn borer damage and stalk traits in corn. Crop Sci. 2004;44:711–717. doi: 10.2135/cropsci2004.7110. [DOI] [Google Scholar]
- 27.Ma D., Xie R., Liu X., Niu X., Hou P., Wang K., Lu Y., Li S. Lodging-related stalk characteristics of maize varieties in China since the 1950s. Crop Sci. 2014;54:2805–2814. doi: 10.2135/cropsci2014.04.0301. [DOI] [Google Scholar]
- 28.Gatch E.W., Munkvold G.P. Fungal species composition in maize stalk in relation to European corn borer injury and transgenic insect protection. Plant Dis. 2002;86:1156–1162. doi: 10.1094/PDIS.2002.86.10.1156. [DOI] [PubMed] [Google Scholar]
- 29.Varsani S., Grover S., Zhou S., Koch K.G., Huang P.C., Kolomiets M.V., Williams W.P., Heng-Moss T., Sarath G., Luthe D.S., et al. 12-Oxo-phytodienoic acid acts as a regulator of maize defense against corn leaf aphid. Plant Physiol. 2019;179:1402–1415. doi: 10.1104/pp.18.01472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.So Y.S., Ji H.C., Brewbaker J.L. Resistance to corn leaf aphid (Rhopalosiphum maidis Fitch) in tropical corn (Zea mays L.) Euphytica. 2010;172:373–381. doi: 10.1007/s10681-009-0044-z. [DOI] [Google Scholar]
- 31.Zhang X., van Doan C., Arce C.C.M., Hu L.F., Gruenig S., Parisod C., Hibbard B.E., Hervé M.R., Nielson C., Robert C.A.M., et al. Plant defense resistance in natural enemies of a specialist insect herbivore. Proc. Natl. Acad. Sci. USA. 2019;116:23174–23181. doi: 10.1073/pnas.1912599116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu L.F., Robert C.A.M., Cadot S., Zhang X., Ye M., Li B.B., Manzo D., Chervet N., Steinger T., van der Heijden M.G.A. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 2018;9:2738. doi: 10.1038/s41467-018-05122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kudjordjie E.N., Sapkota R., Steffensen S.K., Fomsgaard I.S., Nicolaisen M. Maize synthesized benzoxazinoids affect the host associated microbiome. Microbiome. 2019;7:59. doi: 10.1186/s40168-019-0677-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Houseman J.G., Campos F., Thie N.M.R., Philogene B.J.R., Atkinson J., Morand P., Arnason J.T. Effect of the maize-derived compounds DIMBOA and MBOA on growth and digestive processes of European corn borer (Lepidoptera: Pyralidae) J. Econ. Entomol. 1992;85:669–674. doi: 10.1093/jee/85.3.669. [DOI] [Google Scholar]
- 35.Yan F., Xu C., Li S., Lin C., Li J. Effects of DIMBOA on several enzymatic systems in Asian corn borer, Ostrinia furnacalis (Guenee) J. Chem. Ecol. 1995;21:2047–2056. doi: 10.1007/BF02033861. [DOI] [PubMed] [Google Scholar]
- 36.Gutierrez C., Castanera P., Torres V. Wound-induced changes in DIMBOA (2,4-dihydroxy-7-methoxy-2H-1,4 benzoxazin-3 94H)-one concentration in maize plants caused by Sesamia nonagrioides (Lepidoptera: Noctuidae) Ann. Appl. Biol. 1988;113:447–454. doi: 10.1111/j.1744-7348.1988.tb03322.x. [DOI] [Google Scholar]
- 37.Dafoe N.J., Huffaker A., Vaughan M.M., Duehl A.J., Teal P.E., Schmelz E.A. Rapidly induced chemical defenses in maize stems and their effects on short-term growth of Ostrinia nubilalis. J. Chem. Ecol. 2011;37:984–991. doi: 10.1007/s10886-011-0002-9. [DOI] [PubMed] [Google Scholar]
- 38.Dafoe N.J., Thomas J.D., Shirk P.D., Legaspi M.E.L., Vaughan M.M., Huffaker A., Teal P.E., Schelz E.A. European corn borer (Ostrinia nubilalis) induced responses enhance susceptibility in maize. PLoS ONE. 2013;8:e73394. doi: 10.1371/journal.pone.0073394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frey M., Stettner C., Paré P., Schmelz E.A., Tumlinson J.H., Gierl A. An herbivore elicitor activates the gene for indole emission in maize. Proc. Natl. Acad. Sci. USA. 2000;97:14801–14806. doi: 10.1073/pnas.260499897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gierl A., Frey M. Evolution of benzoxazinone biosynthesis and indole production in maize. Planta. 2001;213:493–498. doi: 10.1007/s004250100594. [DOI] [PubMed] [Google Scholar]
- 41.Von Rad U., Huttl R., Lottspeich F., Gierl A., Frey M. Two glucosyltransferases are involved in detoxification of benzoxazinoids in mazie. Plant J. 2001;28:633–642. doi: 10.1046/j.1365-313x.2001.01161.x. [DOI] [PubMed] [Google Scholar]
- 42.Frey M., Huber K., June Park W., Sicker D., Lindberg P., Meely R.B., Simmons C.R., Yalpani N., Gierl A. A 2-oxoglutarate-dependent dioxygenase is integrated in DIMBOA-biosynthesis. Phytochemistry. 2003;62:371–376. doi: 10.1016/S0031-9422(02)00556-3. [DOI] [PubMed] [Google Scholar]
- 43.JonczyK R., Schmidt H., Osterrieder A., Fiesselmann A., Schullehner K., Haslbeck M., Sicker D., Hofmann D., Yalpani N., Simmons C., et al. Elucidation of the final reactions of DIMBOA-glucoside biosynthesis in maize: Characterization of Bx6 and Bx7. Plant Physiol. 2008;146:1053–1063. doi: 10.1104/pp.107.111237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Louis J., Basu S., Varsani S., Castano-Duque L., Jiang V., Williams W.P., Felton G.W., Luthe D.S. Ethylene contributes to maize insect resistance1-mediated maize defense against the phloem sap-sucking corn leaf aphid. Plant Physiol. 2015;169:313–324. doi: 10.1104/pp.15.00958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Christensen S.A., Huffaker A., Kaplan F., Sims J., Zieman S., Doehlemann G., Ji L.X., Schmitz R.J., Kolomiets M.V., Alborn H.T., et al. Maize death acids, 9-lipoxygenase-derived cyclopente(a)nones, display activity as cytotoxic phytoalexins and transcriptional mediators. Proc. Natl. Acad. Sci. USA. 2015;112:11407–11412. doi: 10.1073/pnas.1511131112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Danisman S., Immink R.G.H. Arabidopsis class i and class ii tcp transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 2012;159:1511–1523. doi: 10.1104/pp.112.200303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao X.Q., Stumpe M., Feussner I., Kolomiets M. A novel plastidial lipoxygenase of maize (Zea mays) ZmLOX6 encodes for a fatty acid hydroperoxide lyase and is uniquely regulated by phytohormones and pathogen infection. Planta. 2008;227:491–503. doi: 10.1007/s00425-007-0634-8. [DOI] [PubMed] [Google Scholar]
- 48.Sun L., Yang D.L., Kong Y., Chen Y., Li X.Z., Zeng L.J., Li Q., Wang E.T., He Z.H. Sugar homeostasis mediated by cell wall invertase GRAIN INCOMPLETE FILLING 1 (GIF1) plays a role in pre-existing and induced defence in rice. Mol. Plant Pathol. 2014;15:161–173. doi: 10.1111/mpp.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolouri-Moghaddam M.R., Le Roy K., Xiang L., Rolland F., van den Ende W. Sugar signalling and antioxidant network connections in plant cells. FEBS J. 2010;277:2022–2037. doi: 10.1111/j.1742-4658.2010.07633.x. [DOI] [PubMed] [Google Scholar]
- 50.Essmann J., Schmitz-Thom I., Schön H., Sonnewald S., Weis E., Scharte J. RNA interference-mediated repression of cell wall invertase impairs defense in source leaves of tobacco. Plant Physiol. 2008;147:1288–1299. doi: 10.1104/pp.108.121418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y.Q., Zhang M.N., Du P., Liu H., Zhang Z.X., Xu J., Qin L., Huang B.Y., Zheng Z., Dong W.Z., et al. Transcriptome analysis of pod mutant reveals plant hormones are important regulators in controlling pod size in peanut (Arachis hypogaea L.) Peer J. 2022;10:e12965. doi: 10.7717/peerj.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao X.Q., Niu Y.N., Bai X.D., Mao T.T. Transcriptomic and metabolic profiling reveals a lignin metabolism network involved in mesocotyl elongation during maize seed germination. Plants. 2022;11:1034. doi: 10.3390/plants11081034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao X.Q., Niu Y.N. The combination of conventional QTL analysis, bulked-segregant analysis, and RNA-sequencing provide new genetic insights into maize mesocotyl elongation under multiple deep-seeding environments. Int. J. Mol. Sci. 2022;23:4223. doi: 10.3390/ijms23084223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao X.Q., Shi J., Niu Y.N., Lu P.N., Chen X.J., Mao T.T. 24-epibrassinolide alleviates aluminum toxicity by improving leaf chlorophyll fluorescence and photosynthetic performance and root antioxidant-oxidant balance and ascorbate-glutathione cycle in maize. Russ. J. Plant Physiol. 2022;69:99–108. doi: 10.1134/S1021443722050247. [DOI] [Google Scholar]
- 55.López-Castillo L.M., López-Arciniega J.A.I., Guerrero-Rangel A., Valdés-Rodríguez A., Brieba L.G., García-Lara S., Winkler R. Identification of B6T173 (ZmPrx35) as the prevailing peroxidase in highly insect resistance maize (Zea mays, p84C3) kernels by activity-directed purification. Front. Plant Sci. 2015;6:670. doi: 10.3389/fpls.2015.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng H.J., Wang H., Yang H., Wu J.H., Shi B., Cai R., Xu Y.B., Wu A.Z., Luo L.J. Genetic diversity and molecular evolution of Chinese Waxy maize germplasm. PLoS ONE. 2013;8:e66606. doi: 10.1371/journal.pone.0066606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Farnir F., Coppieters W., Arranz J.J., Berzi P., Cambisano N., Grisart B., Karim L., Marcq F., Moreau L., Mni M., et al. Extensive genome-wide linkage disequilibrium in cattle. Genome Res. 2000;10:220–227. doi: 10.1101/gr.10.2.220. [DOI] [PubMed] [Google Scholar]
- 58.Evanno G., Regnaut S., Goudet J. Detecting the number of cluster of individuals using the software structure: A simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 59.Hill W.G., Robertson A. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- 60.Bradbury P.J., Zhang Z., Kroon D.E., Casstevens T.M., Ramdoss Y., Buckler E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 61.Yu J., Pressoir G., Briggs W.H., Bi I.V., Yamasaki M., Doebley J., McMullen F., Gaut M.D., Nielsen B.S., Holland D.M., et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 62.Zhao X.Q., Fang P., Zhang J.W., Peng Y.L. QTL mapping for six ear leaf architecture traits under water-stressed an well-watered conditions in maize (Zea mays L.) Plant Breed. 2018;137:60–72. doi: 10.1111/pbr.12559. [DOI] [Google Scholar]
- 63.Zhao X.Q., Zhao C., Niu Y.N., Chao W., He W., Wang Y.F., Mao T.T., Bai X.D. Understanding and comprehensive evaluation of cold resistance in the seedlings of multiple maize genotypes. Plants. 2022;11:1881. doi: 10.3390/plants11141881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Handrick V., Robert C.A.M., Ahern K.R., Zhou S.Q., Machado R.A.R., Maag D., Glauser G., Fernandez-Penny F.E., Chandran J.N., Rodgers-Melnik E., et al. Biosynthesis of 8-O-methylated benzoxazinoid defense compounds in maize. Plant Cell. 2016;28:1682–1700. doi: 10.1105/tpc.16.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao M.Z., Ma Z.B., Wang L.L., Tang Z.Q., Mao T., Liang C.B., Gao H., Zhang L.Y., He N., Fu L., et al. SNP-based QTL mapping for panicle traits in the japonica super rice cultivar Liaoxing 1. Crop J. 2020;8:769–780. doi: 10.1016/j.cj.2020.07.002. [DOI] [Google Scholar]
- 66.Zhao X.Q., Zhang J.W., Fang P., Peng Y.L. Comparative QTL analysis for yield components and morphological traits in maize (Zea mays L.) under water-stressed and well-watered conditions. Breed. Sci. 2019;69:621–632. doi: 10.1270/jsbbs.18021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao X.Q., Zhong Y. Genetic dissection of the photosynthetic parameters of maize (Zea mays L.) in drought-stressed and well-watered environments. Russ. J. Plant Physiol. 2021;68:1125–1134. doi: 10.1134/S1021443721060236. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.