Abstract
The C-Repeat Binding Factor (CBF) gene family has been identified and characterized in multiple plant species, and it plays a crucial role in responding to low temperatures. Presently, only a few studies on tree species demonstrate the mechanisms and potential functions of CBFs associated with cold resistance, while our study is a novel report on the multi-aspect differences of CBFs among three tree species, compared to previous studies. In this study, genome-wide identification and analysis of the CBF gene family in Acer truncatum, Acer pseudosieboldianum, and Acer yangbiense were performed. The results revealed that 16 CBF genes (five ApseCBFs, four AcyanCBFs, and seven AtruCBFs) were unevenly distributed across the chromosomes, and most CBF genes were mapped on chromosome 2 (Chr2) and chromosome 11 (Chr11). The analysis of phylogenetic relationships, gene structure, and conserved motif showed that 16 CBF genes could be clustered into three subgroups; they all contained Motif 1 and Motif 5, and most of them only spanned one exon. The cis-acting elements analysis showed that some CBF genes might be involved in hormone and abiotic stress responsiveness. In addition, CBF genes exhibited tissue expression specificity. High expressions of ApseCBF1, ApseCBF3, AtruCBF1, AtruCBF4, AtruCBF6, AtruCBF7, and ApseCBF3, ApseCBF4, ApseCBF5 were detected on exposure to low temperature for 3 h and 24 h. Low expressions of AtruCBF2, AtruCBF6, AtruCBF7 were detected under cold stress for 24 h, and AtruCBF3 and AtruCBF5 were always down-regulated under cold conditions. Taken together, comprehensive analysis will enhance our understanding of the potential functions of the CBF genes on cold resistance, thereby providing a reference for the introduction of Acer species in our country.
Keywords: Acer truncatum, Acer pseudosieboldianum, Acer yangbiense, CBF gene family, cold stress
1. Introduction
Cold stress is a major natural factor that limits plant growth, productivity, and survival, and it determines the geographical distribution of plant species. Low temperature (0–12 °C) will inhibit plant growth and development, and freezing temperature (below 0 °C) will destroy cell membranes and cause cell death [1]. Plants can adapt to low temperatures through inducing the expression of cold tolerance-related genes, synthesizing corresponding protective proteins, and activating the protective enzymes and metabolites, and the process is called cold acclimation [2]. The cold acclimation natural habitat plants have the ability to acclimate completely, whereas plants originating from warmer climatic zones cannot often successfully overwinter, due to poor preparation for cold acclimation [3].
The genus Acer L. belongs to Aceraceae, which are deciduous or evergreen small trees or shrubs with medicinal, ornamental, and economic values. The ability of leaf color change determines the significant ornamental and economic values of Acer, and this ability is limited by diverse temperature zones. In Northeast China, some Acer species, such as Acer pseudosieboldianum, are mainly distributed in the cold temperate zone, and exhibit unique adaptability and cold resistance for tolerating temperatures below 30 °C in winter compared with other Acer species [4,5]. Most Acer species are native to Asia, whereas some species are spread through North America, Europe, and North Africa [6,7]. Over 150 species of Acer germplasm resources have primarily been found in China, accounting for more than half of the world’s Acer resources. The rich resources of germplasm in China are important for further research on the evolutionary history of the Acer species [8]. Acer pseudosieboldianum, Acer yangbiense, and Acer truncatum are three species of Acer endemic to China. A. pseudosieboldianum is endemic to north-east China [9]. A. yangbiense primarily grows in the western valley of Cangshan Mountain and Yunnan Province [10]. The natural distribution area of A. truncatum is mainly concentrated over north China and the north-east of China, and it is the most ubiquitous Acer species [11]. On the basis of distinct geographical environments and climates, various phenotypic differences and growth-specific differences of these three species are exhibited, and they may possess different cold acclimation.
As mentioned, cold acclimation is the inevitable result of a long period of low temperature time, affected by morphological coordination, physiological or biochemical adaptations, and genetic factors. The C-repeat binding factors dehydration-responsive element binding (CBFs/DREB1) proteins have been identified as key transcription factors involved in cold acclimation. They belong to the APETALA2/ethylene-responsive element binding factor (AP2/ERF) transcription factor family, which is further divided into the AP2, DREB, ERF, and Soloist, and related to abscisic acid insensitive 3/viviparous 1 (RAV) groups [12]. Among them, the DREB group consists of six subgroups, of which the A1 subgroup consists of the CBF/DREB1 transcription factors [13]. Studies have performed the genome-wide analysis of CBF/DREB1 genes in several plant species, such as Lolium perenne [14], Taraxacum kok-saghyz [15], Camellia sinensis [16], Eucalyptus grandis [17], Brassica rapa [18] and so on. In previous studies, cold treatment (4 °C) induced the expression of LpCBF3 in Lolium perenne [14], the LeCBF1 gene in Lycopersicon esculentum, and the CpCBF1 and CpCBF2 in Carica papaya were also found to be cold-inducible [19,20]. In Arabidopsis thaliana, the CBF gene family contains six genes, including CBF1/DREB1C, CBF2/DREB1B, CBF3/DREB1A, CBF4/DREB1D, DREB1E/DDF2, and DREB1F/DDF1 [21]. The expression of AtCBF1, AtCBF2, and AtCBF3 is induced under cold stress, whereas the expression of AtCBF4, AtDREB1E, and AtDREB1F is induced under osmotic stress, such as drought and salt [22,23]. Studies have shown that MbCBF2, isolated from Malus baccata, enhances the resistance to cold stress in A. thaliana by increasing proline content, superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) activity [24]. The overexpression of CsCBF1 increases the putrescine (Put) levels in Citrus sinensis, along with remarkably enhancing cold tolerance [25]. Besides low temperature, light quality, photoperiod, and the circadian clock also regulate the CBF gene expression via light-sensitive cis-elements in their promoter region [26].
In this study, we systematically performed genome-wide identification and analysis of the CBF gene family in Acer pseudosieboldianum, Acer yangbiense, and Acer truncatum, including comprehensive analysis of physical and chemical characteristics, chromosomal location, phylogenetic and evolutionary relationship, and conserved motifs. The expression pattern of the CBF genes in different tissues and the expression profile of three Acer CBFs under cold stress were detected. This was the novel report on the multi-aspect differences of CBFs among three tree species compared to previous studies. Our study will enhance our understanding of the CBF genes, which lay a theoretical foundation on the study of the CBFs protein structure and function, and the molecular breeding of resistance to cold in these three Acer species, thereby providing a reference for introducing Acer species in all regions in our country.
2. Results
2.1. Identification of CBF Genes in Three Acer Species
A total of 16 CBF genes were identified from three Acer species, including seven AtruCBF genes, four AcyanCBF genes, and five ApseCBF genes after sequence alignment with A. thaliana. Of these CBFs, AtruCBF7 had the longest protein sequence with 395 amino acids, and the length of CDS was 1185 bp. The protein sequence of AtruCBF5 was the smallest, with 165 amino acids, and the length of CDS was 495 bp. The molecular weight of CBF proteins ranged from 19,008.12 kDa (AtruCBF5) to 43,582.88 kDa (AtruCBF7), and the isoelectric point ranged from 4.77 (ApseCBF2) to 9.79 (AtruCBF4). ApseCBFs and AcyanCBFs were neutral. The results of subcellular localization prediction revealed that most CBF proteins localized in the nucleus, while ApseCBF3, ApseCBF4, AcyanCBF1, AcyanCBF3, and AtruCBF5 proteins also localized in the cytoplasm concurrently (Table 1). These results suggested that significant differences existed in three Acer species CBFs genes.
Table 1.
The identification of CBF genes in three Acer species.
| Gene Name | Gene ID | AA 1 | MW 2 (kDa) | PI 3 | SL 4 | CDS (bp) |
|---|---|---|---|---|---|---|
| ApseCBF1 | Apse002T0243400.1 | 253 | 27,693.81 | 4.9 | Nucleus | 759 |
| ApseCBF2 | Apse002T0243300.1 | 253 | 27,666.73 | 4.77 | Nucleus | 759 |
| ApseCBF3 | Apse011T0099600.1 | 246 | 27,542.91 | 6.68 | Cytoplasm, Nucleus | 738 |
| ApseCBF4 | Apse002T0243600.1 | 235 | 24,831.5 | 5.04 | Cytoplasm, Nucleus | 705 |
| ApseCBF5 | Apse007T0094500.1 | 212 | 22,876.3 | 5.46 | Nucleus | 636 |
| AcyanCBF1 | Acyan11G0087800.1 | 301 | 33,234.08 | 6.03 | Cytoplasm, Nucleus | 903 |
| AcyanCBF2 | Acyan11G0087900.1 | 382 | 42,556.73 | 8.9 | Cytoplasm | 1146 |
| AcyanCBF3 | Acyan02G0289000.1 | 249 | 26,360.25 | 5.42 | Cytoplasm, Nucleus | 747 |
| AcyanCBF4 | Acyan07G0093600.1 | 223 | 23,930.49 | 5.61 | Nucleus | 669 |
| AtruCBF1 | Atru.chr10.1810 | 258 | 27,824.85 | 9.35 | Nucleus | 774 |
| AtruCBF2 | Atru.chr1.1042 | 238 | 27,303.82 | 9.49 | Nucleus | 714 |
| AtruCBF3 | Atru.chr2.1230 | 190 | 20,944.53 | 9.73 | Nucleus | 570 |
| AtruCBF4 | Atru.chr4.1701 | 216 | 24,514.44 | 9.79 | Nucleus | 648 |
| AtruCBF5 | Atru.chr13.731 | 165 | 19,008.12 | 9.37 | Cytoplasm, Nucleus | 495 |
| AtruCBF6 | Atru.chr6.2697 | 291 | 31,912.79 | 7.37 | Nucleus | 873 |
| AtruCBF7 | Atru.chr4.2790 | 395 | 43,582.88 | 6.38 | Nucleus | 1185 |
1 The number of amino acids, 2 Molecular weight, 3 Isoelectric point, 4 Subcellular localization.
2.2. Construction of Phylogenetic Tree
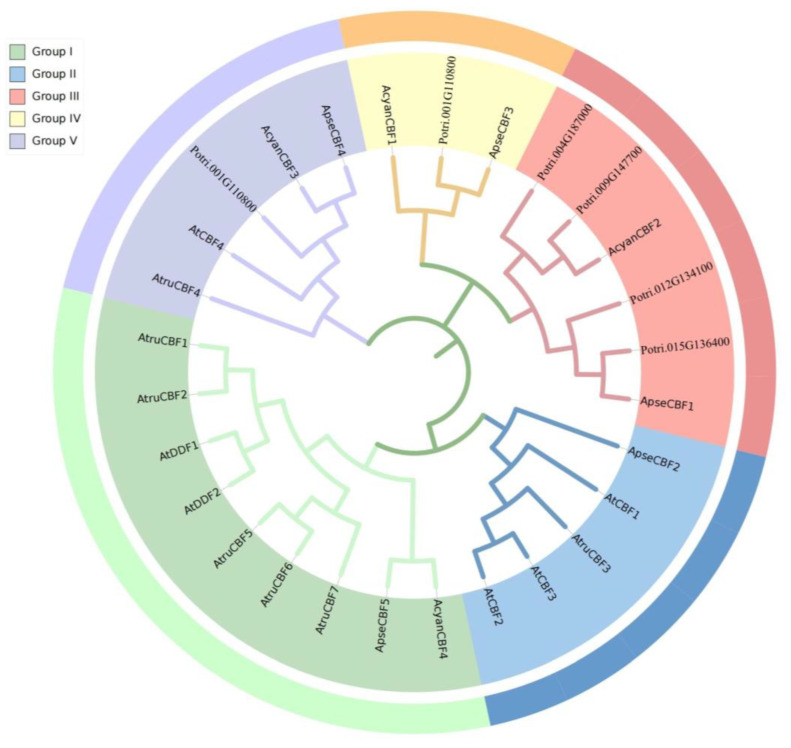
The phylogenetic relationships of the CBF gene family between three Acer species and other plants were analyzed. A total of 28 CBF proteins were used to construct the phylogenetic tree, including seven proteins from A. truncatum (AtruCBFs), four proteins from A. yangbiense (AcyanCBFs), five proteins from A. pseudosieboldianum (ApseCBFs), six proteins A. thaliana (AtCBFs), and six proteins from P. trichocarpa (PtrCBFs). These CBFs were clustered in five groups: Group I, Group II, Group III, Group IV, and Group V. The CBFs from three Acer species were clustered in Group I, Group IV, and Group V. Group I was the largest group, containing one AcyanCBFs, one ApseCBFs, five AtCBFs, AtDDF1, and AtDDF2. Four PtrCBFs were clustered in Group III. Interestingly, all five species are clustered to group V (Figure 1). These results suggested that the evolution relationships of CBFs in Acer species were different with other plant species.
Figure 1.
Phylogenetic tree of the CBF proteins from six species. Different colors represent the groups. At: A. thaliana; Acyan: A. yangbiense; Atru: A. truncatum; Apse: A. pseudosieboldianum; Os: O. sativa; Potri: P. trichocarpa.
2.3. Gene Structure and Conserved Motif of CBF Genes
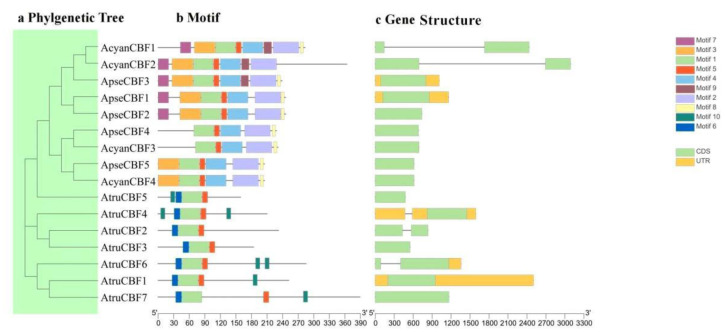
The structure and motif of CBF genes in A. truncatum, A. pseudosieboldianum, and A. yangbiense were analyzed, and these 16 CBFs were ordered according to the phylogenetic tree (Figure 2a). A total of 12 CBFs spanned only one exon, and four CBFs. AcyanCBF1, AcyanCBF2, AtruCBF2, and AtruCBF6 spanned two exons and one intron (Figure 2c). The analysis of conserved motifs showed that the CBF genes had three to eight motifs. Motif 1 and Motif 5 were found from all CBFs. Motif 2 and Motif 4 were found from AcyanCBFs and ApseCBFs, while Motif 6 was only found from AtruCBFs. Further, Motif 3 was found in AcyanCBFs and ApseCBFs, except for ApseCBF4 and AcyanCBF3. Motif 7 was only found in AcyanCBF1, AcyanCBF2, ApseCBF3, ApseCBF1, and ApseCBF2. Motif 10 was only found in AtruCBF1, AtruCBF4, AtruCBF5, AtruCBF6, and AtruCBF7 (Figure 2b). These results suggested that the gene structure and motif of CBFs in three Acer species were relatively conserved.
Figure 2.
The analysis of gene structure and conserved motif. (a) The phylogenetic tree of all CBF proteins in three Acer species. (b) The motif composition of CBF genes in three Acer species. (c) The structure of CBF genes.
2.4. Cis-Acting Elements of CBF Genes
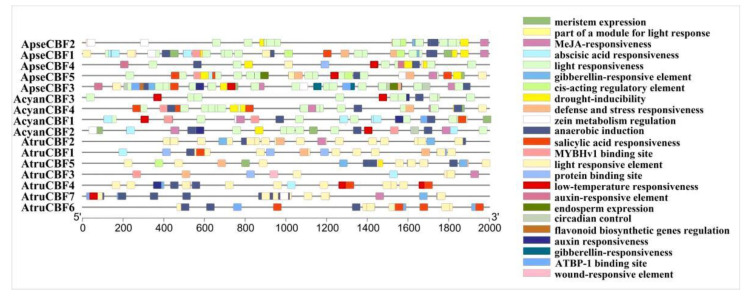
The cis-acting elements of the 16 CBF genes promoters were explored, and a total of 24 types of cis-elements were identified. Low-temperature response elements were found in the promoter of three ApseCBFs genes, including ApseCBF3, ApseCBF4, and ApseCBF5; three AcyanCBFs genes, including AcyanCBF1, AcyanCBF2, AcyanCBF3; and two AtruCBFs genes, including AtruCBF4 and AtruCBF7. The light responsiveness presented in the promoter of all CBF genes with a large number. The promoter of CBF genes also contained elements associated with hormone responsiveness, such as MeJA-responsiveness elements, found in the promoter of ApseCBFs, and gibberellin-responsiveness elements, found in the promoter of ApseCBF3 and AtruCBF6. The promoter of ApseCBF genes had drought inducibility elements, but of all AtruCBFs genes, the drought inducibility elements only presented in the promoter of AtruCBF5 (Figure 3). The detection of these cis-acting elements suggested that the CBF genes in three Acer species played important roles in treating abiotic stress, drought, light and cold stress, etc.
Figure 3.
The number and position of cis-acting elements in three Acer species CBFs promoters. Different color boxes represent different elements.
2.5. Chromosome Location of CBF Genes
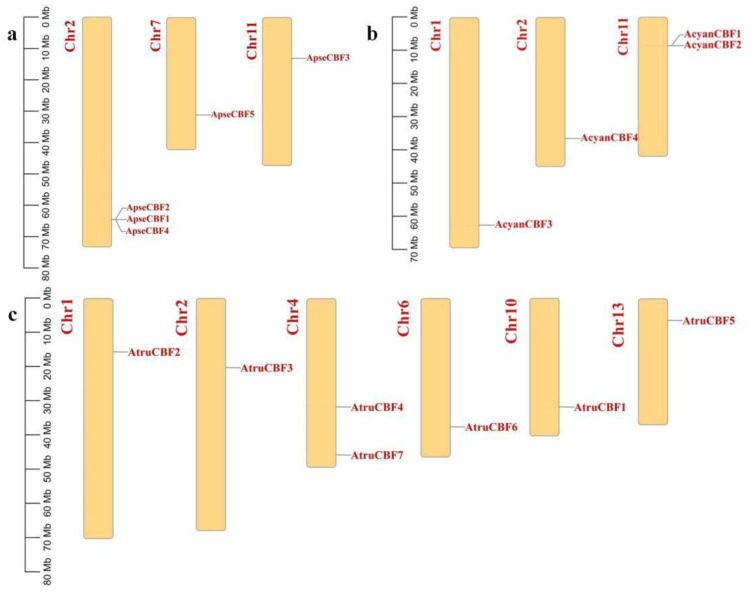
The chromosomal location of CBF genes in three Acer species were analyzed. The ApseCBFs genes were unevenly distributed at one end of three chromosomes in A. pseudosieboldianum, where ApseCBF1, ApseCBF2, ApseCBF4 were located on Chr2, ApseCBF5 was located on Chr7, and ApseCBF3 was located on Chr11 (Figure 4a). The AcyanCBF genes were also unevenly distributed at one end of three chromosomes in A. yangbiense. AcyanCBF3 was located on Chr1, AcyanCBF4 was located on Chr2, and AcyanCBF1 and AcyanCBF2 were located on Chr11 (Figure 4b). The AtruCBF genes dispersed six chromosomes, and each AtruCBF gene was located on one chromosome, except for Chr4 containing AtruCBF4 and AtruCBF7. AtruCBF2, AtruCBF3, AtruCBF6, AtruCBF1, and AtruCBF4 were located on Chr1, Chr2, Chr6, Chr10, and Chr13, respectively (Figure 4c). The CBF gene family was not located on all 13 chromosomes; this difference in gene distribution determined the complexity and diversification of CBFs in three Acer species, providing clues to their evolution.
Figure 4.
(a) The chromosomal location of ApseCBFs in A. pseudosieboldianum. (b) The chromosomal location of AcyanCBFs in A. yangbiense. (c) The chromosomal location of AtruCBFs in A. truncatum.
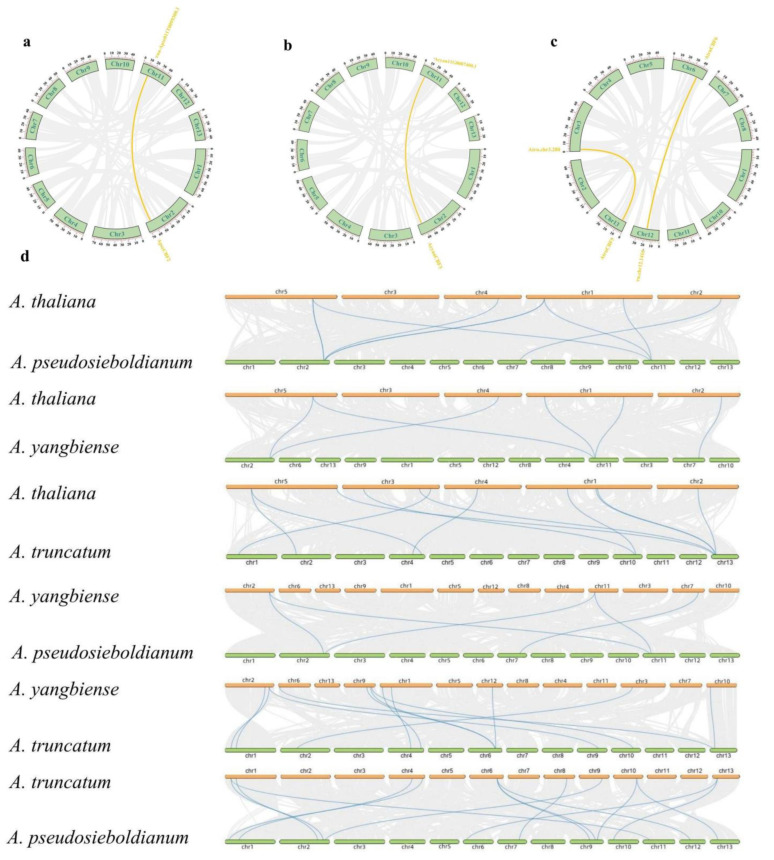
2.6. Synteny Analysis of CBF Genes
The intraspecific and interspecific synteny analysis of CBF genes were explored. One orthologous gene pair located on Chr2 and Chr11 was found in A. pseudosieboldianum and A. yangbiense (Figure 5a,b). Two orthologous gene pairs located on Chr3 and Chr13, Chr6 and Chr12 were found in A. truncatum (Figure 5c). As for the synteny analysis between different species, A. yangbiense and A. truncatum, A. truncatum and A. pseudosieboldianum contained 12 and 14 orthologous gene pairs, indicating they possessed the higher homology. A. yangbiense and A. pseudosieboldianum only contained five orthologous gene pairs, indicating they possessed lowest homology. The results showed that A. pseudosieboldianum and A. truncatum exhibited the highest level of homology. Furthermore, orthologous gene pairs in A. pseudosieboldianum and A. truncatum were mainly distributed on chromosomes 2 and 1. Further, some orthologous gene pairs were also detected between A.thaliana and A. pseudosieboldianum, A.thaliana and A. yangbiense, and A. thaliana and A. truncatum, suggesting these species also exhibited homology (Figure 5d). These results proposed that CBF genes possessed a degree of homology in different Acer species.
Figure 5.
(a) The synteny of ApseCBFs in A. pseudosieboldianum. (b) The synteny of AcyanCBFs in A. yangbiense. (c) The synteny of AtruCBFs in A. truncatum. (d) The synteny analysis of CBFs between A. thaliana and A. pseudosieboldianum, A. thaliana and A. yangbiense, A. thaliana and A. truncatum, A. yangbiense and A. pseudosieboldianum, A. yangbiense and A. truncatum, and A. truncatum and A. pseudosieboldianum. The orthologous gene pairs are highlighted using blue lines.
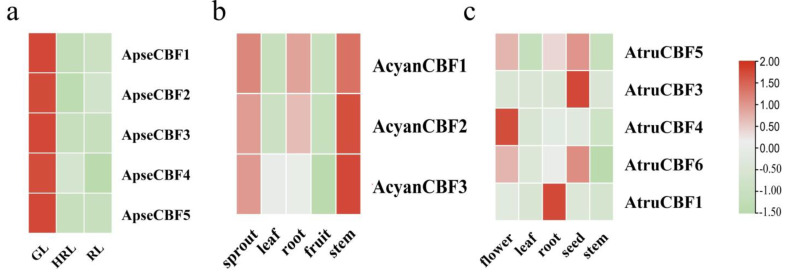
2.7. The Expression of CBF Genes in Different Tissues
The expression of CBF genes in different tissues was explored based on the transcriptomes. In A. pseudosieboldianum, a higher expression of ApseCBFs genes was detected in green leaf (GL) compared to half-red leaf (HRL) and red leaf (RL) (Figure 6a). In A. yangbiense, the expressions of AcyanCBF1, AcyanCBF2, and AcyanCBF3 were up-regulated in the stem and sprout, whereas these genes were down-regulated in the leaf and fruit (Figure 6b). In A. truncatum, the expressions of AtruCBF5 and AtruCBF6 were up-regulated in the flower and seed, while the expressions of AtruCBF3, AtruCBF4, and AtruCBF1 were up-regulated in seed, flower, and root, respectively (Figure 6c). The results revealed that the CBF genes possessed tissue-specific expression.
Figure 6.
The expression pattern of the CBF genes in different tissues. (a) The expression of five ApseCBF genes in green leaf (GL), half-red leaf (HRL), red leaf (RL) of A. pseudosieboldianum. (b) The expression of three AcyanCBF genes in sprout, leaf, root, fruit, and stem of A. yangbiense. (c) The expression of five AtruCBF genes in flower, leaf, root, seed, and stem of A. truncatum.
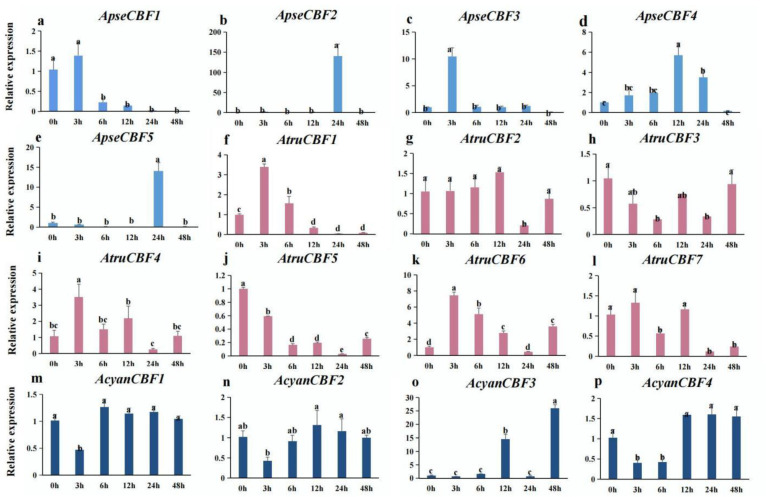
2.8. Expression of CBF Genes in Three Acer Species under Low-Temperature Conditions
Previous studies have shown that CBF genes play an important role in responding to cold stress [24,25]. Therefore, the expression of CBF genes in three Acer species under low temperatures was detected. The results showed that low temperatures could induce the expression of CBF genes (Figure 7). The expression of ApseCBF1, ApseCBF3, AtruCBF1, AtruCBF4, AtruCBF6, and AtruCBF7 was up-regulated on exposure to low temperature for 3 h, and the differences were significant except for ApseCBF1 and AtruCBF7 (p < 0.05) (Figure 7a,c,f,i,k,l). ApseCBF3, ApseCBF4, and ApseCBF5 were significantly highly expressed under cold for 24 h (p < 0.05) (Figure 7b,d,e), while AtruCBF2, AtruCBF6, and AtruCBF7 were significantly lowly expressed at 24 h (p < 0.05) (Figure 7g,k,l). Moreover, the expressions of AtruCBF3 and AtruCBF5 were always down-regulated during the low temperature conditions (Figure 7h,j). The expressions of AcyanCBF1 and AcyanCBF2 exhibited no significant differences under cold conditions, except for at 3 h (Figure 7m,n), while a significantly higher expression of AcyanCBF3 was detected on exposure to low temperature for 12 h and 48 h (Figure 7o). These results suggested that CBF genes in three Acer species have specific expression patterns when exposed to low-temperature conditions for different time durations.
Figure 7.
(a–e) The expression of CBF genes in A. pseudosieboldianum under cold stress. (f–l) The expression of CBF genes in A. truncatum under cold stress. (m–p) The expression of CBF genes in A. yangbiense under cold stress. The Y-axis and X-axis represent the gene expression level detected by qRT-PCR and treatment time, respectively. The values are the mean ± SD of three technical replicates. The letter indicates significant differences (p < 0.05).
3. Discussion
Acer is a medicinal, ornamental, and economic tree species. The Acer species mentioned in our study, A. pseudosieboldianum, A. yangbiense, and A. truncatum, are distributed across different areas of China. Presently, the studies on genome assembly of A. pseudosieboldianum, A. yangbiense, and A. truncatum heve been reported, which provide references for exploring the genome-wide identifications and functions of genes in three species [8,27,28]. CBF transcription factors, also known as DREB1 proteins, belong to the AP2/ERF family. They play an important role in mediating plant responses to biotic and abiotic stress, including high salt, drought, and low temperature. Particularly, CBFs can enhance the resistance to cold stress in several tree species, such as Eucalyptus gunnii, Malus × domestica, and Betula pendula [29,30,31]. On the basis of their genome, our study performed genome-wide identification and analysis of CBFs in three Acer species, which will lay the foundation for further study of CBF gene functions in cold resistance.
Five, four, and seven CBF genes were identified from A. pseudosieboldianum, A. yangbiense, and A. truncatum. Most CBF proteins are localized predominantly in the nucleus (Table 1), in accordance with their biological roles as transcription factors. The study on identifying the gene family based on the whole genome is significant for understanding the origin, evolution, and differentiation of gene families [32]. These results were similar to the report on CsCBFs [16]. According to the phylogenetic tree analysis, the results showed that the CBF family members of the three species of Acer, P. trichocarpa, and A. thaliana did not form a separate cluster of evolutionary branches, indicating that there was a certain degree of homology between the CBF gene families of several species. At the same time, the CBF gene family may have undergone great differentiation in evolution (Figure 1) [15]. Although CBF homologs from A. thaliana, A. pseudosieboldianum, and A. yangbiense were clustered in the same group, they were divided into separate subgroups, indicating that the duplications of CBFs in eudicot plants were independent events, and the duplication and divergence occurred after speciation [33]. The analysis of gene structure and conserved motif showed that genes clustered on the same branch exhibited similarity, such as similar exon number, motif number, and motif position (Figure 2a–c). The certain conserved motifs played important functional and/or structural roles in active proteins [34]. All ApseCBFs, most AcyanCBFs, and AtruCBFs were single-exon structures, while AcyanCBF1, AcyanCBF2, AtruCBF2, AtruCBF6 contained one intron (Figure 2c). The results showed that most members of the CBF family in three Acer species were intron deletion genes, which were consistent with CBFs in Taraxacum koksaghyz [15]. The loss of introns might shorten the time required for gene transcription to translation, thereby accelerating gene expression and functional protein production to adapt to the changes of the plant and environment [35].
Most ApseCBFs, AcyanCBFs, and AtruCBFs were unevenly distributed at one end of the chromosomes in three Acer species, while ApseCBF1/2/4, AcyanCBF1/2, AtruCBF4/7 were located on one chromosome (Figure 4a–c). The difference in gene distribution determined the complexity and diversification of CBFs in three Acer species, which might be caused by the differences in the structure and size of the chromosomes. The intraspecific and interspecific synteny analysis of CBFs could also be used to indicate the homology. Interestingly, A. yangbiense and A. truncatum, A. truncatum and A. pseudosieboldianum had higher homology, while A. yangbiense and A. pseudosieboldianum had lower homology (Figure 5d). The distribution areas of A. yangbiense and A. pseudosieboldianum were distinct; one was distributed in Northeast China, the other was distributed in Yunnan Province [9,10]. The regional differences might lead to the separation of plants and gene origin, divergence, and evolution [36].
The gene promoters are upstream of the transcriptional start, which contains plenty of cis-acting elements, and controls the transcription of genes [37]. The promoter polymorphisms of CBFs affect the expression of CBF genes, and affect the expression of related response genes in A. thaliana [38,39,40]. The studies have also shown that AtCBF2 negatively regulated AtCBF3 and AtCBF1, while AtCBF4 functioned in drought stress tolerance [22,41]. In our study, low-temperature, drought, light, and plant hormone-responsive elements were identified from the promoter region of ApseCBFs, AcyanCBFs, and AtruCBFs genes. For example, the low-temperature responsiveness cis-acting elements were found in the promoter of ApseCBF4, ApseCBF5, ApseCBF3, AcyanCBF1, AcyanCBF2, AtruCBF4, AtruCBF7, and the expression of these genes could be significantly induced by cold stress, especially ApseCBF4 at 12 h, ApseCBF5 at 24 h, ApseCBF3 at 3 h, AcyanCBF1 at 6 h, AcyanCBF2 at 12 h, AtruCBF4 at 3 h, AtruCBF7 at 3 h (Figure 3 and Figure 7). It is speculated that some transcription activators could specifically bind to and activate the promoters of CBFs on exposure to low temperature, thereby inducing the expression of these mentioned genes; therefore they may play crucial roles in cold resistance [14,42]. Other cis-acting elements, such as drought-inducibility elements in the promoters of ApseCBFs, AcyanCBF2/4, AtruCBF5; defense and stress responsiveness; and wound-responsive elements were found (Figure 3). These elements might help in activating CBFs to cope with other abiotic stresses. The expression patterns of CBFs under cold stress were explored. A high expression of some CBF genes on exposure to low temperature for 3 to 12 h was detected, while an increase in expression of ApseCBF4, AcyanCBF3, and AtruCFB2 on exposure to low temperature for 12 h, AcyanCBF2, and AcyanCBF4 on exposure to low temperature for 12 to 48 h was observed (Figure 7). The results indicated that some CBF genes might function during the early stages of response to cold stress, and that genes such as AcyanCFB2/3, and AcyanCBF4 might function during the late stages of response to cold stress. Previous studies have also demonstrated that the CBF genes respond to cold stress in a time-dependent manner in Secale cereale L. [43] and Camellia sinensis [16]. However, no significantly negative regulations were found in three Acer species between CBF2 and CBF1/3 from gene expression patterns under low temperature, so we will explore this relationship in the future. Further, the tissue-specific expression of CBF genes was exhibited (Figure 6a–c); these results were also reported in Eucalyptus grandis and Punica granatum [17,44]. In summary, our study excavated some CBFs significantly induced by low temperature in three Acer species, which provided a reference for further gene function research and molecular regulation mechanisms in cold resistance. The results might direct the introduction of Acer species.
4. Materials and Methods
4.1. Plant Materials
The three-year-old A. truncatum, A. yangbiense and A. pseudosieboldianum were obtained from the Northeast Forestry University greenhouse (126°38′8.92″ E, 45°43′20.64″ N), Harbin, Heilongjiang Province, China. Acer species were exposed to low temperatures (i.e., 4 °C) in an intelligent light incubator (ToppYunnong Technology Co., Ltd., Hangzhou, China) for 0 h, 3 h, 6 h, 12 h, 24 h, and 48 h. The functional leaves (the third to fifth leaves from the main branches) were collected, and total RNA was extracted to analyze the expression pattern of A. pseudosieboldianum CBFs (ApseCBFs), A. yangbiense CBFs (AcyanCBFs), and A. truncatum CBFs (AtruCBFs) genes.
4.2. Retrieving the CBF Gene Family Sequences
To perform genome-wide analysis of the CBF genes from three Acer species, the whole genome sequences were directly obtained according to previous studies [8,27,28]. The six known CBF transcription factor family genes from A. thaliana were selected as the query objects, and the protein sequences were retrieved using the Arabidopsis Information Resource (TAIR) (https://www.arabidopsis.org/browse/genefamily/index.jsp, accessed on 12 December 2021) [45]. The CBF genes in A. truncatum, A. yangbiense and A. pseudosieboldianum were identified using the BLAST by Toolbox for Biologists (TBtools) v 1.087 (e-value < 1 × 10−5) based on A. thaliana [46]. Each A. thaliana gene was successfully matched with multiple CBF genes, and the alignment sequence IDs of candidate CBFs were obtained eventually after eliminating the repeated values and blanks. The candidate CBF genes were further manually analyzed using Batch CD-Search in the National Centre for Biotechnology database (NCBI) (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi, accessed on 12 December 2021) to detect the presence of the CBF domain, and 16 candidate CBFs were identified. The biochemical properties, such as the molecular weight (MW) and isoelectric point (pI), were determined using the Compute pI/Mw tool on the ExPASy (https://web.expasy.org/protparam, accessed on 15 December 2021). The subcellular localization was analyzed using the WoLF PSORT tool (https://wolfpsort.hgc.jp/, accessed on 15 December 2021).
4.3. Analyses of Gene Structure and Protein Motif Composition
The structures of candidate CBF genes in A. truncatum, A. pseudosieboldianum, and A. yangbiense were identified and visualized using the TBtools software [47]. MEME, a web-based tool (http://meme-suite.org/tools/meme, accessed on 20 December 2021), was used to explore the conserved motif in A. truncatum, A. pseudosieboldianum, and A. yangbiense. The parameters were set at a maximum of 10 motifs. TBtools was further used to visualize the motif composition [48].
4.4. The Analyses of Chromosomal Location and Collinearity
The chromosome locations of CBFs in three Acer species were analyzed, and they were mapped on 13 chromosomes (named Chr 1 to Chr 13) according to their physical positions (bp). The McScan software was used to perform the collinearity analysis, and homology between ApseCBFs, AcyanCBFs, AtruCBFs and AtCBFs was evaluated with default parameters. These results were visualized using TBtools.
4.5. The Analysis of Phylogenetic Tree
The evolutionary relationship of CBF genes in A. thaliana, A. pseudosieboldianum, A. yangbiense, A. truncatum, and Populus trichocarpa was analyzed. The phylogenetic tree was constructed using the Neighbor-joining method by MEGA software, the Bootstrap method value was set to 1000, and other parameters were set to default. Interactive Tree of Life (iTOL) (https://itol.embl.de/, accessed on 22 December 2021) was used to beautify the phylogenetic tree.
4.6. The Analysis of Cis-Acting Elements
The 2000 bp sequences upstream of CBF genes coding sequences (CDSs) were extracted as promoter sequences using TBtools. The cis-acting elements were identified using the Plant CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html).
4.7. The Analysis of Gene Expression
The RNA-seq data of A. pseudosieboldianum, A. yangbiense, and A. truncatum were obtained from NCBI (accession numbers were PRJNA736515, PRJNA524417, and PRJNA665613), respectively. After quality control, alignment, and quantitative analysis, the expression levels of CBF genes were represented using fragment per kilobase per million mapped reads (FPKM). The gene expression patterns of green leaf (GL), half-red leaf (HRL), and red leaf (RL) in A. pseudosieboldianum; sprout, leaf, root, fruit, and stem in A. yangbiense; and flower, leaf, root, seed, stem in A. truncatum were explored.
Total RNA was extracted from the functional leaves of three Acer species under cold stress (4 °C for 0, 3, 6, 12, 24, and 48 h) using RNAprep Pure Plant Kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. After detecting the integrity and quality of total RNA, the cDNA was synthesized using PrimeScriptTM RT reagent Kit with gDNA Eraser (TaKaRa, Beijing, China). A total of 16 CBF genes were selected to explore the gene expression using qRT-PCR. The primer sequences were listed in Supplementary Table S1, and 18S was used as an internal reference gene. The qRT-PCR was performed on the ABI 7500 Real-Time PCR system (Applied Biosystems, Carlsbad, CA, USA), using TB Green® Premix Ex TaqTM II (Tli RNaseH Plus) (TaKaRa, Beijing, China) with three technical replicates. Amplification system and procedure were carried out according to Li [32]. The relative gene expression levels were calculated using the 2−ΔΔCT method [49].
5. Conclusions
In conclusion, we have identified seven AtruCBF genes, four AcyanCBF genes, and five ApseCBF genes from A. truncatum, A. yangbiens, and A. pseudosieboldianum. These CBF genes, clustered in five subgroups based on phylogenetic relationships, mainly contained conserved Motif 1 and Motif 5, and 12 CBF genes only spanned one exon. The cis-acting elements in the promoters of CBF genes were involved in hormone, light, drought, and low-temperature responsiveness. The CBFs were unevenly distributed at chromosomes in three Acer species, mostly at Chr2 and Chr11. One, one, and two orthologous gene pairs were found in A. pseudosieboldianum, A. yangbiense, and A. truncatum. A. yangbiense and A. truncatum, A. truncatum and A. pseudosieboldianum possessed high homology, while A. yangbiense and A. pseudosieboldianum possessed the lowest homology. In addition, high expressions of ApseCBF1, ApseCBF3, AtruCBF1, AtruCBF4, AtruCBF6, AtruCBF7, and ApseCBF3, ApseCBF4, ApseCBF5 were detected on exposure to low temperature for 3 h and 24 h. Low expressions of AtruCBF2, AtruCBF6, AtruCBF7 were detected under cold stress for 24 h, and AtruCBF3 and AtruCBF5 were always down-regulated under cold conditions. These results provided a meaningful direction for gene function research on the cold-resistance of A. truncatum, A. pseudosieboldianum, and A. yangbiens, which is favorable for the future introduction of these three Acer species.
Acknowledgments
Thanks to the members of the College of Forestry and Grassland Science at Jilin Agricultural University and the State Key Laboratory of Tree Genetics and Breeding at Northeast Forestry University for their practical assistance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24032088/s1.
Author Contributions
Q.Z., K.C., L.L. and R.H. wrote the manuscript. Y.L., H.Y. and G.Q. performed the experiments, the plant sample collection, and RNA extraction as such. Q.Z. and K.C. analyzed the data, identified the gene families, and constructed the phylogenetic trees as such. X.Z. designed the experiments and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by the Heilongjiang Province Applied Technology Research and Development Plan Project (No. GA20B402), Fundamental Research Funds for the Central Universities (Northeast Forestry University) (No. 2572020DR01).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Kidokoro S., Shinozaki K., Yamaguchi-Shinozaki K. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 2022;27:922–935. doi: 10.1016/j.tplants.2022.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Hu Y., Zhang H., Gu B., Zhang J. The transcription factor VaMYC2 from Chinese wild Vitis amurensis enhances cold tolerance of grape (V. vinifera) by up-regulating VaCBF1 and VaP5CS. Plant Physiol. Biochem. 2022;192:218–229. doi: 10.1016/j.plaphy.2022.10.011. [DOI] [PubMed] [Google Scholar]
- 3.Jankauskienė J., Mockevičiūtė R., Gavelienė V., Jurkonienė S., Anisimovienė N. The application of auxin-like compounds promotes cold acclimation in the oilseed rape plant. Life. 2022;12:1283. doi: 10.3390/life12081283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun B., Liang M., Yang Y., Xu H. A primary analysis on cold resistance of Acer pseudosieboldianum (pax) kom in Heilongjiang Province. Territ. Nat. Resour. Study. 2013;3:88–89. [Google Scholar]
- 5.Liu C., Tsuda Y., Shen H., Hu L., Saito Y., Ide Y. Genetic structure and hierarchical population divergence history of Acer mono var. mono in South and Northeast China. PLoS ONE. 2014;9:e87187. doi: 10.1371/journal.pone.0087187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Renner S.S., Beenken L., Grimm G.W., Kocyan A., Ricklefs R.E. The evolution of dioecy, heterodichogamy, and labile sex expression in Acer. Evolution. 2007;61:2701–2719. doi: 10.1111/j.1558-5646.2007.00221.x. [DOI] [PubMed] [Google Scholar]
- 7.Renner S.S., Grimm G.W., Schneeweiss G.M., Stuessy T.F., Ricklefs R.E. Rooting and dating maples (Acer) with an uncorrelated-rates molecular clock: Implications for North American/Asian Disjunctions. Syst. Biol. 2008;7:795–808. doi: 10.1080/10635150802422282. [DOI] [PubMed] [Google Scholar]
- 8.Ma Q., Sun T., Li S., Wen J., Zhu L., Yin T., Yan K., Xu X., Li S., Mao J., et al. The Acer truncatum genome provides insights into nervonic acid biosynthesis. Plant J. 2020;104:662–678. doi: 10.1111/tpj.14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Y.F., Zhao D.H., Zhang J.Q., Chen J.S., Li J.L., Weng Z., Rong L.P. De novo transcriptome sequencing and anthocyanin metabolite analysis reveals leaf color of Acer pseudosieboldianum in autumn. BMC Genom. 2021;22:383. doi: 10.1186/s12864-021-07715-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao L., Sun W., Yang J. Development and characterization of microsatellite markers in the critically endangered species Acer yangbiense (Aceraceae) Am. J. Bot. 2011;98:e247–e249. doi: 10.3732/ajb.1100142. [DOI] [PubMed] [Google Scholar]
- 11.Cao Y., Zhang R., Chi B., Li H. Analysis of growth and physiological characteristics of different provenances of Acer truncatum. Liaoning For. Sci. Technol. 2020;1:17–20, 31, 72. [Google Scholar]
- 12.Xiao L., Ren J.Z., Li Q., Yang B., Liu Z.J., Chen R.B., Zhang L. Genome-wide analysis of AP2/ERF superfamily in Isatis indigotica. J. Integr. Med. 2022;21:77–78. doi: 10.1016/j.joim.2022.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Dietz K.J., Vogel M.O., Viehhauser A. AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma. 2010;245:3–14. doi: 10.1007/s00709-010-0142-8. [DOI] [PubMed] [Google Scholar]
- 14.Wang D., Cui B., Guo H., Liu Y., Nie S. Genome-wide identification and expression analysis of the CBF transcription factor family in Lolium perenne under abiotic stress. Plant Signal Behav. 2022;17:2086733. doi: 10.1080/15592324.2022.2086733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H., Gong Y., Sun P., Chen S., Ma C. Genome-wide identification of CBF genes and their responses to cold acclimation in Taraxacum koksaghyz. PeerJ. 2022;10:e13429. doi: 10.7717/peerj.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Z., Ban Q.Y., Hao J., Zhu X.X., Cheng Y.H., Mao J.L., Lin M.L., Xia E.H., Li Y.Y. Genome-wide characterization of the C-repeat Binding Factor (CBF) gene family involved in the response to abiotic stresses in tea plant (Camellia sinensis) Front. Plant Sci. 2020;11:921. doi: 10.3389/fpls.2020.00921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao P.B., Azar S., SanClemente H., Mounet F., Dunand C., Marque G., Marque C., Teulières C. Genome-wide analysis of the AP2/ERF family in Eucalyptus grandis: An intriguing over-representation of stress-responsive DREB1/CBF genes. PLoS ONE. 2015;10:e0121041. doi: 10.1371/journal.pone.0121041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S.C., Lim M.H., Yu J.G., Park B.S., Yang T.J. Genome-wide characterization of the CBF/DREB1 gene family in Brassica rapa. Plant Physiol. Biochem. 2012;61:142–152. doi: 10.1016/j.plaphy.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X., Fowler S.G., Cheng H., Lou Y., Thomashow M.F., Stockinger E.J., Thomashow M.F. Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J. 2010;39:905–919. doi: 10.1111/j.1365-313X.2004.02176.x. [DOI] [PubMed] [Google Scholar]
- 20.Maurya N.K., Goswami A.K., Singh S.K., Prakash J., Kumari A., Chinnusamy V., Talukdar A., Pradhan S., Kumari A. Studies on expression of CBF1 and CBF2 genes and anti-oxidant enzyme activities in papaya genotypes exposed to low temperature stress. Sci. Hortic. 2019;261:108914. doi: 10.1016/j.scienta.2019.108914. [DOI] [Google Scholar]
- 21.Akhtar M., Jaiswal A., Taj G., Jaiswal J.P., Qureshi M.I., Singh N.K. DREB1/CBF transcription factors: Their structure, function and role in abiotic stress tolerance in plants. J. Genet. 2012;91:385–395. doi: 10.1007/s12041-012-0201-3. [DOI] [PubMed] [Google Scholar]
- 22.Haake V., Cook D., Riechmann J.L., Pineda O., Thomashow M.F., Zhang J.Z. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 2002;130:639–648. doi: 10.1104/pp.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakashima K., Ito Y., Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and Grasses. Plant Physiol. 2009;149:88–95. doi: 10.1104/pp.108.129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X., Liang X., Li W., Yao A., Liu W., Wang Y., Yang G., Han D. Isolation and functional analysis of MbCBF2, a Malus baccata (L.) Borkh CBF transcription factor gene, with functions in tolerance to cold and salt stress in transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2022;23:9827. doi: 10.3390/ijms23179827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song J., Wu H., He F., Qu J., Wang Y., Li C., Liu J.H. Citrus sinensis CBF1 functions in cold tolerance by modulating putrescine biosynthesis through regulation of arginine decarboxylase. Plant Cell Physiol. 2022;3:19–29. doi: 10.1093/pcp/pcab135. [DOI] [PubMed] [Google Scholar]
- 26.Maibam P., Nawkar G.M., Park J.H., Sahi V.P., Lee S.Y., Kang C.H. The influence of light quality, Circadian rhythm, and photoperiod on the CBF-mediated freezing tolerance. Int. J. Mol. Sci. 2013;14:11527–11543. doi: 10.3390/ijms140611527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J., Wariss H.M., Tao L., Zhang R., Yun Q., Hollingsworth P., Dao Z., Luo G., Guo H., Ma Y., et al. De novo genome assembly of the endangered Acer yangbiense, a plant species with extremely small populations endemic to Yunnan Province, China. Gigascience. 2019;8:giz085. doi: 10.1093/gigascience/giz085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X., Cai K.W., Han Z.M., Zhang S.K., Sun A.R., Xie Y., Han R., Guo R.X., Tigabu M., Sederoff R., et al. Chromosome-level genome assembly for Acer pseudosieboldianum and highlights to mechanisms for leaf color and shape change. Front. Plant Sci. 2022;13:850054. doi: 10.3389/fpls.2022.850054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navarro M., Ayax C., Martinez Y., Laur J., El Kayal W., Marque C., Teulières C. Two EguCBF1 genes overexpressed in Eucalyptus display a different impact on stress tolerance and plant development. Plant Biotechnol. J. 2011;9:50–63. doi: 10.1111/j.1467-7652.2010.00530.x. [DOI] [PubMed] [Google Scholar]
- 30.Wisniewski M., Norelli J., Artlip T. Overexpression of a peach CBF gene in apple: A model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front. Plant Sci. 2015;6:85. doi: 10.3389/fpls.2015.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welling A., Palva E.T. Involvement of CBF transcription factors in winter hardiness in birch. Plant Physiol. 2008;147:1199–1211. doi: 10.1104/pp.108.117812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X., Cai K., Pei X., Li Y., Hu Y., Meng F., Song X., Tigabu M., Ding C., Zhao X. Genome-wide identification of NAC transcription factor family in Juglans mandshurica and their expression analysis during the fruit development and ripening. Int. J. Mol. Sci. 2021;22:12414. doi: 10.3390/ijms222212414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiong Y., Fei S.Z. Functional and phylogenetic analysis of a DREB/CBF-like gene in perennial ryegrass (Lolium perenne L.) Planta. 2006;224:878–888. doi: 10.1007/s00425-006-0273-5. [DOI] [PubMed] [Google Scholar]
- 34.Yamasaki K., Kigawa T., Seki M., Shinozaki K., Yokoyama S. DNA-binding domains of plant-specific transcription factors: Structure, function, and evolution. Trends Plant Sci. 2013;18:267–276. doi: 10.1016/j.tplants.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 35.Shu Y., Liu Y., Zhang J., Song L., Guo C. Genome-wide analysis of the AP2/ERF superfamily genes and their responses to abiotic stress in Medicago truncatula. Front. Plant Sci. 2016:1247. doi: 10.3389/fpls.2015.01247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y.J., Cao Y., Wang J., Xiong Z. Transcriptome sequencing of Pinus kesiya var. langbianensis and comparative analysis in the Pinus phylogeny. BMC Genom. 2018;19:725. doi: 10.1186/s12864-018-5127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doelling J.H., Pikaard C.S. The minimal ribosomal RNA gene promoter of Arabidopsis thaliana includes a critical element at the transcription initiation site. Plant J. 1995;8:683–692. doi: 10.1046/j.1365-313X.1995.08050683.x. [DOI] [PubMed] [Google Scholar]
- 38.McKhann H.I., Gery C., Bérard A., Lévêque S., Zuther E., Hincha D.K., De Mita S., Brunel D., Téoulé E. Natural variation in CBF gene sequence, gene expression and freezing tolerance in the versailles core collection of Arabidopsis thaliana. BMC Plant Biol. 2008;8:105–118. doi: 10.1186/1471-2229-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang J., Zhang H., Sun T., Shi Y., Wang J., Zhang B., Wang Z., Zhou Y., Gu H. Natural variation of C-Repeat-Binding Factor (CBFs) genes is a major cause of divergence in freezing tolerance among a group of Arabidopsis thaliana populations along the Yangtze River in China. New Phytol. 2013;199:1069–1080. doi: 10.1111/nph.12335. [DOI] [PubMed] [Google Scholar]
- 40.Gehan M.A., Park S., Gilmour S.J., An C., Lee C.M., Thomashow M.F. Natural variation in the C-Repeat Binding Factor cold response pathway correlates with local adaptation of Arabidopsis ecotypes. Plant J. 2015;84:682–693. doi: 10.1111/tpj.13027. [DOI] [PubMed] [Google Scholar]
- 41.Novillo F., Alonso J.M., Ecker J.R., Salinas J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2004;101:3985–3990. doi: 10.1073/pnas.0303029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X.D., Zhuang K.Y., Liu Z.M., Yang D.Y., Ma N.N., Meng Q.W. Overexpression of a novel NAC-type tomato transcription factor, SlNAM1, enhances the chilling stress tolerance of transgenic tobacco. J. Plant Physiol. 2016;204:54–65. doi: 10.1016/j.jplph.2016.06.024. [DOI] [PubMed] [Google Scholar]
- 43.Jung W.J., Seo Y.W. Identification of novel C-repeat binding factor (CBF) genes in rye (Secale cereale L.) and expression studies. Gene. 2019;684:82–94. doi: 10.1016/j.gene.2018.10.055. [DOI] [PubMed] [Google Scholar]
- 44.Liu L., Zheng S., Zheng J. Identification and expression analysis of CBF (C-repeat Binding Factor) gene family in Pomegranate (Punica granatum) Acta Agric. Boreali-Occident. Sin. 2022;31:1154–1167. [Google Scholar]
- 45.Li X., Guo C., Ahmad S., Wang Q., Yu J., Liu C., Guo Y. Systematic analysis of MYB family genes in potato and their multiple roles in development and stress responses. Biomolecules. 2019;9:317. doi: 10.3390/biom9080317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen C.J., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y.H., Xia R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 47.Cao S., Zhang J., Cheng H., Aslam M., Lv H., Dong W., Hu A., Guo M., Liu Q., Qin Y. Identification and evolutionary analysis of FAD2 gene family in green plants. Trop. Plant Biol. 2021;14:239–250. doi: 10.1007/s12042-020-09276-x. [DOI] [Google Scholar]
- 48.Bailey T.L., Johnson J., Grant C.E., Noble W.S. The MEME Suite. Nucleic Acids Res. 2015;43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are contained within the article and Supplementary Materials.