Abstract
Dysregulated endocrine hormones (EHs) contribute to tumorigenesis, but how EHs affect the tumor immune microenvironment (TIM) and the immunotherapy of non–small cell lung cancer (NSCLC) is still unclear. Here, endogenous ouabain (EO), an adrenergic hormone, is elevated in patients with NSCLC and closely related to tumor pathological stage, metastasis, and survival. EO promotes the suppression of TIM in vivo by modulating the expression of immune checkpoint proteins, in which programmed cell death protein ligand 1 (PD-L1) plays a major role. EO increases PD-L1 transcription; however, the EO receptor Na- and K-dependent adenosine triphosphatase (Na, K-ATPase) α1 interacts with PD-L1 to trigger the endocytic degradation of PD-L1. This seemingly contradictory result led us to discover the mechanism whereby EO cooperates with Na, K-ATPase α1 to finely control PD-L1 expression and dampen tumoral immunity. In conclusion, the Na, K-ATPase α1/EO signaling facilitates immune escape in lung cancer, and manipulation of this signaling shows great promise in improving immunotherapy for lung adenocarcinoma.
The Na, K-ATPase α1/EO signaling in lung adenocarcinoma facilitates tumor immune escape by fine-tuning PD-L1 expression.
INTRODUCTION
Lung cancer is the leading cause of cancer-related deaths worldwide. Most lung cancers are non–small cell lung cancer (NSCLC). After diagnosis, the 5-year survival rate of patients with NSCLC is less than 15%. Immune checkpoint blockers like monoclonal antibodies (Abs) against programmed cell death protein 1 (PD-1) and its ligand (PD-L1) have recently demonstrated improved clinical benefits in NSCLC (1). However, the overall response rate to PD-1/PD-L1 blockade rarely exceeds 40% (2), with the main obstacle being the tumor immunosuppressive microenvironment (TIME) (3). Tumor PD-L1 expression significantly affects the clinical efficacy of PD-1/PD-L1 immunotherapy and drug resistance (4, 5). Clarifying the factors affecting PD-L1 expression in TIME is essential for improving PD-1/PD-L1–based immunotherapy.
Endocrine hormones (EHs) significantly shape TIME in cancer (6); however, the role of EH in TIM of NSCLC remains little understood. In endocrine-related cancers, endocrine therapy has become an important treatment option (7). NSCLC is nonhormonally driven cancer, but increasing evidence demonstrated that EHs such as estrogen are important in NSCLC cell growth and therapy (8). Various EH receptors have further been identified on the cell membranes of NSCLC cells (9, 10). Hence, the importance of EH in NSCLC development and the relevant personalized treatment cannot be underestimated.
Endogenous ouabain (EO) is a newly found EH mainly produced by the adrenal glands (11). After binding to the receptor Na- and K-dependent adenosine triphosphatase (Na, K-ATPase), low concentrations of serum EO (ranging from low nanomoles to picomoles, varying between different cells types) can initiate signaling events without significantly affecting the ion transport activity Na, K-ATPase (11, 12). Thus, Na, K-ATPase is also a hormone receptor. EO-mediated signaling is mainly associated with cardiovascular and renal diseases (11). However, it is completely unknown whether EO is involved in cancer development. Accumulating evidence has suggested that Na, K-ATPase could be a potential target for cancer therapy (13). Na, K-ATPase is mainly composed of α and β subunits, and its main isoform in tumors is α1β1 (14). Chemical inhibitors of Na, K-ATPase, also known as cardiac glycosides (CGs), have been shown to have antitumor effects in vitro and in vivo by inhibiting the ion transport activity of Na, K-ATPase and thereby causing intracellular Ca2 + and Na+ disturbance and some are undergoing clinical trials (13). However, factors such as narrow therapeutic window and limited in vivo efficacy have hindered the development of CGs as anticancer drugs (15). In addition, the presence of endogenous Na, K-ATPase ligands has long been overlooked when assessing the antitumor efficacy of CGs. Since the discovery of EO, scientists have predicted that they may have many endogenous Na, K-ATPase ligands in mammals that are chemically similar to, and share Na, K-ATPase receptors with, CGs (11). However, it is unclear whether these endogenous Na, K-ATPase ligands influence the clinical efficacy and safety of CGs in vivo. Without this knowledge, it is difficult to determine whether CGs may be used in cancer treatment.
Here, we described that serum EO was elevated in patients with NSCLC or immunologically intact tumor-bearing mice. Unlike the antitumor effects of CGs, EO was found to promote the development of NSCLC. In cooperation with Na, K-ATPase, EO plays an important role in reprogramming tumor immunity toward an immunosuppressive phenotype by finely controlling PD-L1 expression, on tumor or suppressive immune cells, at both transcriptional and posttranslational levels in TIME. The Na, K-ATPase/EO signaling pathway therefore represents an endocrine pathway that can be exploited by lung cancer cells to facilitate immune escape. On the basis of this finding, EO antagonism and the combined use of CGs or cinobufacini injections with anti–PD-L1 immunoglobulin G (IgG) shows great promise for the immunotherapy of NSCLC.
RESULTS
Low expression of the Na, K-ATPase α1 subunit in NSCLC correlates with poor prognosis
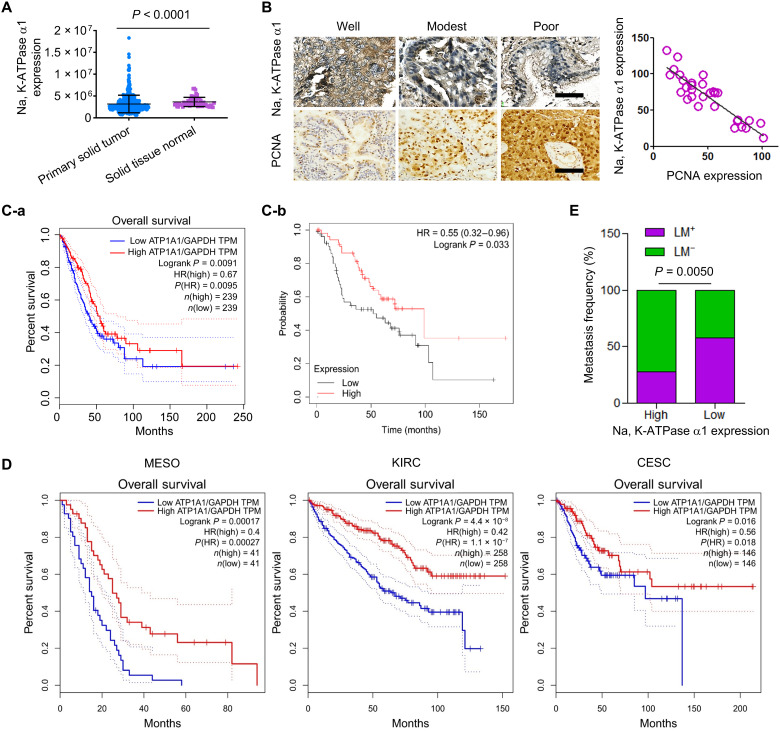
The Tumor Immune Estimation Resource (TIMER) database analysis showed that the overall expression of the Na, K-ATPase α1 subunit was significantly decreased in a variety of cancers including lung adenocarcinoma (fig. S1A). Consistent results were obtained by using the DriverDBv3 database, in which a total of 489 patients with lung adenocarcinoma were enrolled (Fig. 1A). Furthermore, when patients with NSCLC were divided into three groups on the basis of their tumor stages, the protein expression of Na, K-ATPase α1 was significantly lower in late-stage NSCLC (grade 3) than in immediate-stage NSCLC (grade 2) (P < 0.0001; fig. S1B). The expression of Na, K-ATPase α1 decreased with the progression of NSCLC. Those data were confirmed in clinical tumor tissues with different degrees of differentiation; Na, K-ATPase α1 protein expression was significantly decreased in poorly differentiated NSCLC when compared to that of modestly differentiated NSCLC (Fig. 1B, left; and fig. S1C). There is a notable negative correlation between expressions of Na, K-ATPase α1 and proliferating cell nuclear antigen (PCNA) (R = −0.7721) in tumor tissue (Fig. 1B, right). A low Na, K-ATPase α1 expression (ATP1A1) level corresponded with a decrease in overall survival (OS) in The Cancer Genome Atlas (TCGA) (Fig. 1C-a) or BioPortal databases (Fig. 1C-b). Similar results were found in mesothelioma, renal clear cell carcinoma (16) and cervical adenocarcinoma (Fig. 1D). Patients with NSCLC with low Na, K-ATPase α1 expression had a significantly increased incidence of lymphatic metastasis compared with those with high Na, K-ATPase α1 expression (P = 0.005; Fig. 1E).
Fig. 1. Low expression of the Na, K-ATPase α1 in NSCLC correlates with poor prognosis.
(A) Analysis of Na, K-ATPase α1 expression in normal lung tissue or solid tumor of lung adenocarcinoma using DriverDBv3 database [DriverDBV3 (cmu.edu.tw)]. n (tumor) = 489 and n (normal) = 56. (B) Protein expressions of Na, K-ATPase α1 and proliferating cell nuclear antigen (PCNA) in tumor tissues of patients with NSCLC with different degrees of differentiation (left), as examined by immunohistochemical analysis. The correlation between Na, K-ATPase α1 and PCNA expression in tumor tissues of patients with NSCLC (n = 30). (C-a) Analysis of the association between Na, K-ATPase α1 and overall survival (OS) in patients with NSCLC in The Cancer Genome Atlas (TCGA) database. n (high) = 239 and n (low) = 239. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. TPM, Transcripts Per Kilobase of exon model per Million mapped reads. (C-b) Analysis of the association between Na, K-ATPase α1 and OS in patients with advanced NSCLC in the Kaplan-Meier plotter database. n (high) = 53 and n (low) = 52. (D) Analysis of the association between Na, K-ATPase α1 and OS in patients with mesothelioma (MESO) [n (high) = 41 and n (low) = 41], kidney renal clear cell carcinoma (KIRC) [n (high) = 258 and n (low) = 258], and cervical adenocarcinoma (CESC) [n (high) = 146 and n (low) = 146] in TCGA database. (E) Lymphatic metastasis (LM) frequency in patients with NSCLC with different expression levels of Na, K-ATPase α1.
Serum EO is elevated in clinical lung adenocarcinoma and Lewis lung cancer mice
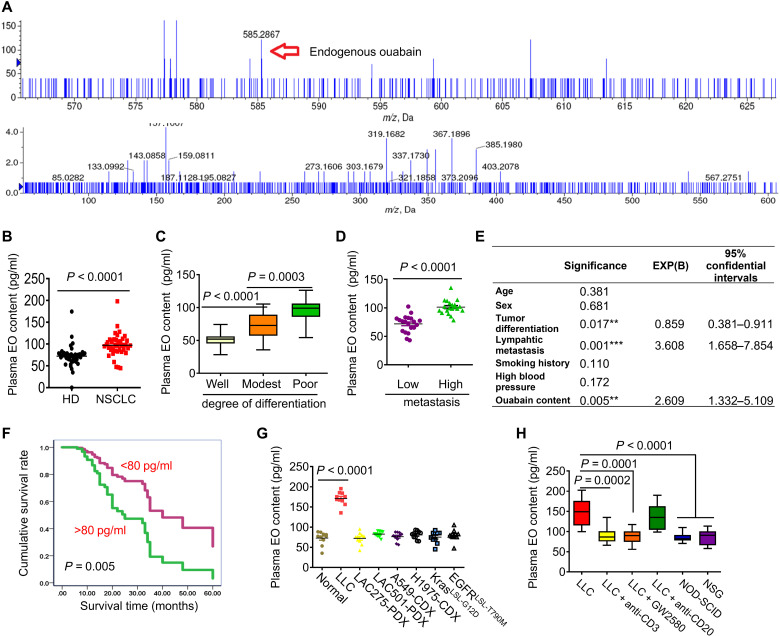
Na, K-ATPase activity in mammals is finely regulated by its endogenous ligands, but only a few of them, such as EO, have been identified (17). It is unknown whether EO is present in patients with NSCLC. By using an AB SCIEX 4600 high-performance liquid chromatography coupled with tandem mass spectrometry (HPLC-MS/MS) instrument, an endogenous compound with a similar molecular weight (585.2867 Da) and fragment ions (403.2078, 337.1730 Da, etc.) as the plant ouabain was found to be present in the plasma of patients with NSCLC (Fig. 2A and fig. S2A). EO is therefore present in the sera of patients with NSCLC and is identical to plant ouabain. The serum content of EO was further quantified by AB SCIEX 5600 HPLC-MS/MS and was significantly higher in patients with NSCLC than in healthy donors (HDs) (P < 0.0001; Fig. 2B). Similar results were obtained by an enzyme-linked immunosorbent assay (ELISA) (fig. S2B). Digoxin is another endogenous Na, K-ATPase ligand that is chemically similar to ouabain; however, endogenous digoxin was not elevated in patients with NSCLC (fig. S2C). The plasma EO levels were significantly higher in poorly differentiated NSCLC than in well or modestly differentiated NSCLC (Fig. 2C), which was contrary to that of Na, K-ATPase α1 protein expression (Fig. 1B). Patients with higher metastasis occurrences showed increased plasma EO levels than those with lower metastasis (P < 0.0001; Fig. 2D). A Cox regression analysis revealed that the level of plasma EO was one of the independent risk factors influencing the survival of patients with NSCLC (Fig. 2E). Patients with higher plasma EO levels (>80 pg/ml) had a shorter survival time than those with lower plasma EO (Fig. 2F). To test whether the elevation of plasma EO also occurred in mouse tumor models, we collected plasma from various mouse models of lung adenocarcinoma with immunocompetent, immunodeficient, or Kras and EGFR gene mutant backgrounds (Fig. 2G). Elevated plasma EO was only detected in immunologically intact Lewis lung cancer (LLC) tumor models. To determine the underlying cause of this, we depleted immune cells in LLC mice using their neutralizing Abs. The increase in plasma EO was significantly abolished in mice after T cell or macrophage depletion (Fig. 2H). LLC cells were then inoculated into T and B cell–deficient nonobese diabetic–severe combined immunodeficient (NOD-SCID) or NOD-SCID IL-2 receptor γ null (NSG) mice with impaired T cell, B cell, and macrophage function. Plasma EO again failed to increase in both strains of mice (Fig. 2H). Thus, immune cells, especially T cells and macrophages, are required for EO production in LLC-bearing mice. Notably, despite immune integrity, plasma EO was not increased in KrasLSL-G12D or EGFRLSL-T790M transgenic mice (Fig. 2G), likely because a single-gene mutation in EGFR or Kras is not sufficient to replicate tumor heterogeneity in the tumor microenvironment (18). LLC mice may therefore be the most appropriate tumor-bearing models when investigating the role of EO in NSCLC.
Fig. 2. Serum EO is elevated in clinical lung adenocarcinoma and LLC mice.
(A) Top: Representative HPLC-MS/MS chromatogram and multiple reaction monitoring (MRM) chromatogram (bottom) of EO in plasma of patients with NSCLC. m/z, mass/charge ratio. (B) Quantitative analysis of EO in the plasma of healthy donors (n = 48) and patients with NSCLC (n = 41) by HPLC-MS/MS. (C) Comparison of plasma EO content in patients with NSCLC with different degrees of differentiation (n = 72). (D) Comparison of plasma EO content in patients with NSCLC with different metastasis frequencies (n = 40). (E) Cox regressive analysis of the risk factors influencing the survival of patients with NSCLC. **P<0.01, ***P<0.001. (F) Comparison of the survival of patients with NSCLC with different plasma EO content (n=80). (G) Measurement of plasma EO in various lung adenocarcinoma mice models (n=10). (H) Measurement of plasma EO in NOD-SCID or NSG immunodeficient mice inoculated with LLC cells, or in T cells-depleted mice using anti-CD3 neutralizing Ab, or in B cells-depleted mice using anti-CD20 neutralizing Ab, or in macrophages-depleted mice using GW2580 (n=10).
EO from the adrenal glands promotes lung cancer development
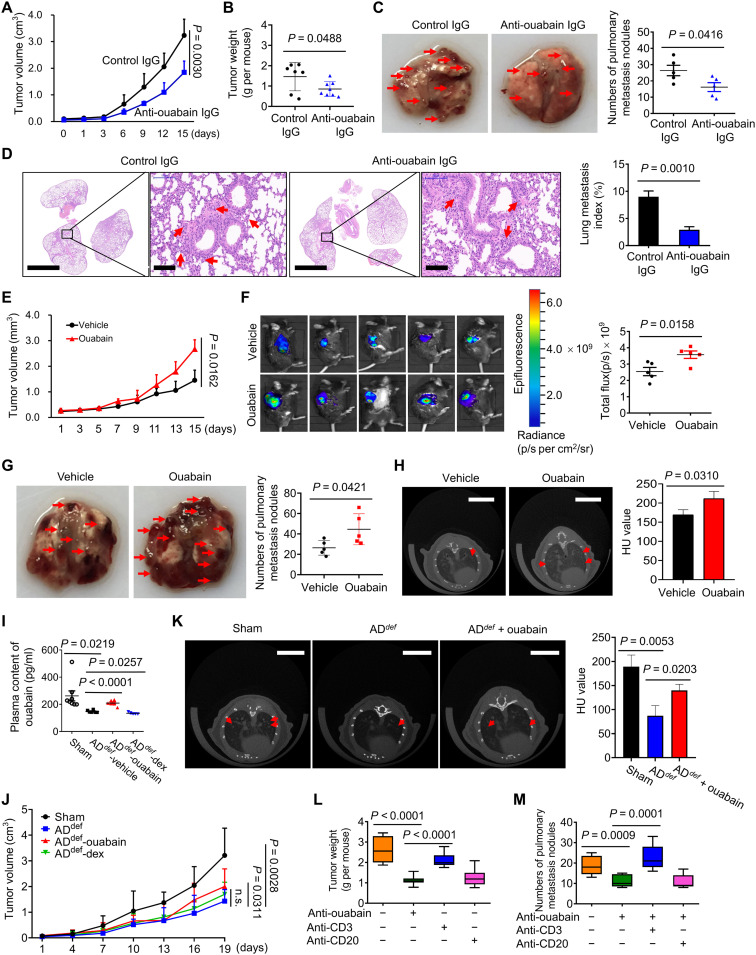
We prepared a rabbit polyclonal anti-ouabain IgG to block the function of EO in vivo. This Ab has not exhibited any cross-reaction with other CGs, and meanwhile, there was no obvious nonspecific binding of this Ab in tissue staining experiments (fig. S2D). The Ab effectively reduced plasma EO (P < 0.0001; fig. S2E) and significantly inhibited LLC tumor growth and weight (Fig. 3, A and B). Similar results were also obtained by rostafuroxin, a well-established chemical blocker for EO (fig. S3A) (19). LLC mice developed spontaneous lung metastasis (20), which was significantly reduced by anti-ouabain IgG, as evidenced by pulmonary metastasis nodules (Fig. 3C), and hematoxylin and eosin staining (Fig. 3D). To further confirm it, we developed nanobodies against ouabain. Details on the development of nanobodies will be reported separately. As shown in fig. S3B, two nanobodies targeting different epitopes of ouabain significantly inhibited the LLC tumor growth. Hence, EO in vivo facilitates tumor development. The LLC mice were further challenged with plant ouabain at 0.1 mg/kg to mimic the effects of EO overproduction, not only because ouabain at this concentration did not cause notable toxicity on mouse hearts, livers, and kidneys (fig. S3, C and D) but also because this dose achieved an average of 115 pg/ml plasma ouabain in mice, falling within the physiological concentration range of EO (fig. S3E). Under this condition, plant ouabain significantly promoted LLC tumor growth (Fig. 3, E and F) and pulmonary metastasis (Fig. 3, G and H, and fig. S3F). Ouabain-induced tumor growth was largely suppressed in mice after treatment with anti-ouabain IgG (fig. S3G) or rostafuroxin (fig. S3A), suggesting the specificity of ouabain’s effect.
Fig. 3. EO promotes lung cancer development in vivo.
(A) Effect of anti-ouabain IgG treatment in vivo on tumor growth (A) and weight (B) (n = 7 to 8). (C) Representative photos of pulmonary metastasis nodules and the metastatic nodules were indicated in red arrows. The numbers of pulmonary metastasis nodules were counted and plotted in right (n = 5). (D) Microscopic metastatic lesions in lungs of mice treated with either control IgG or anti-ouabain IgG, as examined by hematoxylin and eosin staining and quantified by using Image-Pro Plus Software in right (n = 5). Scale bars, 2000 μm; magnified scale bars, 100 μm. (E) Effect of ouabain treatment on LLC tumor growth (n = 5). (F) Representative fluorescence images of the in vivo tumor growth of mice injected with a red fluorescent protein (RFP)–labeled LLC cells, the quantitative analysis was performed in right (n = 5). (G) Representative photos of pulmonary metastasis nodules in control or ouabain-treated groups. The numbers of metastatic nodules were counted and plotted in right (n = 5). (H) Representative microcomputed tomography imaging of mice lung from the vehicle or anti-ouabain treatment group 30 days after inoculation with LLC cells, and the quantitative analysis is shown in right (n = 5). Scale bars, 5 mm. HU, Hounsfiled Unit. (I) The adrenal glands were surgically removed from C57BL/6 mice to produce AGdef mice. The tumor-bearing AGdef mice were challenged with ouabain or dexamethasone to detect the content of ouabain in plasma (n = 5 to 8). (J) The tumor-bearing AGdef mice were challenged with ouabain or dexamethasone to observe their effects on tumor growth (n = 5 to 8). (K) Representative microcomputed tomography imaging of LLC tumor-bearing AGdef mice lung after treatment with ouabain, and the quantification result is shown in right (n = 5 to 8). Scale bars, 5 mm. (L) Effects of T cells or B cells depletion on anti–ouabain-induced tumor growth inhibition and tumor metastasis suppression (M) (n = 9). n.s., not significant.
Adrenal glands are major sites for EO production (11). Bilateral adrenalectomy is a relatively safe method for the treatment of Cushing’s syndrome in humans (21), and it is also a commonly used experimental method to detect the biological function of adrenal-derived hormones (22). Here, in adrenalectomized (AGdef) mice, the plasma EO was significantly reduced (P < 0.05; Fig. 3I). LLC tumor cells were therefore inoculated into adrenalectomized mice to establish the AGdef-LLC tumor model (fig. S4A). Tumor growth was significantly inhibited compared to that in sham-treated mice (Fig. 3J), but the supplementation of AGdef-LLC mice with ouabain significantly restored tumor growth (P = 0.0311). As a control, dexamethasone did not exert this effect. Likewise, LLC metastasis was reduced in AGdef mice but increased following ouabain supplementation (Fig. 3K). We further found that the decrease in tumor growth (Fig. 3L) and pulmonary metastasis (Fig. 3M) by anti-ouabain IgG treatment was largely reversed when T cells, but not B cells, were depleted in LLC mice.
EO leads to an intratumoral immunosuppressive microenvironment
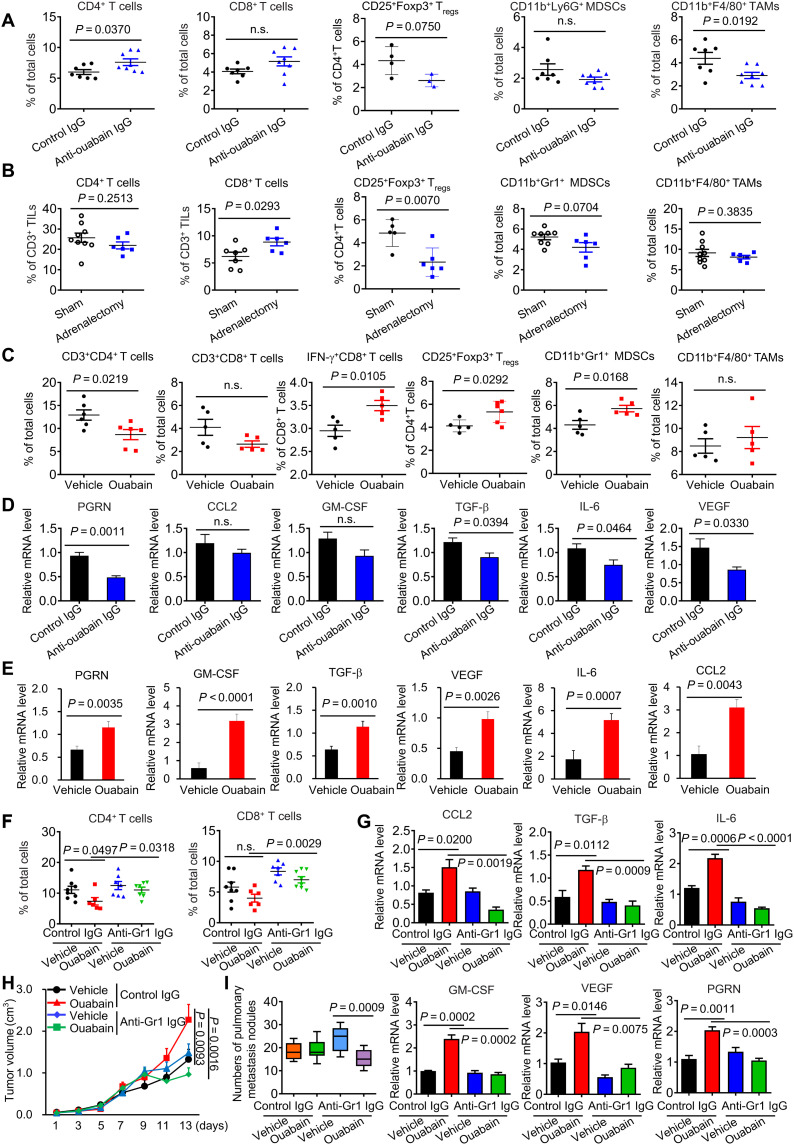
Given the critical role of immune cells in EO production and function, we assessed the possible link between EO and tumor immunity. In LLC mice treated with anti-ouabain IgG, the proportion and the absolute number of CD4+ T cells were significantly increased relative to the control group (Fig. 4A and table S6), but the proportion of CD11b+F4/80 tumor-associated macrophages (TAMs) was significantly reduced. In AGdef-LLC mice, the ratio of CD8+ T cells was increased, but the PD-1+CD3+ T cells decreased (Fig. 4B). Ouabain supplementation in AGdef mice significantly increased PD-1+CD3+ T cells and CD11b+Gr1+ Myeloid-derived suppressor cells (MDSCs) (fig. S4B). In normal mice, ouabain treatment led to a significant reduction in CD3+CD4+ in tumors, but the ratios of IFN-γ+CD8+ T cells, CD11b+Gr1+ MDSCs, and CD25+Foxp3+ regulatory T cells (Tregs) were all increased (Fig. 4C). The expression of immunosuppressive marker genes including progranulin (PGRN), vascular endothelial growth factor (VEGF), transforming growth factor–β (TGF-β), and interleukin-6 (IL-6) was significantly lower in the tumors of the anti-ouabain IgG-treated group than in the vehicle group (Fig. 4D). Similar results were obtained in AGdef mice (fig. S4C). Conversely, ouabain treatment significantly increased the expression of those immunosuppressive cytokines (Fig. 4E). In AGdef mice, ouabain but not dexamethasone supplementation restored immunosuppressive cytokines expression that had been down-regulated following adrenal gland removal (fig. S4C). Because the absolute number of MDSCs is larger than that of Tregs in ouabain-treated tumors (table S6), we sought to examine whether MDSCs contribute to ouabain-mediated immunosuppression; anti-Gr1 IgG was used to deplete MDSCs in vivo. Ouabain-induced reduction in CD4+T cells was significantly reversed by the treatment (Fig. 4F). Similar results occurred in immunosuppressive cytokines expression in tumors (Fig. 4G), tumor growth (Fig. 4H), and metastasis (Fig. 4I).
Fig. 4. EO promotes the formation of tumor immunosuppressive microenvironment.
(A) Percentage of CD4+ T cells, CD8+ T cells, CD4+CD25+Foxp3+ regulatory T cells (Tregs), CD11b+Ly6G+ MDSCs, and CD11b+F4/80+ macrophages in the tumor tissues of the anti–ouabain-treated group, as measured by flow cytometry (n = 3 to 8). (B) Percentage of CD4+ T cells, CD8+ T cells, CD4+CD25+Foxp3+ Tregs, CD11b+Ly6G+ MDSCs, and CD11b+F4/80+ macrophages in the tumor tissues of sham or bilateral adrenalectomy-treated group, as measured by flow cytometry (n = 6 to 8). (C) Percentage of CD3+CD4+ T cells, CD3+CD8+ T cells, IFN-γ+CD8+ T cells, CD4+CD25+Foxp3+ Tregs, CD11b+Ly6G+ MDSCs, and CD11b+F4/80+ macrophages in the tumor tissues of the ouabain-treated group (n = 5 to 6). (D) The quantitative polymerase chain reaction (qPCR) analysis of PGRN, GM-CSF, VEGF, TGF-β, CCL2, and IL-6 mRNA expressions in the tumor tissues of the anti–ouabain-treated group (n = 7 to 8). (E) The qPCR analysis of PGRN, GM-CSF, VEGF, TGF-β, CCL2, and IL-6 expressions in the tumor tissues of the ouabain-treated group (n = 5 to 6). (F) Percentage of CD4+ T cells and CD8+ T cells in the tumor tissues of control IgG, ouabain, anti-Gr1 IgG, or anti-Gr1 IgG + ouabain–treated group (n = 6 to 8). (G) The qPCR analysis of PGRN, GM-CSF, VEGF, TGF-β, CCL2, and IL-6 mRNA expressions in the tumor tissues of control IgG, ouabain, anti-Gr1 IgG, or anti-Gr1 + ouabain–treated groups (n = 6 to 8). Effect of MDSCs depletion by anti-Gr1 IgG on ouabain-induced tumor growth (H) and pulmonary metastasis (I) (n = 6 to 8). TILs, tumor-infiltrating lymphocytes; GM-CSF, granulocyte-macrophage colony-stimulating factor; PGRN, progranulin.
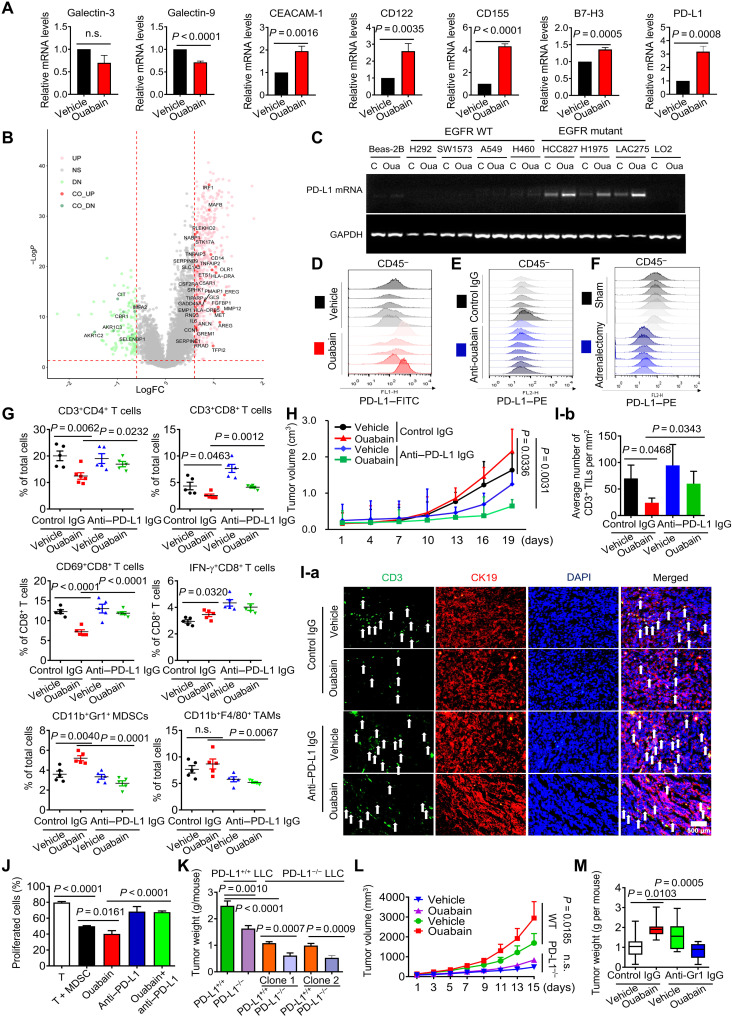
EO modulates immune checkpoint protein expression in tumor and nontumor cells
To characterize the role of ouabain in tumor immunosuppression, we established a cell coculture system with human peripheral blood mononuclear cells (PBMCs) and H1975 cells (fig. S5A). Ouabain not only protected H1975 cells from the killing effect of PBMCs but also enhanced the killing effect of H1975 cells on PBMCs (fig. S5, B to D). Ouabain significantly up-regulated PD-L1, CEACAM-1, CD122, CD155, and B7-H3 gene expression, all of which exert negative regulatory effects on tumor immunity (Fig. 5A) (1, 23). Ouabain also increased gene expression of some costimulatory proteins and positive activators of antigen presentation (fig. S6A). Among those immune-related proteins, PD-L1 is a well-established target for tumor immunotherapy in NSCLC. PD-L1 expression in tumors is closely related to many other genes, including MET (24), IRF1 (25), and so on. When the association of those PD-L1–related genes with ouabain-responsive genes was analyzed, approximately 37 PD-L1–related genes were found to be up- or down-regulated by ouabain (Fig. 5B and table S5), suggesting that PD-L1 may play an important role in ouabain-mediated tumor immunosuppression. Ouabain treatment substantially increased the expression of the PD-L1 gene in a variety of human NSCLC cell lines, especially those carrying EGFR mutations including HCC827, H1975, and LAC275 (Fig. 5C), but had no notable effect in EGFR wild-type (WT) cell lines (A549 and H460) or normal cell types. Consistent results were also obtained in immunoblot analysis (fig. S6B). Furthermore, ouabain time- and dose-dependently increased PD-L1 protein expression in EGFR mutant LAC275 cells but failed to affect EGFR WT SW1573 cells (fig. S6C), suggesting that EGFR status largely determines the cells’ responsiveness to ouabain-mediated PD-L1 up-regulation. In vivo, ouabain treatment significantly increased PD-L1 expression on the cell membrane in CD45− LLC tumor cells (Fig. 5D and fig. S6, D and E). The expression of PD-L1 was significantly reduced when EO production was blocked by anti-ouabain IgG (Fig. 5E and fig. S6F) or in AGdef mice (Fig. 5F and fig. S6G). PD-L1 up-regulation by ouabain occurred not only in lung cancer cells but also in CD11b+F4/80 TAMs (fig. S6H) and CD11b+Gr1+ MDSCs (fig. S6I). Notably, ouabain-induced PD-L1 up-regulation in tumor and nontumor cells was almost completely suppressed by anti-ouabain IgG in vivo (fig. S7, A to G). Ouabain-mediated increases in suppressive cytokines expression were also significantly alleviated by anti-ouabain IgG (fig. S7H).
Fig. 5. PD-L1 plays a critical role in EO-mediated tumor immune escape.
(A) LAC275 cells were treated with ouabain at 50 nM for 6 hours, and mRNA expression was measured by qPCR. (B) The genes whose expression was positively or negatively correlated with PD-L1 were shown. LogFC, log fold change. (C) Cells were treated with ouabain at 50 nM for 6 hours, and the PD-L1 mRNA level was detected by semiquantitative PCR analysis. WT, wild type. (D to F) Representative histograms of PD-L1 surface protein expression on each tumor of LLC mice in different treatment groups [(D), n = 5; (E), n = 7 to 8; (F), n = 6 to 8]. FITC, fluorescein isothiocyanate. (G) Percentage of immune cells in the tumor tissues of different treatment groups (n = 5). (H) Effect of ouabain in combination with anti–PD-L1 IgG on LLC tumor growth (n = 5 to 7). (I-a) The immunofluorescent staining for CD3 (green), CK19 (red), and 4′,6-diamidino-2-phenylindole (DAPI) (blue) in the tumor tissues of different treatment groups. Scale bar, 500 μm. White arrows indicate the tumor-infiltrating CD3+ T cells. (I-b) Quantification of CD3+ T cell number per mm2 (n = 3 to 5). (J) Coculture of activated CD3+ T cells with bone marrow–derived cell under treatment with ouabain, anti–PD-L1 IgG, or both. CD3+T cell proliferation was measured by carboxyl fluorescein succinimidyl ester (CFSE) staining. (K) PD-L1+/+ or PD-L1−/− LLC cells of two clones were inoculated into PD-L1+/+ or PD-L1−/− mice to produce tumor-bearing mice, the mice were challenged with ouabain at 0.1 mg/kg tumor weight was measured (n = 9), and the tumor growth was plotted [(L), n = 9]. (M) PD-L1−/−cells were inoculated into PD-L1+/+ mice, and then the mice were challenged with anti-Gr1 IgG at 250 μg per mouse for 7 days and further treated with ouabain for a successive 12 days; the tumors weight was measured (n = 6 to 8).
PD-L1 up-regulation in tumor and nontumor cells plays a critical role in EO-induced tumor immune escape
The PD-L1 monoclonal blocking Ab was administered to LLC mice starting the day after tumor formation. PD-L1 Ab could eliminate the decrease in CD3+CD4+, CD3+CD8+, and activated CD69+CD8+ T cells induced by ouabain (Fig. 5G). PD-L1 Ab further reversed the ouabain-induced up-regulation of immunosuppressive cytokines expression (fig. S6J), inhibited ouabain-induced tumor growth (Fig. 5H), and improved intratumoral T lymphocyte infiltration (Fig. 5I), indicating that ouabain-mediated changes in immune infiltrate are critically dependent on PD-L1 expression. In the cell coculture system as above mentioned (fig. S5A), silencing of PD-L1 expression or using PD-L1 Ab attenuated the killing effect of H1975 on PBMCs in the presence of ouabain (fig. S5, E to G). In addition, in a coculture system with MDSCs and activated CD3+ T cells (fig. S8A), ouabain-mediated CD3+ T cell proliferation suppression was significantly attenuated in the presence of PD-L1 Ab treatment (Fig. 5J and fig. S8, B to D).
We thus generated PD-L1−/− LLC cells by CRISPR-Cas9. The PD-L1−/− LLC cells of two different clones were inoculated into C57BL6 mice to produce tumor-bearing models. PD-L1−/− LLC tumor growth was suppressed after ouabain treatment as opposed to the PD-L1+/+ LLC group (P < 0.0001; Fig. 5K). PD-L1 expression in tumor cells, therefore, contributed to ouabain-mediated tumor growth. The PD-L1+/+ LLC cells were then inoculated into PD-L1−/− or PD-L1+/+ mice. Ouabain-induced tumor growth in PD-L1+/+ mice was significantly attenuated compared to PD-L1−/− mice (Fig. 5, K and L), suggesting that other nontumor cells also participate in this process. To verify this, we used anti-Gr1 IgG to deplete MDSCs in PD-L1+/+ mice inoculated with PD-L1−/− LLC cells. PD-L1−/− LLC tumor growth under ouabain treatment was significantly alleviated in the presence of anti-Gr1 Ab (P = 0.0005; Fig. 5M). Notably, we also observed that anti-ouabain IgG failed to suppress tumor growth in PD-L1−/− LLC mice (fig. S8E). This result, together with that in Fig. 5L, indicated that PD-L1 is critical for ouabain/EO-induced tumor growth. Together, PD-L1 up-regulation in tumor and nontumor cells contributed to ouabain-induced immunosuppression and tumor development.
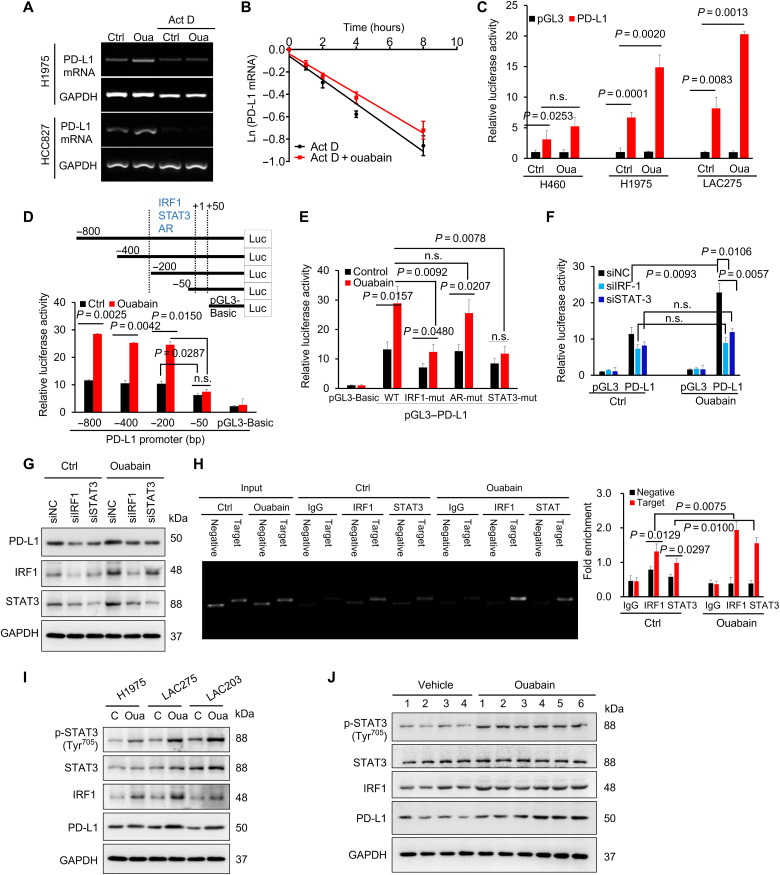
Ouabain activates PD-L1 gene transcription
The ouabain-induced increase in PD-L1 mRNA expression was substantially inhibited when actinomycin D was used to halt global gene transcription (Fig. 6A); however, ouabain failed to notably affect the half-life of PD-L1 mRNA (Fig. 6B). We further cloned the human PD-L1 promoter into the luciferase reporter construct (Fig. 6C). Ouabain significantly increased the PD-L1 promoter activity in H1975 and LAC275 cells but not H460 cells. Truncated variants of the full-length PD-L1 promoter were constructed (Fig. 6D). The ouabain-mediated activation of the PD-L1 promoter was completely abolished in the −50 to +50–base pair (bp) truncated variant, suggesting that an ouabain-responsive cis-element may be present in the PD-L1 promoter between −200 and −50 bp. Bioinformatics analysis revealed the presence of interferon regulatory factor 1 (IRF1), androgen receptor (AR), and signal transducer and activator of transcription 3 (STAT3) binding sites (−200 to −50) within this region. PD-L1 promoter variants with deletions of the IRF1, AR, or STAT3 binding sites were constructed. The ouabain-induced up-regulation of PD-L1 gene promoter activity was largely suppressed when the STAT3 or IRF1 binding sequences were deleted (Fig. 6E). Ouabain further failed to increase PD-L1 promoter activity (Fig. 6F) and PD-L1 protein expression (Fig. 6G) in LAC275 cells, in which IRF1 and STAT3 expression was silenced. Chromatin immunoprecipitation (ChIP) assays confirmed that lysates of ouabain-treated cells exhibited increased IRF1 and STAT3 binding to the TGTTTCACTTTCT and TTCACCGAAG sequences (Fig. 6H). Ouabain increased STAT3 phosphorylation and the protein expression of IRF1 in H1975, LAC275, and LAC203 cells (Fig. 6I). Similar results were found in the LLC tissues (Fig. 6J). Last, to understand why EGFR mutant lung cancer cells are more sensitive to ouabain-mediated PD-L1 up-regulation than EGFR WT cells, we observed a significant increase in epidermal growth factor receptor (EGFR) phosphorylation in ouabain-treated EGFR mutant cells (H1975, LAC275, and LAC203 cells) but not in EGFR WT cells (H460) (fig. S9A). Furthermore, ouabain-induced PD-L1 gene transcription in LAC275 cells can be significantly blocked by erlotinib, an EGFR tyrosine kinase inhibitor (fig. S9B).
Fig. 6. Ouabain activated the PD-L1 gene promoter through signal transducer and activator of transcription 3 and interferon regulatory factor 1.
(A) PCR analysis of PD-L1 mRNA expression in lung cancer cells after the treatment of actinomycin D (Act D; 8 μg/ml) or ouabain (50 nM) or both for 4 hours. (B) LAC275 cells were treated with actinomycin D (8 μg/ml) or ouabain + actinomycin D (8 μg/ml) for different times. The mRNA levels were measured with qPCR analysis, and the half-life of PD-L1 mRNA was calculated. (C) LAC275, H460, and H1975 cells were transfected with the human PD-L1 full-length promoter-luciferase construct and were incubated without or with ouabain (50 nM) for 6 hours. the transcriptional activity of PD-L1 was determined by dual-luciferase reporter assay. (D) LAC275 cells were transfected with the deletion mutants of the human PD-L1 promoter, and the luciferase activity of each construct was determined. bp, base pair. (E) The PD-L1 promoter constructs with mutations in specific IRF1, AR, and STAT3 sites were transfected into cells; and the luciferase activity was measured. (F) Effect of interferon regulatory factor 1 (IRF1) and signal transducer and activator of transcription 3 (STAT3) knockdown on PD-L1 luciferase reporter activity. siNC, siRNA negative control. (G) The PD-L1 protein expression in LAC275 cells after IRF1 or STAT3 expression was silenced. (H) Binding of IRF1 and STAT3 into PD-L1 promoter, as examined by ChIP-qPCR assay. (I) Protein expressions levels of STAT3, phosphor (p)–STAT3, IRF1, and PD-L1 in lung cancer cells after treatment with ouabain (50 nM) for 8 hours. (J) Protein expression levels of STAT3, phosphor-STAT3, IRF1, and PD-L1 in tumor tissues from groups treated with vehicle or ouabain (0.1 mg kg−1).
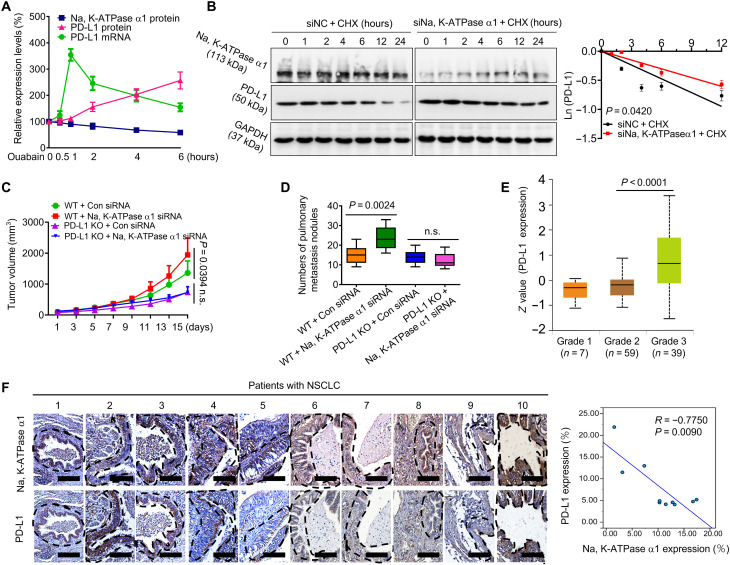
Na, K-ATPase α1 negatively regulates PD-L1 protein expression in lung adenocarcinoma
EO promotes immune escape by up-regulating PD-L1. However, it is unclear how EO is inversely correlated with Na, K-ATPase α1 expression in NSCLC. We found that the effect of ouabain on PD-L1 transcription was indeed dependent on Na, K-ATPase α1 (fig. S10, A to C); however, the protein expression of Na, K-ATPase α1 gradually decreased with the duration of ouabain treatment (Fig. 7A and fig. S10D). Conversely, the protein expression of PD-L1 continuously increased. To determine why, we noted that silencing Na, K-ATPase α1 could also notably increase the protein expression of PD-L1 even without ouabain treatment (fig. S10E). Na, K-ATPase α1 silencing failed to increase PD-L1 promoter activity; however, in a cycloheximide chase experiment, PD-L1 protein degradation was significantly delayed in Na, K-ATPase α1–silenced cells compared with control cells (Fig. 7B). Na, K-ATPase α1 therefore negatively regulates PD-L1 protein stability. In support of it, Na, K-ATPase α1 overexpression decreased membrane PD-L1 abundance (fig. S10F). Silencing Na, K-ATPase α1 expression, however, led to increased PD-L1 expression in LLC mice (fig. S10, G and H). When LLC mice were intratumorally delivered with lentivirus expressing Na, K-ATPase α1 short hairpin RNAs (shRNAs), tumor growth (Fig. 7C) and metastasis (Fig. 7D) were increased; however, this effect was largely attenuated in the absence of PD-L1 expression.
Fig. 7. Na, K-ATPase α1 negatively regulates PD-L1 protein expression in lung adenocarcinoma.
(A) LAC275 cells were treated with ouabain (50 nM) for 0.5, 1, 2, 4, and 6 hours; after treatments, Na, K-ATPase α1 protein, PD-L1 protein, and PD-L1 mRNA expression were examined by Western blot or qPCR analysis, respectively. (B) Left: LAC275 cells were transfected with small interfering RNA (siRNA) against Na, K-ATPase α1 or control siRNA and then treated with cycloheximide (CHX; 20 μg/ml) for different times, the indicated proteins were detected by Western blotting, and the half-life of PD-L1 protein was determined in right. (C) The PD-L1+/+(WT) or PD-L1−/−(KO) (knockout) LLC cells were inoculated into C57/BL6 to produce tumor-bearing mice; after tumor formation, the lentiviral Na, K-ATPase α1 shRNAs or control shRNAs were intratumorally delivered into mice, tumor size was monitored every 2 days, and the pulmonary metastasis of each group was quantified in (D) (n = 9). (E) Analysis of PD-L1 expression in patients with NSCLC with different pathological grades using the University of Alabama at Birmingham cancer data analysis Portal (UALCAN) database. (F) Left: Immunohistochemical analysis of the Na, K-ATPase α1 and PD-L1 protein expression in tumor tissues from patients with NSCLC. Scale bars, 200 mm. Right: Pearson correlation between the PD-L1 and Na, K-ATPase α1 expression in tumor tissue (n = 10).
The data from TCGA showed that, in patients with NSCLC, the protein expression of Na, K-ATPase α1 was inversely correlated with PD-L1 at almost all stages (Fig. 7E and fig. S1B). With the progression of NSCLC, the expression of PD-L1 had a notable upward trend. In fresh tumor tissues, the expression of Na, K-ATPase α1 and PD-L1 showed a significant negative correlation, with a correlation coefficient of 0.775 (Fig. 7F). However, this correlation did not occur in paracancerous tissues.
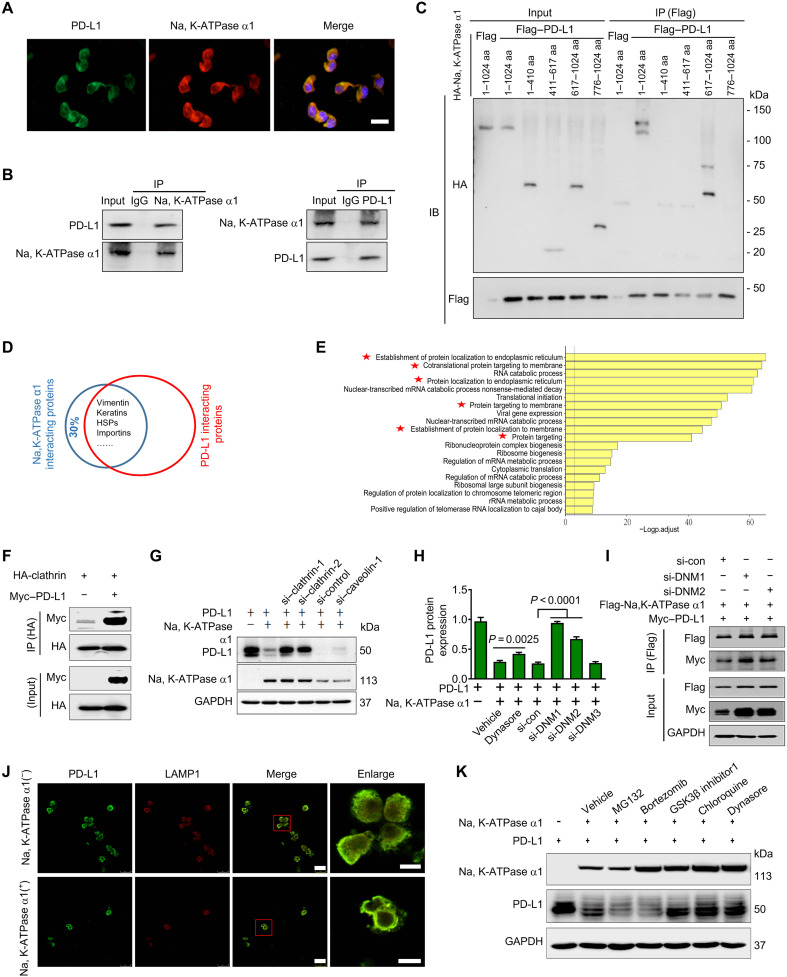
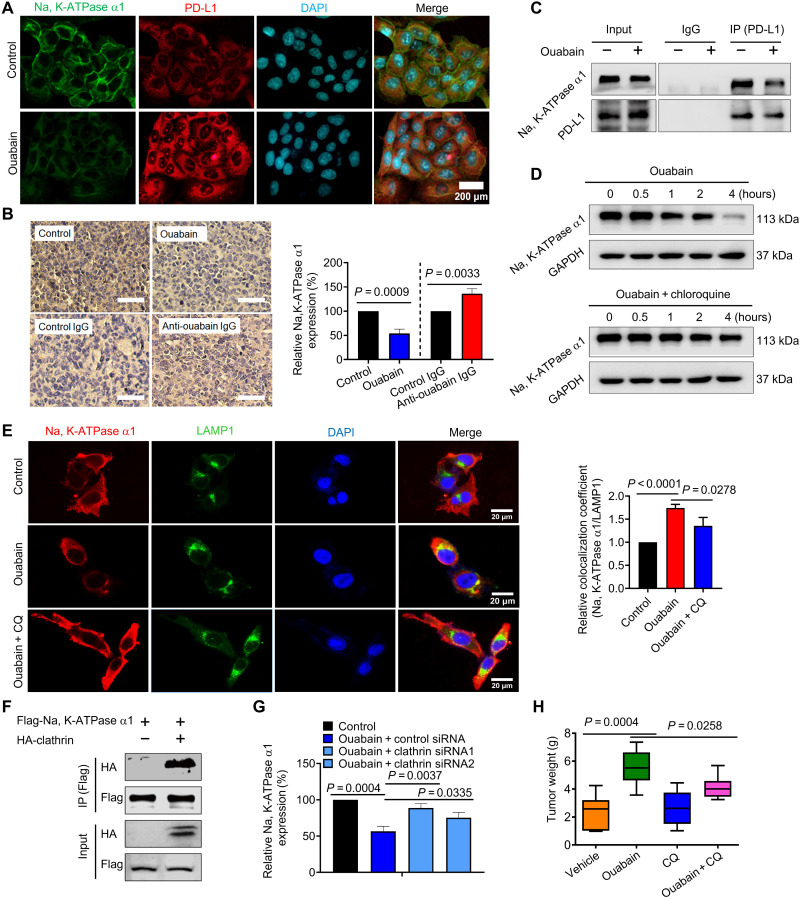
Na, K-ATPase α1 interacts with PD-L1 and promotes PD-L1 endocytosis
Na, K-ATPase α1 and PD-L1 were colocalized in the cytoplasm and cell membrane, indicating a potential interaction between them (Fig. 8A). Endogenous Na, K-ATPase α1 was found to interact with PD-L1 and vice versa (Fig. 8B). PD-L1/Na, K-ATPase α1 interaction was also detected in human embryonic kidney–293 T cells where they were coexpressed (fig. S11A). By constructing deletion mutants of Na, K-ATPase α1 and PD-L1, Na, K-ATPase α1 was found to interact with PD-L1 each other through their cytoplasmic domains (Fig. 8C and fig. S11B).
Fig. 8. Na, K-ATPase α1 interacts with PD-L1 and promotes PD-L1 endocytosis.
(A) The immunofluorescence analysis of the cellular location of Na, K-ATPase α1 and PD-L1 in LAC275 cells. Scale bar, 100 μm. (B) The interaction between endogenous Na, K-ATPase α1 and PD-L1. (C) The interaction between Na, K-ATPase α1 deletion mutant variants and PD-L1 in 293T cells. IB, immunoblot; aa, amino acids. (D) The shared proteins that interact with both Na, K-ATPase α1 and PD-L1 by immunoprecipitation mass spectrometry (IP-MS) analysis. HSP, Heat shock proteins. (E) The gene oncology enrichment analysis of the shared proteins in (D). (F) The interaction between HA-tagged clathrin and Myc-tagged PD-L1 in LAC275 cells. HA, hemagglutinin. (G) LAC275 cells were transfected with siRNAs against clathrin or caveolin-1, further cotransfected with Na, K-ATPase α1 and PD-L1 for 24 hours. Na, K-ATPase α1 and PD-L1 protein expressions were examined. (H) LAC275 cells were transfected with siRNAs against dynamin-1 (si-DNM1), dynamin-2, or dynamin-3, and further cotransfected with Na, K-ATPase α1 and PD-L1 for 24 hours. PD-L1 protein expression was examined. Cells were also treated with dynasore for 1 hour before harvesting. (I) LAC275 cells were transfected with siRNAs against dynamin-1 and dynamin-2, and further cotransfected with Flag-tagged Na, K-ATPase α1 and Myc-tagged PD-L1 for 24 hours, Na, K-ATPase α1/PD-L1 interaction was detected by immunoblotting. (J) The lysosomal colocalization of PD-L1 and LAMP1. LAMP1 was used to stain the lysosome. Scale bars, 25 μm; magnified scale bars, 5 μm. (K) After transfection with PD-L1 and Na, K-ATPase α1 for 24 hours, LAC275 cells were treated with different chemical inhibitors for 1 to 5 hours, and protein expressions of Na, K-ATPase α1 and PD-L1 were examined.
Notably, the immunoprecipitation MS experiments revealed that nearly 70% of Na, K-ATPase α1–interacting proteins can also interact with PD-L1, suggesting that Na, K-ATPase α1 may form a protein complex with PD-L1 (Fig. 8D and data files S1 and S2) (26). Most of those shared interacting proteins are associated with membrane protein targeting (Fig. 8E). For example, both Na, K-ATPase α1 and PD-L1 can bind to vimentin, Rab, and adenosine 5′-diphosphate ribosylation factor 1 family proteins that are closely related to vesicular trafficking (fig. S12A) (26, 27). We therefore speculated whether Na, K-ATPase α1 could trigger the internalization and degradation of PD-L1. Clathrin-mediated endocytosis (CME) is a common way for membrane proteins to internalize and transmit signals (28). Proteins that undergo CME contain sequences that can be recognized by adaptor protein (AP) complex such as AP-2 (28, 29). Human PD-L1 contains such characteristic sequences (fig. S11C), and they are conserved across species (fig. S11D). PD-L1 was further found to interact with clathrin (Fig. 8F and fig. S11E). Na, K-ATPase α1–induced PD-L1 down-regulation was substantially prevented in clathrin-silenced cells (Fig. 8G). There is growing evidence that dynamin-1 is highly expressed in lung cancer to trigger rapid and deregulated CME (30, 31). Here, Na, K-ATPase α1–mediated PD-L1 down-regulation was largely attenuated in cells after treatment with dynasore, a dynamin inhibitor (Fig. 8H), or in cells with reduced expression of dynamin-1 and dynamin-2 but not dynamin-3. The Na, K-ATPase α1/PD-L1 interaction was strengthened in cells after silencing dynamin-1 or dynamin-2 (Fig. 8I). Last, the Na, K-ATPase α1–induced PD-L1 down-regulation was substantially prevented in dynamin-1−/− cells relative to the controls (fig. S11F). To examine whether Na, K-ATPase α1 delivered PD-L1 into the lysosome for degradation, we found, in immunofluorescence analysis, that in the presence of Na, K-ATPase α1 overexpression, PD-L1 was densely colocalized with lysosomes (Fig. 8J). The Na, K-ATPase α1–mediated down-regulation of PD-L1 was greatly suppressed in the presence of the lysosomal inhibitor chloroquine (CQ), dynasore, or glycogen synthase kinase 3β (GSK3β) inhibitor but not proteasomal inhibitors including MG132 and bortezomib (Fig. 8K).
Ouabain inhibits the Na, K-ATPase α1/PD-L1 interaction and promotes the degradation of Na, K-ATPase α1 subunit to enhance PD-L1 expression
Ouabain-induced Na, K-ATPase–dependent PD-L1 transcription and PD-L1–dependent immune escape, but Na, K-ATPase promotes PD-L1 degradation, and there is a negative correlation between EO and Na, K-ATPase expression. We therefore hypothesized that sustained ouabain treatment may intentionally induce Na, K-ATPase α1 degradation to maintain PD-L1 expression. As anticipated, ouabain yielded notably reduced Na, K-ATPase α1 expression but enhanced PD-L1 expression in LAC275 cells (Fig. 9A). The in vivo administration of ouabain also resulted in a significant reduction of Na, K-ATPase α1 expression in LLC tumor tissues. Anti-ouabain IgG, however, improved Na, K-ATPase α1 expression in most cases (Fig. 9B). We further found that ouabain inhibited the interaction between endogenous Na, K-ATPase α1 and PD-L1 (Fig. 9C). The ouabain-induced down-regulation of Na, K-ATPase α1 was greatly prevented by the lysosomal inhibitor CQ, suggesting that ouabain triggers the lysosome-mediated degradation of Na, K-ATPase α1 (Fig. 9D). An immunofluorescence analysis showed that a large amount of Na, K-ATPase α1 colocalized with lysosomal associated membrane protein 1 (LAMP1) in ouabain-treated cells, an effect that was prevented by CQ treatment (Fig. 9E). As with PD-L1, Na, K-ATPase α1 interacted with clathrin (Fig. 9F). The ouabain-mediated down-regulation of Na, K-ATPase α1 was further largely suppressed in cells after clathrin expression had been silenced (Fig. 9G). Last, the tumor-promoting effect of ouabain was significantly suppressed when LLC mice were treated with ouabain in combination with CQ (Fig. 9H).
Fig. 9. Ouabain inhibits Na, K-ATPase α1/PD-L1 interaction and promotes lysosomal degradation of Na, K-ATPase α1.
(A) LAC275 cells were treated with ouabain (50 nM) for 6 hours, and the cellular expression and distribution of Na, K-ATPase α1 and PD-L1 were detected by immunofluorescence analysis. (B) LLC mice were challenged with ouabain or anti-ouabain IgG, then immunohistochemical analysis was performed to examine Na, K-ATPase α1 expression in tumor tissues, and the quantification results are shown in right (n = 3 to 5). (C) The interaction between endogenous Na, K-ATPase α1/PD-L1 in LAC275 cells after treatment with ouabain (50 nM). (D) LAC275 cells were treated with ouabain alone or ouabain + CQ at different times, and then Na, K-ATPase α1 protein expression was examined. (E) Left: LAC275 cells were treated with ouabain (50 nM) or ouabain + CQ for 12 hours, and then the lysosomal location of Na, K-ATPase α1 (red) was detected by immunofluorescence. Scale bars, 20 μm. Right: Colocalization coefficient (Na, K-ATPase α1/LAMP1) was analyzed by ImageJ software. (F) The interaction between HA-tagged clathrin and Flag-tagged Na, K-ATPase α1 in LAC275 cells. (G) After transfection with control siRNA or siRNAs against clathrin, LAC275 cells were treated with ouabain at 50 nM for 8 hours, and membrane Na, K-ATPase α1 abundance was detected by flow cytometry analysis. (H) LLC mice were challenged with ouabain (0.1 mg/kg), CQ, or both for a successive 15 days, and after experiments, tumors were removed and weighed (n = 7 to 8).
Na, K-ATPase α1/EO signaling influences NSCLC immunotherapy
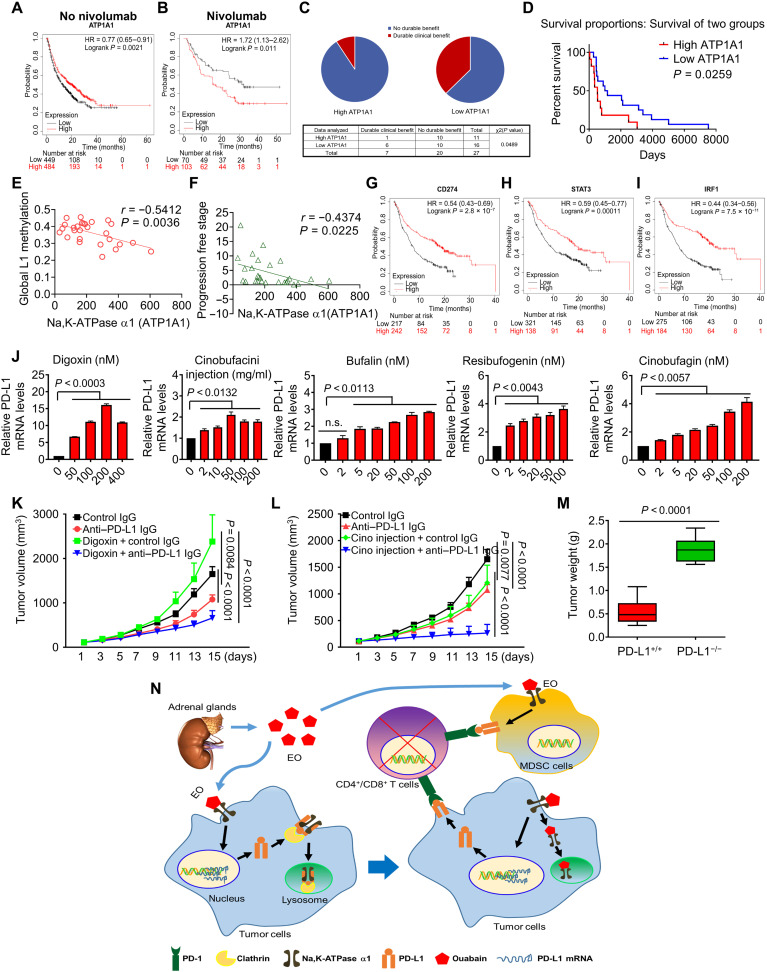
Because Na, K-ATPase α1 negatively regulates PD-L1 expression, we analyzed the relationship between Na, K-ATPase α1 expression and patient response to anti–PD-1/PD-L1 immunotherapy using the Kaplan-Meier plotter database (32). Patients with nine different types of cancer were included in the analysis. As expected, patients with low levels of Na, K-ATPase α1 expression have relatively poor OS compared with patients with high expression (Fig. 10A). However, among nivolumab-treated patients, those with low Na, K-ATPase α1 expression showed better responsiveness and survival than those with high expression (P = 0.011; Fig. 10B). To be more specific for lung adenocarcinoma, we analyzed the data in GSE135222, where patient reactivity to anti–PD-1/PD-L1 IgG was documented. The result indicated that the Na, K-ATPase α1–low-expression group obtained a more significant clinical benefit relative to the high-expression group (Fig. 10C). In addition, the survival time of patients with low Na, K-ATPase α1 expression was significantly longer than that of patients with high expression (Fig. 10D). On the basis of the data in GSE135222, the report showed that patients with NSCLC with increased genomic global L1 methylation benefited more from anti–PD-1/PD-L1 therapy (33). Consistently, Na, K-ATPase α1 expression in patients with NSCLC was inversely associated with global L1 methylation (Fig. 10E) and progression-free stage (Fig. 10F). Ouabain-mediated PD-L1 up-regulation is mainly dependent on STAT3 and IRF1. Notably, patients with NSCLC with high expression of PD-L1(CD274) (Fig. 10G), STAT3 (Fig. 10H), or IRF1 (Fig. 10I) all benefited from anti–PD-L1 immunotherapy more than those with low expression.
Fig. 10. Na, K-ATPase α1/EO signaling in the immunotherapy of lung adenocarcinoma.
(A) The survival of patients with cancer with different expression levels of Na, K-ATPase α1, as analyzed by data in the Kaplan-Meier plotter database. (B) The survival of patients with cancer with different expression levels of Na, K-ATPase α1 after nivolumab treatment. Patients with bladder cancer, esophageal adenocarcinoma, glioblastoma, hepatocellular carcinoma, head and neck squamous cell carcinoma, melanoma, NSCLC, small cell lung cancer, and urothelial cancer were included in the analysis (A and B). Clinical benefit (C) and survival (D) of NSCLC patients with the different expression levels of Na, K-ATPase α1 after anti–PD-1/PD-L1 therapy; the data are obtained from GSE135222. Correlation of Na, K-ATPase α1 expression level in patients with NSCLC with global L1 methylation (E) and progression-free stage (F). The survival of patients with different expression levels of PD-L1 (CD274) (G), STAT3 (H), or IRF1 (I) after anti–PD-1 IgG therapy. The patients included are the same as that in (A) and (B). (J) LAC275 cells were treated with different dosages of digoxin, cinobufacini injection, bufalin, cinobufagin, and resibufogenin for 6 hours; PD-L1 mRNA expression was detected by qPCR analysis. LLC mice were challenged with digoxin (K) and cinobufacini injection (L) in combination with anti–PD-L1 IgG for a successive 15 days, and tumor size was monitored every 2 days (n = 7 to 8). (M) PD-L1+/+ and PD-L1−/− LLC cells were inoculated into C57/BL6 mice to produce a tumor-bearing model and then challenged with cinobufacini injection in combination with anti–PD-L1 IgG, and after experiments, tumors were removed and weighed (n = 9). (N) Na, K-ATPase α1/EO signaling promotes tumor immune escape in lung adenocarcinoma by fine-tuning PD-L1 expression.
Structural analogs of ouabain, also known as CGs, are abundantly present in natural products including digoxin in digitalis and bufalin in toad venom (fig. S12B) (13, 34). The cinobufacini injection is refined from the dried skin extract of the toad Bufo gargarizans in modern Chinese medicine. The major CGs in cinobufacini injections are bufalin, cinobufagin, and resibufogenin (fig. S12B). In the clinical treatment of NSCLC, cinobufacini injections are often used in combination with chemotherapy, radiotherapy, or targeted therapy, and the synergistic effects are remarkable (35, 36). However, it is unclear whether cinobufacini injections can enhance the effect of PD-L1–targeted immunotherapy.
We found that digoxin, cinobufacini injections, and their three major CGs significantly enhanced PD-L1 mRNA expression in NSCLC (Fig. 10J). As with ouabain, digoxin treatment at 0.1 mg/kg in vivo increased tumor growth; however, when it was used in combination with anti–PD-L1 IgG, a significant reduction in tumor growth was observed (Fig. 10K). Unlike digoxin, cinobufacini injections (Fig. 10L) and their three CGs (fig. S12, C to E) had a significant inhibitory effect on LLC tumor growth when used alone. LLC tumor growth was further significantly inhibited when they were combined with anti–PD-L1 IgG, especially in the group in which cinobufacini injections had been combined with anti–PD-L1 IgG (Fig. 10L). The antitumor synergy between anti–PD-L1 IgG and the cinobufacini injections was greatly diminished in PD-L1–deficient LLC cells (P < 0.0001; Fig. 10M).
DISCUSSION
To the best of our knowledge, this is the first study to report the EO presence in lung adenocarcinoma and describe its role in the regulation of cancer immunity. The serum EO content is negatively correlated with Na, K-ATPase α1 expression in tumors from patients with NSCLC or LLC mice. EO is further able to promote tumor immune escape by inducing PD-L1 expression in tumor and suppressive tumor–associated immune cells. Ouabain induces Na, K-ATPase α1–dependent PD-L1 transcription; Na, K-ATPase α1,however, posttranslationally induces the endocytic degradation of PD-L1. This seemingly contradictory result led us to discover the mechanism whereby ouabain cooperates with Na, K-ATPase α1 to finely control PD-L1 expression in cancer cells. Ouabain promotes cellular PD-L1 supply through rapid transcriptional activation, and with the accumulation of translated PD-L1 protein, membrane Na, K-ATPase α1 interacts with PD-L1 and induces its internalization and degradation, preventing PD-L1 from entering the cell membrane. This may have been a feedback mechanism activated by cells to maintain PD-L1 expression within a reasonable range. However, this benefit is counteracted by continued ouabain treatment, which inhibits the interaction between Na, K-ATPase α1 and PD-L1 and induces the lysosomal degradation of Na, K-ATPase α1. Although the transcription rate of PD-L1 is halted, more translated PD-L1 proteins enter the cell membrane. By taking advantage of these subtle mechanisms, ouabain supports the attack of CD4+/CD8+ T cells by tumor or tumor suppressor immune cells (Fig. 10K).
As a hormone-like substance, EO is similar to cortisol in structure, production, release, and metabolism. However, contrary to the known roles of cortisol in human physiology, pathology, and pharmacology, the biological function of EO is far from understood. Here, we confirmed that serum EO in patients with NSCLC or LLC mice is identical to plant ouabain by HPLC-MS/MS and ELISA methods. Additional functional evidence is as follows: (i) The anti-ouabain IgG significantly reduced serum EO and activated tumor immunity in LLC mice. (ii) After the adrenalectomies in the LLC mice to reduce EO production, tumor immunity was activated, but this was reversed by ouabain supplementation. (iii) Low concentrations of ouabain can mimic almost all the effects of EO on lung cancer development, and this can be significantly suppressed by anti-ouabain IgG or rostafuroxin. (iv) EO is specifically elevated in NSCLC, although digoxin has a similar chemical structure and protumorigenic effect as that of ouabain, and endogenous digoxin was not detected in NSCLC. Therefore, serum EO is present and has a biological function in lung adenocarcinoma. In this regard, it is necessary to emphasize that, although EO is chemically identical to plant ouabain, they are conceptually different. EO is an endogenously produced substance in mammals, and studies on the biological function of EO should be carried out under specific physiological or pathological circumstances. Plant ouabain exhibits antitumor activity in vitro, but this does not mean that EO has a similar in vivo effect. Ouabain has to be used at relatively high concentrations (micromole or millimole) in vitro to inhibit Na, K-ATPase ion-transporting activity and has a cytotoxic effect on transformed cells; however, the physiological concentration of EO is normally at the level of picomol, such a low concentration of EO does not significantly affect the ion-transporting activity of Na, K-ATPase but uses Na, K-ATPase as scaffold proteins to initiate signals events, leading to Src activation and cell proliferation.
In the present study, ouabain increased the expression of immunosuppressive genes including IL6, VEGF, CCL2, TGF-β, PGRN, and GM-CSF and increased the expression of some immunosuppressive checkpoint genes including PD-L1, CEACAM-1, CD155, CD122, and B3-H4. A previous study also showed that ouabain reduced the expression of checkpoint protein 2′3′-dioxygenase 1 in A549 cells, although whether this effect also occurs in vivo is unknown (37). The immunosuppressive effect of ouabain is therefore not caused by PD-L1 alone but rather the combined effect of multiple tumor immune–related proteins. However, because ouabain-mediated immunosuppression was greatly attenuated by anti–PD-L1 IgG treatment, we hypothesize that PD-L1 may be the most important checkpoint protein in EO/ouabain-mediated immunosuppression. Ouabain-mediated cytokine expression occurs primarily at the transcriptional level. Here, ouabain triggers a rapid PD-L1 transcription, and this effect is mainly mediated by increased IRF1 expression and phosphorylation of EGFR and STAT3 but not intracellular [Na+] increase (fig. S13). Notably, both STAT3 and IRF1 are signaling molecules downstream of EGFR. EGFR signaling is one of the most important oncogenic pathways in NSCLC. In addition to its role in tumor growth, EGFR signaling also significantly modulates tumor immunity by up-regulating PD-L1, as confirmed both in vitro and in vivo (38). In addition, PD-L1 was expressed at a significantly higher level in NSCLC cell lines with mutant EGFR than in cells with WT EGFR (39). Although the mechanism of this effect remains unclear, there is increasing evidence that PD-L1 expression is closely related to EGFR mutation in cancer cells. After binding with ouabain, Na, K-ATPase α1 can promote Src-EGFR interaction and lead to EGFR transactivation (40). In this study, Na, K-ATPase α1 was also found to interact with many EGFR-related proteins. Ouabain preferentially increased EGFR phosphorylation and PD-L1 transcription in EGFR mutant lung cancer cells but not EGFR WT cells. Ouabain-mediated PD-L1 transcription can also be significantly suppressed by EGFR tyrosine kinase inhibitor. Together, we speculated that the transcription machinery of the PD-L1 gene may be open and active in EGFR overactivated mutant cells, which makes them more sensitive to ouabain-induced EGFR phosphorylation than WT cells.
In addition to transcription, ouabain can also regulate gene expression at other levels. In previous studies, ouabain was found to integrate multiple posttranscriptional mechanisms to control the expression of genes with AU-rich elements (AREs) in their 3′ untranslated regions (3′UTRs), such as TNF-α (41). The AREs are also abundantly distributed in the 3′UTR of immunosuppressive cytokines including IL-6, VEGF, chemokine (C-C motif) ligand 2 CCL2), granulocyte-macrophage colony-stimulating factor (GM-CSF), and TGF-β, as mentioned in this study. Ouabain increased the gene expression of those cytokines, which could be a result of regulation at the posttranscriptional level. A recent study suggests that membrane PD-L1 protein can translocate into the nucleus to participate in the transcription of cytokine genes (26). Ouabain-mediated increase in cytokines expression was significantly suppressed by anti–PD-L1 IgG, and, in addition, ouabain also triggered a small fraction of PD-L1 into the nucleus in lung cancer cells. Hence, the possibility that the nuclear function of PD-L1 is involved in ouabain-mediated cytokines gene expression cannot be excluded. In summary, Na, K-ATPase/ouabain signaling modulates cytokine expression at multiple levels. This property makes it possible for ouabain to act as a multifunctional hormone in a variety of physiological or pathological processes. Here, ouabain promoted immunosuppression in lung cancer by increasing PD-L1 expression; ouabain could also reverse sepsis-induced immunosuppression by up-regulating tumor necrosis factor–α (TNF-α) (41) or human leukocyte antigen DR (HLA-DR) expression (42). Therefore, the specific role of ouabain in immunoregulation depends on the concrete immune context and cytokine expression.
The fact that EO is elevated in NSCLC or tumor-bearing mice is intriguing. It may, however, be related to adrenal gland–mediated energy homeostasis. As tumors develop, Na, K-ATPase activity in cancerous cells needs to be up-regulated to meet the rapid demand for substance exchange, lactate production, and glycolysis (16). These processes require large cellular adenosine 5´-triphosphate (ATP) supplies. Adrenal glands can sense abnormal ATP consumption and release EO to down-regulate Na, K-ATPase activity. Unfortunately, this energy balance is ultimately exploited by cancer cells to facilitate immune escape. On the other hand, increasing evidence revealed that Hypothalamic-pituitary-adrenal (HPA) axis is activated in cancer (43), resulting in dysregulation of adrenal gland–derived hormones such as cortisol. Those dysregulated cortisols are reported to affect tumor growth and immunity (44). The release of EO is regulated by the HPA axis and adrenocorticotropic hormone (ACTH) (11). Scientists have observed that ACTH was present in cancer tissue extracts and the blood of patients with lung cancer (45), and furthermore, this phenomenon is unique to lung cancer (46). In agreement with this finding, we observed that serum ACTH was elevated in LLC mice but failed to increase in EGFRT790M mice, in which serum EO was not elevated (fig. S14). Given this, we speculated that serum EO elevation in lung cancer could be a result of HPA activation and ACTH release. Notably, the immune system is also involved in EO secretion in mice. Adrenal glands are closely linked with tumor immunity in lung cancer. About 70 years ago, scientists have found that the adrenal glands are the primary metastatic sites of NSCLC (47). Surgical removal of adrenal glands results in long-term survival in isolated adrenal metastases (48). There is evidence of retroperitoneal lymphatic channels in adrenal metastases (49). On the other hand, endocrine-related adverse events have been reported with PD-L1–based immunotherapy, and the adrenal glands are susceptible to immune checkpoint blockade (50). Here, serum EO increase in LLC mice was blocked by macrophages or T cell depletion. Meanwhile, tumor immunity was activated in LLC mice after adrenalectomy, and ouabain rescue significantly reversed it. Together, it seems reasonable to speculate that lung cancer may recruit certain immune cells or produce cytokines to promote the release of EO from the adrenal gland. Furthermore, there may be EO-mediated cross-talk between endocrine and immunity in lung adenocarcinoma that sustains suppression of tumor immunity. This hypothesis can be investigated in future studies to gain deeper insight. Here, serum EO failed to increase in either KrasLSL-G12D or EGFRLSL-T790M model with intact immunity and adrenal glands. We speculate that serum EO may be induced by tumor heterogeneity due to mutations in a large number of pro- or antioncogenic genes rather than a single gene of EGFR or KRAS. Report has shown that genome-wide tumor mutational burden contributes more to the complexity and heterogeneity of the tumor microenvironment than a single-gene mutation (51). On the other hand, we are unsure whether Kras or EGFR mutation in cancer compromises the cross-talk among the tumor microenvironment, immune cells, and endocrine glands that leads to EO production. This concern will be addressed in future studies.
In this study, the results of Ab neutralization and bilateral adrenalectomy both support the suppressive effect of EO on tumor immunity, but their effects on immune cell populations are not entirely consistent. To understand this difference, we suggest that, although removal of bilateral adrenal glands can block the production of EO by adrenal glands, this approach also reduces the production of other adrenergic hormones that may have effects on tumor immunity. This does not mean that the data obtained with adrenalectomy are not meaningful, as ouabain rescue significantly increased adrenalectomy-suppressed tumor growth, highlighting that EO, among adrenergic hormones, is of great significance in the formation of suppressive tumor immunity. Similarly, the results of anti-ouabain IgG and plant ouabain on immune cell populations were not completely consistent, which may be due to the fact that, although anti-ouabain IgG has a high neutralizing effect on EO, it does not completely neutralize all EO in serum.
Na, K-ATPase α1 triggers clathrin-dependent PD-L1 endocytosis. This study, including others, showed that Na, K-ATPase is associated with proteins involved in vesicular trafficking and cytoskeletal rearrangement (52). Therefore, the Na, K-ATPase α1/PD-L1 interaction may enhance the local aggregation of vesicular trafficking–related proteins around PD-L1, thereby increasing the occurrence of PD-L1 endocytosis. On the other hand, CME is tightly regulated by the kinase pathway. Na, K-ATPase controls the activities of c-Src, EGFR, and phosphatidylinositol 3-kinase (PI3K) (53). In this study, Na, K-ATPase α1–mediated down-regulation of PD-L1 was prevented by inhibition of dynamin activity or GSK3β inhibitor. Dynamin activity is controlled by GSK3β and PI3K/Akt activity (54, 55). Na, K-ATPase α1–mediated PD-L1 endocytosis is also closely related to kinase activation, which warrants further analysis.
Although EO promotes immune escape in lung adenocarcinoma, targeting the EO/Na, K-ATPase signaling pathway shows great prospects in cancer therapy. Clinical data show that, although patients with NSCLC with low Na, K-ATPase α1 expression have relatively poor survival, they benefit more from anti–PD-1/PD-L1 therapy. Moreover, this phenomenon is not limited to lung adenocarcinoma, suggesting that Na, K-ATPase α1 expression is useful in predicting patient response to PD-1/PD-L1 therapy. Besides, the antihypertensive drug rostafuroxin significantly reduced tumor burden and activated tumor immunity in lung adenocarcinoma by blocking the EO/Na, K-ATPase interaction. Digoxin is already used to treat cardiovascular diseases, and it has significant antitumor effects (56, 57). Here, digoxin combined with anti–PD-L1 IgG resulted in a significant suppression in tumor burden. Because digoxin is a clinically used drug, digoxin may be superior to ouabain in combination with the PD-L1 Ab in NSCLC immunotherapy, which deserves further investigation. This study further confirmed that cinobufacini injections are superior to digoxin in combined immunotherapy. Cinobufacini injections have a broad spectrum of antitumor effects and have been widely used in clinical cancer treatment in China (58). They are the first traditional Chinese medicine injections approved for antitumor clinical trials sponsored by MD Anderson Cancer Center and the National Cancer Institute (NCT00837239, completed). Clinical evidence has shown that cinobufacini injections can significantly improve the efficacy of a variety of anticancer drugs, including chemotherapy, radiotherapy, and tyrosine kinase–targeted therapy. They can also improve the adverse reactions, such as leukopenia, caused by chemotherapy and improve the quality of life of patients (36, 59). Therefore, cinobufacini injections combined with PD-1/PD-L1 blocker are a potential clinical treatment for NSCLC that warrant further study.
In conclusion, the Na, K-ATPase/EO signaling pathway promotes immune escape in NSCLC by regulating the PD-1/PD-L1 axis. EO is an adrenal gland–derived hormone that negatively regulates tumor immunity in vivo. As we attempted circumventing Na, K-ATPase/EO–induced immunosuppression, it will be reasonable to develop strategies that target Na, K-ATPase/EO signaling to improve PD-1/PD-L1–based immunotherapy.
MATERIALS AND METHODS
Study design
The purpose of this study was to explore the regulatory role of Na, K-ATPase in tumor immune microenvironment that has never been described. These objectives were addressed by (i) analyzing the clinical significance of Na, K-ATPase α1 expression in patients with NSCLC, (ii) confirming EO presence in patients with NSCLC and its clinical significance, identifying suitable tumor-bearing mice models that can mimic the elevated serum EO in clinical lung adenocarcinoma, (iii) evaluating the effects of EO on lung cancer development and tumor immune microenvironment, (iv) exploring the mechanism underlying EO-mediated tumor immune escape, (v) investigating the molecular mechanism underlying the delicate control of PD-L1 expression by Na,K-ATPase α1/EO signaling, and (vi) examining the role of Na, K-ATPase α1/EO signaling in the immunotherapy of lung adenocarcinoma.
Institutional Review Board and Institutional Animal Care and Use Committee (IACUC) guidelines were followed with human participants or animal subjects. Mice were randomly assigned to experimental and control groups, but investigators were not blinded. No data were excluded from studies in this manuscript. Pathology analysis was performed in a blinded fashion. All procedures were performed following institutional protocols approved by the Animal Ethics Committee of Nanjing University. A total of 10 biopsy samples from patients with NSCLC, blood samples from 80 patients with NSCLC, and 48 HDs were selected for analysis and retrospectively reviewed in agreement with the Ethics Committee of the Jiangsu Cancer Hospital (no: 2018608), Nanjing, China.
Patients and peripheral blood and tissue samples
The study on the specimens and patients’ information was approved by the Ethics Committee of the Jiangsu Cancer Hospital (no: 2018608). Written informed consent was provided by all study participants before sampling. Data from patients who had received surgery biopsy with histological diagnosis of NSCLC at the Jiangsu Cancer Hospital, Nanjing, China, from November 2018 to January 2019 were retrospectively collected in this study. Clinicopathological data of the patients with NSCLC, including age, sex, tumor size, regional lymph node status, tumor, lymph node, metastasis (TNM) stage, and differentiation, were obtained. None of the patients had received ouabain or its analog treatment before peripheral blood samples were collected. Peripheral blood (2 ml) from healthy volunteers and patients with NSCLC was drawn from the cubital vein and was collected in tubes containing K2-EDTA (1.8 mg/ml). The blood samples were centrifuged at 3000 rpm for 10 min to isolate plasma. From June 2018 to June 2019, 10 consecutive cases of fresh NSCLC tissues were prospectively collected during the operation. The tissue mass was used for immunohistochemistry detection of PD-L1 or Na, K-ATPase α1 expression.
Measurement of plasma content of EO by HPLC-MS/MS analysis
Calibrator samples were prepared by spiking a pool of EDTA plasma from leftover samples with a solution of ouabain in methanol (20%) to obtain nine concentration levels: 1, 5, 10, 25, 50, 100, 200, 500, and 1000 pg/ml. Calibrator samples were aliquoted (500 μl) and stored at −20°C. In the same way, quality control (QC) samples were prepared in three levels: low QC level, 5.0 pg/ml; medium QC level, 50.0 pg/ml; and high QC level, 500.0 pg/ml. The QC materials were aliquoted (500 μl) and stored at −20°C. An internal standard (IS) dexamethasone stock solution (C = 100 ng/ml) was prepared in methanol, and an IS working solution was prepared in methanol (20%), which were stored at −20° and 4°C, respectively.
Two microliters of IS working solution was added to 500 μl of calibrator samples, QC materials, or plasma samples and mixed thoroughly for 5 min at room temperature (RT). Samples were diluted with 500 μl of 0.1% trifluoroacetic acid in water, mixed thoroughly for 5 min at RT, and centrifuged at 3000 rpm for 30 min at 4°C. Samples were extracted applying solid phase extraction using Copure HLB Solid Phase Extraction (30 mg/ml; COHLB130; Shenzhen Comma Biotechnology Co. Ltd., Shenzhen, China), which were conditioned with 1 ml of acetonitrile and equilibrated with 1 ml of water. Supernatants were loaded onto the columns, which were subsequently washed with 1 ml of water. Analytes were eluted with 2× 600 μl of 25% acetonitrile. The combined eluate was evaporated to dryness using a Bachofer SpeedVac Concentrator (Bachofer GmbH, Reutlingen, Germany). Residues were reconstituted using 100 μl of 5% methanol and mixed thoroughly for 5 min, and the resulting solutions were centrifuged at 10,000 rpm for 3 min at RT. Supernatants were transferred into HPLC vials with a small volume insert and analyzed using HPLC-MS/MS (TripleTOF 4600 or 5600, AB SCIEX, USA).
Cell lines and animals
LAC275 and LAC203 cell lines are primary lung adenocarcinoma cells isolated from patients with NSCLC, and the cultures were established in our laboratory. The sequence analysis confirmed that EGFR in the LAC275 genome has a mutation in A837T. Human HCC827 (ATCC-CRL-2868), H1975 (ATCC-CRL-5908), A549 (ATCC-CRM-CCL-185), H460 (ATCC-HTB-177), LO2 (CRL-12461), BEAS-2B (ATCC-CRL-9609), and murine LLC cell lines (ATCC-CRL-1642) are all obtained from the American Type Culture Collection (ATCC). Among them, HCC827 cells are EGFR mutant cells with E746_A750 deletion, and H1975 cells are EGFR mutant cells with L858R/T790M mutation. A549 and H460 cells have no EGFR mutation. The EGFR gene in murine LLC cells used in this study has several mutations including R528L, as confirmed by sequence analysis. CRISPR-Cas9 technology was used to produce PD-L1 gene knockout (KO) LLC cells or dynamin-1 gene KO LAC275 cells. The CRISPR-Cas9 single guide RNA (sgRNA) design tool developed by Zhang’s laboratory (http://crispr.mit.edu/) was used to design sgRNAs for Cas9 nuclease targeting dynamin-1 in the human genome and PD-L1 in the mouse genome. The target sequences in the exons of genes of interest were selected following analysis of each gene. The 20-nucleotide target sequences (PD-L1 sgRNA sequence, 5′-GTATGGCAGCAACGTCACGA-3′ and dynamin-1 sgRNA sequence, 5′-GCATAAGCATGTCTCGGATC-3′), including the protospacer adjacent motif (5′-NGG-3′), were selected for their predicted high score and reduced off-target effects. After transfection with PX459 plasmid expressing Cas9 (Addgene) and optimized sgRNAs and transient selection with puromycin (Invitrogen), cells were single-cell seeded into 96-well plates without puromycin. To select KO clones, the expression of PD-L1 and dynamin-1 was determined by flow cytometry or Western blot analysis. PD-L1−/− mice, KrasLSL-G12D, and EGFRLSL-T790M transgenic mice were provided by Model Animal Center (Shanghai, China). NOD-SCID and NSG mice were provided by Nanjing KMK Pharmaceutical Co. (China).
Construction of Na, K-ATPase α1–shRNA–expressing lentiviral vector and transduction
shRNAs targeting Na, K-ATPase α1 mRNA were synthesized from Sangon Biotech (Shanghai, China): sh-Na, K-ATPase α1 sense (5′-3′)TCATGTGGTTTGACAATCAATTCAAGAGATTGATTGTCAAACCACATGTTTTTTC and antisense (5′-3′)TCGAGAAAAAACATGTGGTTTGACAATCAATCTCTTGAATTGATTGTCAAACCACATGA. A negative sequence that shared no homology with the mammalian genome was used as a control. Successful cloning of these sequences into the pLentilox 3.7 lentivector was confirmed by Sanger sequencing. The lentiviral system (Addgene) including the packaging vectors pCMV-VSV-G and pCMV-ΔR8.2 was used in this study. We used a 293T cell for the production of pseudoviral particles according to the manufacturer’s instructions (lenti-pseudoviral particles; GeneBiology, Shanghai, China). After 48 hours, lentiviral particles were collected and concentrated from the supernatant using a lentivirus concentration solution [Yeasen Biotechnology (Shanghai) Co. Ltd.]. pLentilox3.7-sh-Na, K-ATPase α1 was used for the animal study.
Murine tumor-bearing model and the drug schemes
Three- to 4-week-old male C57BL/6 mice were purchased from the Model Animal Research Center of Nanjing University (Nanjing, China). Animal care protocol and experiments were approved by the Animal Committee at Nanjing University (no: IACUC-2201005) and conformed to the Regulations for the Administration of Affairs Concerning Experimental Animals (China). To improve the spontaneous metastatic activity of LLC cells, we isolated metastatic nodules from the lung of LLC mice and obtained LLC cells with strong metastatic activity through culture and purification. LLC cells (5 × 105 or 5 × 106) were subcutaneously inoculated into shaved lateral flanks of C57BL/6 mice with or without bilateral adrenalectomy. Seven days later, the mice were randomly divided into two or four groups. Two groups: vehicle [phosphate-buffered saline (PBS), intraperitoneally] or sham, ouabain (0.1 mg/kg, ip) or rostafuroxin [10 mg/kg, intragastric gavage (i.g.)], or anti-ouabain Ab (100 μg per mouse, ip) or bilateral adrenalectomy. Four groups: vehicle (PBS, ip), ouabain (0.1 mg/kg, ip), anti–PD-L1 Ab (200 μg per mouse, ip) or anti-Gr1 Ab (250 μg per mouse, ip), anti–PD-L1 Ab (200 μg per mouse, ip) or anti-Gr1 Ab (250 μg per mouse, ip) + ouabain (0.1 mg/kg, ip). All the treatments were carried out according to the experimental design. The tumor size was estimated following the formula length × width2/2 and was recorded every day. At the end of the experiment, the animals were euthanized, and the tumors were dissected and subjected to subsequent experiments.
Bilateral adrenalectomy mice
The surgical removal of adrenal glands from mice was performed as previously described with minor modification (22). Male C57BL/6J mice (6 to 8 weeks old) were anesthetized with ether and were placed prostrate on the operating table. Mice were disinfected with iodophors after back hair removal. The upper edges of the kidneys were exposed after cutting the mouse skin from the mid side of the back (the lower edge of the ribs) and the muscles of both sides. Then, bilateral adrenal glands were bluntly separated and removed after ligation. Both muscles and skin were sequentially sewn and sterilized with iodophors. The mice were put back into a squirrel cage after regaining consciousness and provided food and water ad libitum. Mice in the sham group received the same surgery except for bilateral adrenal glands removal. Subsequent experiments performed on these mice were resumed 2 weeks later.
Correlation of PD-L1–related genes with ouabain-regulated genes
The RNA sequencing data were downloaded from TCGA (https://tcga-data.nci.nih.gov/tcga/) by TCGAbiolinks packages (60) and the tumor samples between samples with high PD-L1 expression and samples with low PD-L1 expression were compared by Limma package (61). The differential genes related to PD-L1 are defined by |log2FC| > 0.6 and P < 0.05. LAC275 cells treated by ouabain were subject to microarray analysis. The differential genes induced by ouabain are defined by |log2FC| > 0.6. In the volcano plot, the genes whose expression was positively correlated with PD-L1 were defined as “UP” genes; among them, the genes that were able to be up-regulated by ouabain in LAC275 cells were further defined as “CO_UP” genes. On the contrary, the genes whose expression was negatively correlated with PD-L1 were defined as “DN” genes; among them, the genes that were able to be down-regulated by ouabain in LAC275 cells were further defined as “CO_DN” genes. The genes whose expression is not significantly correlated with PD-L1 were defined as “NS” genes.
Protein immunoprecipitation and MS
LAC275 cells after transfection with hemagglutinin (HA)–tagged Na, K-ATPase α1 were lysed with radioimmunoprecipitation assay (RIPA) buffer [50 mM tris-Cl (pH 8.0), 150 mM NaCl, and 1% NP-40]. The lysates were centrifuged, and supernatants were transferred to preequilibrated HA-agarose beads and incubated at 4°C overnight. The beads with precipitated proteins were washed with RIPA buffer five times and collected by SDS loading buffer. Pull-down protein complexes were resolved on a 10% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gel. Protein bands were stained with instant Coomassie blue staining and excised, destained, and digested with sequencing-grade trypsin. The peptide mass and peptide fragment mass were measured by TripleTOFTM5600 LC-MS/MS (AB SCIEX, USA), followed by identifying the matched proteins to the NCBI databases. The PD-L1–interacting protein database was obtained from a previous study (24). The shared proteins that can immunoprecipitate with both Na, K-ATPase α1 and PD-L1 were analyzed and provided in data file S2.
Luciferase reporter assays
LAC275 cells (1.0 × 104/ml) were plated in 24-well plates 24 hours before transfection. The pGL3-PD-L1 promoter-Luc, an IS plasmid pRL-SV40 (Promega, WI, USA), and control vector pGL3-Basic were cotransfected into LAC275 cells with the use of a PolyJet reagent. The next day, the medium was changed, ouabain (50 nM) was added, and 6 hours later, measurements of firefly and Renilla luciferase activities in cell lysates were carried out with the use of the dual-luciferase reporter assay system according to the manufacturer’s instructions (Promega, WI, USA).
ChIP assay
LAC275 cells were fixed with 1% formaldehyde at 37°C for 10 min and subsequently washed twice with ice-cold PBS containing protease inhibitors. Cells were incubated in a lysis buffer [1% SDS, EDTA (10 mM), tris-HCl (50 mM; pH 8.1)] for 10 min on ice and sonicated to shear genomic DNA. The lysate was centrifuged for 10 min at 13,000 rpm at 4°C. The supernatant was diluted in a ChIP dilution buffer [0.01% SDS, 1% Triton X-100, EDTA (2 mM), tris-HCl (16.7 mM; pH 8.1), NaCl (167 mM), and protease inhibitors]. Anti-STAT3, anti-IRF1, or IgG (negative control) Abs were added to the supernatant and incubated overnight at 4°C with rotation. Protein A–agarose slurry was added and incubated at 4°C for 1 hour with constant rotation. Agarose beads were collected by centrifugation and washed, and Ab-bound chromatin was released from the agarose beads. DNA was purified by phenol/chloroform extraction and isoamyl alcohol precipitation. Binding was detected by polymerase chain reaction.
Protein coimmunoprecipitation
Tissue and whole-cell lysates were extracted by homogenizing tissues or cell pellets in RIPA buffer. For coimmunoprecipitation, lysates containing 500 μg of total protein were incubated with specific Abs (2 μg) for 18 hours at 4°C with constant rotation. After that, 50 μl of protein G agarose beads was added to incubate for 3 hours. The beads were washed five times with lysis buffer. The precipitated proteins were resuspended in 30 μl of SDS sample buffer and boiled at 95°C for 10 min. Proteins were separated on an 8% SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (62). The membranes were blocked in 5% bovine serum albumin (BSA; Bio-Rad, Hercules, CA, USA) for 1 hour at RT and incubated with primary Abs against Na, K-ATPase α1, PD-L1, HA-tag (Cell Signaling Technology, Danvers, MA, USA), or Flag-tag (Sigma-Aldrich) at 4°C overnight. The membranes were then incubated with horseradish peroxidase–conjugated secondary Abs, and the protein bands were detected by enhanced chemiluminescence reagents (GE Healthcare Life Sciences, Pittsburgh, PA, USA).
Colocalization of Na, K-ATPase α1 or PD-L1 with lysosome by confocal imaging
LAC275 cells were cultured on coverslips overnight; after transfection with Na, K-ATPase α1, PD-L1, or control plasmids for 48 hours, cells were fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.3% Triton X-100 in PBS for 10 min, and then blocked with 5% BSA in phosphate buffered solution with Tween® 20 (PBST) for 1 hour. The primary Abs were diluted in the blocking buffer, and cells were incubated with primary Abs (1:500) or LAMP1 (1:200) in chamber slides at 4°C overnight. After washing three times with PBST for 5 min, the cells were incubated with the fluorescent-conjugated secondary Ab (diluted in PBST, 1:800) for 1 hour at RT, followed by washing three times with PBS, each time for 5 min. The slides were mounted with resin and imaged on a confocal microscope (Leica TCS SP8-MP). The colocalization among Na, K-ATPase α1, PD-L1, and LAMP1-marked lysosome was determined by immunofluorescence.
Coculture of CD3+ T cells with MDSCs and measurement of lymphocyte proliferation
To purify CD3+ T cells, erythrocyte-depleted splenocytes from naïve C57BL/6 mice were incubated with anti–CD3-FITC (fluorescein isothiocyanate) Abs and isolated by cell sorting on a BD Biosciences FACSAria II to >90% purity. To purify MDSCs, single-cell suspensions from the bone marrow of tumor-bearing mice were incubated with IL-4 (40 ng/ml) and GM-CSF (40 ng/ml), followed by cell sorting using a BD Biosciences FACSAria II after cell staining with FITC-conjugated anti–Gr-1 Ab and phycoerythrin (PE)-conjugated anti-CD11b Ab. The purity of the MDSC population was higher than 90%. Carboxyl fluorescein succinimidyl ester (CFSE) labeling was used to detect the proliferation of CD3+ T cells. Briefly, 2 × 106 CD3+ T cells were cocultured with MDSCs at ratios of 1:0, 1:1, 5:1, and 20:1 in 24-well plates (Costar, Lowell, MA, USA) and stimulated with anti-CD3 (5 μg/ml) and anti-CD28 (2 μg/ml) monoclonal Abs (both from Invitrogen, Carlsbad, CA, USA) for 3 days in RPMI 1640 supplemented with 10% fetal bovine serum (Gibco Life Technologies, Carlsbad, CA, USA). As the CFSE signal was diluted with each cell division, cells exhibiting low fluorescence intensity of CFSE were considered to have proliferated. Samples were analyzed using a flow cytometer (Becton Dickinson, San Jose, CA, USA), and data were analyzed with the FlowJo (FlowJo, Tree Star Inc.) software.
Statistical analysis
Data were presented as the means ± SEM. All data were analyzed using appropriate statistical analysis methods with the Statistical Package for the Social Sciences software (version 23.0) or GraphPad Prism (version 8.0), as presented in the figure legends. For data with Gaussian distribution, parametric statistical analysis was performed using the two-tailed Student’s t test for two groups. For multiple comparisons, one-way analysis of variance (ANOVA) was applied with least significant difference post hoc analysis for data meeting homogeneity of variance or with Dunnett’s’s T3 analysis for data not assuming equal variances. Linear regression analysis was performed to assay the correlations between Na, K-ATPase α1 expression and PCNA expression in patients with NSCLC. In all cases, differences were considered significant at P < 0.05.
Acknowledgments
Funding: This project was sponsored by the National Natural Science Foundation of China (NSFC 81673462, 82073916, and 91540119 to W.Y.; 81873002 to F.Y.; and 81874452 to Y.C.) and the Key Development Project of Jiangsu Province (BE2017712 to W.Y.).
Author contributions: Conceptualization: W.Y. Methodology: K.Y., W.Y., Z.L., Y.C., F.Y., C.F., T.Z., X.L., L.C., C.R., L.F., Y.Q., X.J., J.Z., Y.W., P.Z., and L.M. Investigation: K.Y., Z.L., Y.C., X.L., X.J., J.Z., L.F., and W.Y. Visualization: K.Y. and Z.L. Supervision: W.Y. and Y.C. Writing (original draft): W.Y. and K.Y. Writing (review and editing): W.Y. and K.Y.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Materials and Methods
Figs. S1 to S14
Tables S1 to S6
References
Other Supplementary Material for this : manuscript includes the following:
Data files S1 to S3
REFERENCES AND NOTES
- 1.G. Morad, B. A. Helmink, P. Sharma, J. A. Wargo, Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell 184, 5309–5337 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.J. M. Pitt, M. Vetizou, R. Daillere, M. P. Roberti, T. Yamazaki, B. Routy, P. Lepage, I. G. Boneca, M. Chamaillard, G. Kroemer, L. Zitvogel, Resistance mechanisms to immune-checkpoint blockade in cancer: Tumor-intrinsic and -extrinsic factors. Immunity 44, 1255–1269 (2016). [DOI] [PubMed] [Google Scholar]
- 3.R. S. Herbst, D. Morgensztern, C. Boshoff, The biology and management of non-small cell lung cancer. Nature 553, 446–454 (2018). [DOI] [PubMed] [Google Scholar]
- 4.M. J. Grant, R. S. Herbst, S. B. Goldberg, Selecting the optimal immunotherapy regimen in driver-negative metastatic NSCLC. Nat. Rev. Clin. Oncol. 18, 625–644 (2021). [DOI] [PubMed] [Google Scholar]
- 5.M. Wu, Q. Huang, Y. Xie, X. Wu, H. Ma, Y. Zhang, Y. Xia, Improvement of the anticancer efficacy of PD-1/PD-L1 blockade via combination therapy and PD-L1 regulation. J. Hematol. Oncol. 15, 24 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.N. Acharya, A. Madi, H. Zhang, M. Klapholz, G. Escobar, S. Dulberg, E. Christian, M. Ferreira, K. O. Dixon, G. Fell, K. Tooley, D. Mangani, J. Xia, M. Singer, M. Bosenberg, D. Neuberg, O. Rozenblatt-Rosen, A. Regev, V. K. Kuchroo, A. C. Anderson, Endogenous glucocorticoid signaling regulates CD8+ T cell differentiation and development of dysfunction in the tumor microenvironment. Immunity 53, 658–671.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.A. B. Hanker, D. R. Sudhan, C. L. Arteaga, Overcoming endocrine resistance in breast cancer. Cancer Cell 37, 496–513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.H. Katcoff, A. S. Wenzlaff, A. G. Schwartz, Survival in women with NSCLC: The role of reproductive history and hormone use. J. Thorac. Oncol. 9, 355–361 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.B. Ricciuti, S. E. Dahlberg, A. Adeni, L. M. Sholl, M. Nishino, M. M. Awad, Immune checkpoint inhibitor outcomes for patients with non-small-cell lung cancer receiving baseline corticosteroids for palliative versus nonpalliative indications. J. Clin. Oncol. 37, 1927–1934 (2019). [DOI] [PubMed] [Google Scholar]
- 10.A. G. Schwartz, A. S. Wenzlaff, G. M. Prysak, V. Murphy, M. L. Cote, S. C. Brooks, D. F. Skafar, F. Lonardo, Reproductive factors, hormone use, estrogen receptor expression and risk of non small-cell lung cancer in women. J. Clin. Oncol. 25, 5785–5792 (2007). [DOI] [PubMed] [Google Scholar]
- 11.A. Y. Bagrov, J. I. Shapiro, O. V. Fedorova, Endogenous cardiotonic steroids: Physiology, pharmacology, and novel therapeutic targets. Pharmacol. Rev. 61, 9–38 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.J. M. Hamlyn, R. Ringel, J. Schaeffer, P. D. Levinson, B. P. Hamilton, A. A. Kowarski, M. P. Blaustein, A circulating inhibitor of (Na+ + K+)ATPase associated with essential hypertension. Nature 300, 650–652 (1982). [DOI] [PubMed] [Google Scholar]
- 13.I. Prassas, E. P. Diamandis, Novel therapeutic applications of cardiac glycosides. Nat. Rev. Drug Discov. 7, 926–935 (2008). [DOI] [PubMed] [Google Scholar]
- 14.T. Mijatovic, I. Roland, E. Van Quaquebeke, B. Nilsson, A. Mathieu, F. Van Vynckt, F. Darro, G. Blanco, V. Facchini, R. Kiss, The alpha1 subunit of the sodium pump could represent a novel target to combat non-small cell lung cancers. J. Pathol. 212, 170–179 (2007). [DOI] [PubMed] [Google Scholar]
- 15.C. T. Durlacher, K. Chow, X. W. Chen, Z. X. He, X. Zhang, T. Yang, S. F. Zhou, Targeting Na(+)/K(+) -translocating adenosine triphosphatase in cancer treatment. Clin. Exp. Pharmacol. Physiol. 42, 427–443 (2015). [DOI] [PubMed] [Google Scholar]
- 16.M. Banerjee, X. Cui, Z. Li, H. Yu, L. Cai, X. Jia, D. He, C. Wang, T. Gao, Z. Xie, Na/K-ATPase Y260 phosphorylation-mediated Src regulation in control of aerobic glycolysis and tumor growth. Sci. Rep. 8, 12322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.J. M. Hamlyn, M. P. Blaustein, S. Bova, D. W. DuCharme, D. W. Harris, F. Mandel, W. R. Mathews, J. H. Ludens, Identification and characterization of a ouabain-like compound from human plasma. Proc. Natl. Acad. Sci. U.S.A. 88, 6259–6263 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.N. D. Marjanovic, M. Hofree, J. E. Chan, D. Canner, K. Wu, M. Trakala, G. G. Hartmann, O. C. Smith, J. Y. Kim, K. V. Evans, A. Hudson, O. Ashenberg, C. B. M. Porter, A. Bejnood, A. Subramanian, K. Pitter, Y. Yan, T. Delorey, D. R. Phillips, N. Shah, O. Chaudhary, A. Tsankov, T. Hollmann, N. Rekhtman, P. P. Massion, J. T. Poirier, L. Mazutis, R. Li, J. H. Lee, A. Amon, C. M. Rudin, T. Jacks, A. Regev, T. Tammela, Emergence of a high-plasticity cell state during lung cancer evolution. Cancer Cell 38, 229–246.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.P. Ferrari, Rostafuroxin: an ouabain-inhibitor counteracting specific forms of hypertension. Biochim. Biophys. Acta 1802, 1254–1258 (2010). [DOI] [PubMed] [Google Scholar]
- 20.I. Henneke, S. Greschus, R. Savai, M. Korfei, P. Markart, P. Mahavadi, R. T. Schermuly, M. Wygrecka, J. Sturzebecher, W. Seeger, A. Gunther, C. Ruppert, Inhibition of urokinase activity reduces primary tumor growth and metastasis formation in a murine lung carcinoma model. Am. J. Respir. Crit. Care Med. 181, 611–619 (2010). [DOI] [PubMed] [Google Scholar]
- 21.K. Ritzel, F. Beuschlein, A. Mickisch, A. Osswald, H. J. Schneider, J. Schopohl, M. Reincke, Clinical review: Outcome of bilateral adrenalectomy in Cushing’s syndrome: A systematic review. J. Clin. Endocrinol. Metab. 98, 3939–3948 (2013). [DOI] [PubMed] [Google Scholar]
- 22.L. Hoffman-Goetz, J. Quadrilatero, J. Boudreau, J. Guan, Adrenalectomy in mice does not prevent loss of intestinal lymphocytes after exercise. J. Appl. Physiol. (1985) 96, 2073–2081 (2004). [DOI] [PubMed] [Google Scholar]
- 23.P. Kucan Brlic, T. Lenac Rovis, G. Cinamon, P. Tsukerman, O. Mandelboim, S. Jonjic, Targeting PVR (CD155) and its receptors in anti-tumor therapy. Cell. Mol. Immunol. 16, 40–52 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.M. Saigi, J. J. Alburquerque-Bejar, A. Mc Leer-Florin, C. Pereira, E. Pros, O. A. Romero, N. Baixeras, A. Esteve-Codina, E. Nadal, E. Brambilla, M. Sanchez-Cespedes, MET-oncogenic and JAK2-inactivating alterations are independent factors that affect regulation of PD-L1 expression in lung cancer. Clin. Cancer Res. 24, 4579–4587 (2018). [DOI] [PubMed] [Google Scholar]
- 25.H. Sato, A. Niimi, T. Yasuhara, T. B. M. Permata, Y. Hagiwara, M. Isono, E. Nuryadi, R. Sekine, T. Oike, S. Kakoti, Y. Yoshimoto, K. D. Held, Y. Suzuki, K. Kono, K. Miyagawa, T. Nakano, A. Shibata, DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat. Commun. 8, 1751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Y. Gao, N. T. Nihira, X. Bu, C. Chu, J. Zhang, A. Kolodziejczyk, Y. Fan, N. T. Chan, L. Ma, J. Liu, D. Wang, X. Dai, H. Liu, M. Ono, A. Nakanishi, H. Inuzuka, B. J. North, Y. H. Huang, S. Sharma, Y. Geng, W. Xu, X. S. Liu, L. Li, Y. Miki, P. Sicinski, G. J. Freeman, W. Wei, Acetylation-dependent regulation of PD-L1 nuclear translocation dictates the efficacy of anti-PD-1 immunotherapy. Nat. Cell Biol. 22, 1064–1075 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.C. D’Souza-Schorey, P. Chavrier, ARF proteins: Roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7, 347–358 (2006). [DOI] [PubMed] [Google Scholar]
- 28.M. Mettlen, P. H. Chen, S. Srinivasan, G. Danuser, S. L. Schmid, Regulation of clathrin-mediated endocytosis. Annu. Rev. Biochem. 87, 871–896 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. S. Bonifacino, L. M. Traub, Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395–447 (2003). [DOI] [PubMed] [Google Scholar]
- 30.R. Ramachandran, S. L. Schmid, The dynamin superfamily. Curr. Biol. 28, R411–R416 (2018). [DOI] [PubMed] [Google Scholar]
- 31.B. Antonny, C. Burd, P. De Camilli, E. Chen, O. Daumke, K. Faelber, M. Ford, V. A. Frolov, A. Frost, J. E. Hinshaw, T. Kirchhausen, M. M. Kozlov, M. Lenz, H. H. Low, H. McMahon, C. Merrifield, T. D. Pollard, P. J. Robinson, A. Roux, S. Schmid, Membrane fission by dynamin: What we know and what we need to know. EMBO J. 35, 2270–2284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.S. A. Kovacs, B. Gyorffy, Transcriptomic datasets of cancer patients treated with immune-checkpoint inhibitors: A systematic review. J. Transl. Med. 20, 249 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.H. Jung, H. S. Kim, J. Y. Kim, J. M. Sun, J. S. Ahn, M. J. Ahn, K. Park, M. Esteller, S. H. Lee, J. K. Choi, DNA methylation loss promotes immune evasion of tumours with high mutation and copy number load. Nat. Commun. 10, 4278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.R. Kumavath, S. Paul, H. Pavithran, M. K. Paul, P. Ghosh, D. Barh, V. Azevedo, Emergence of cardiac glycosides as potential drugs: Current and future scope for cancer therapeutics. Biomolecules. 11, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.F. Zhang, Y. Yin, T. Xu, Cinobufotalin injection combined with chemotherapy for the treatment of advanced NSCLC in China: A PRISMA-compliant meta-analysis of 29 randomized controlled trials. Medicine (Baltimore). 98, e16969 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.L. L. Li, Y. X. Su, Y. Mao, P. Y. Jiang, X. L. Chu, P. Xue, B. H. Jia, S. J. Zhu, The effectiveness and safety of cinobufotalin injection as an adjunctive treatment for lung cancer: A meta-analysis of randomized controlled trials. Evid. Based Complement. Alternat. Med. 2021, 2021, 8852261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.M. A. Shandell, A. L. Capatina, S. M. Lawrence, W. J. Brackenbury, D. Lagos, Inhibition of the Na(+)/K(+)-ATPase by cardiac glycosides suppresses expression of the IDO1 immune checkpoint in cancer cells by reducing STAT1 activation. J. Biol. Chem. 298, 101707 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.E. A. Akbay, S. Koyama, J. Carretero, A. Altabef, J. H. Tchaicha, C. L. Christensen, O. R. Mikse, A. D. Cherniack, E. M. Beauchamp, T. J. Pugh, M. D. Wilkerson, P. E. Fecci, M. Butaney, J. B. Reibel, M. Soucheray, T. J. Cohoon, P. A. Janne, M. Meyerson, D. N. Hayes, G. I. Shapiro, T. Shimamura, L. M. Sholl, S. J. Rodig, G. J. Freeman, P. S. Hammerman, G. Dranoff, K. K. Wong, Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 3, 1355–1363 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.K. Azuma, K. Ota, A. Kawahara, S. Hattori, E. Iwama, T. Harada, K. Matsumoto, K. Takayama, S. Takamori, M. Kage, T. Hoshino, Y. Nakanishi, I. Okamoto, Association of PD-L1 overexpression with activating EGFR mutations in surgically resected nonsmall-cell lung cancer. Ann. Oncol. 25, 1935–1940 (2014). [DOI] [PubMed] [Google Scholar]
- 40.M. Haas, H. Wang, J. Tian, Z. Xie, Src-mediated inter-receptor cross-talk between the Na+/K+-ATPase and the epidermal growth factor receptor relays the signal from ouabain to mitogen-activated protein kinases. J. Biol. Chem. 277, 18694–18702 (2002). [DOI] [PubMed] [Google Scholar]
- 41.C. Dan, B. Jinjun, H. Zi-Chun, M. Lin, C. Wei, Z. Xu, Z. Ri, C. Shun, S. Wen-Zhu, J. Qing-Cai, Y. Wu, Modulation of TNF‐α mRNA stability by human antigen R and miR181s in sepsis-induced immunoparalysis. EMBO Mol. Med. 7, 140–157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.L. Chen, M. Zhang, X. Wang, Y. Liu, J. Bian, D. Yan, W. Yin, Cardiac steroid ouabain transcriptionally increases human leukocyte antigen DR expression on monocytes. Steroids 175, 108915 (2021). [DOI] [PubMed] [Google Scholar]
- 43.K. van Norren, J. T. Dwarkasing, R. F. Witkamp, The role of hypothalamic inflammation, the hypothalamic-pituitary-adrenal axis and serotonin in the cancer anorexia-cachexia syndrome. Curr. Opin. Clin. Nutr. Metab. Care 20, 396–401 (2017). [DOI] [PubMed] [Google Scholar]
- 44.M. H. Ahmad, M. A. Rizvi, M. Fatima, A. C. Mondal, Pathophysiological implications of neuroinflammation mediated HPA axis dysregulation in the prognosis of cancer and depression. Mol. Cell. Endocrinol. 520, 111093 (2021). [DOI] [PubMed] [Google Scholar]
- 45.G. Gewirtz, R. S. Yalow, Ectopic ACTH production in carcinoma of the lung. J. Clin. Invest. 53, 1022–1032 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.L. F. Ayvazian, B. Schneider, G. Gewirtz, R. S. Yalow, Ectopic production of big ACTH in carcinoma of the lung. Its clinical usefulness as a biologic marker. Am. Rev. Respir. Dis. 111, 279–287 (1975). [DOI] [PubMed] [Google Scholar]
- 47.H. L. Abrams, R. Spiro, N. Goldstein, Metastases in carcinoma; analysis of 1000 autopsied cases. Cancer 3, 74–85 (1950). [DOI] [PubMed] [Google Scholar]
- 48.O. Mercier, E. Fadel, M. de Perrot, S. Mussot, F. Stella, A. Chapelier, P. Dartevelle, Surgical treatment of solitary adrenal metastasis from non-small cell lung cancer. J. Thorac. Cardiovasc. Surg. 130, 136–140 (2005). [DOI] [PubMed] [Google Scholar]
- 49.C. M. Raynaud, O. Mercier, P. Dartevelle, F. Commo, K. A. Olaussen, V. de Montpreville, F. Andre, L. Sabatier, J. C. Soria, Expression of chemokine receptor CCR6 as a molecular determinant of adrenal metastatic relapse in patients with primary lung cancer. Clin. Lung Cancer 11, 187–191 (2010). [DOI] [PubMed] [Google Scholar]
- 50.M. Sznol, M. A. Postow, M. J. Davies, A. C. Pavlick, E. R. Plimack, M. Shaheen, C. Veloski, C. Robert, Endocrine-related adverse events associated with immune checkpoint blockade and expert insights on their management. Cancer Treat. Rev. 58, 70–76 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Q. Jia, W. Wu, Y. Wang, P. B. Alexander, C. Sun, Z. Gong, J. N. Cheng, H. Sun, Y. Guan, X. Xia, L. Yang, X. Yi, Y. Y. Wan, H. Wang, J. He, P. A. Futreal, Q. J. Li, B. Zhu, Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat. Commun. 9, 5361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.W. J. Nelson, P. J. Veshnock, Ankyrin binding to (Na+ + K+)ATPase and implications for the organization of membrane domains in polarized cells. Nature 328, 533–536 (1987). [DOI] [PubMed] [Google Scholar]
- 53.Z. Li, Z. Xie, The Na/K-ATPase/Src complex and cardiotonic steroid-activated protein kinase cascades. Pflugers Arch. 457, 635–644 (2009). [DOI] [PubMed] [Google Scholar]
- 54.E. L. Clayton, N. Sue, K. J. Smillie, T. O’Leary, N. Bache, G. Cheung, A. R. Cole, D. J. Wyllie, C. Sutherland, P. J. Robinson, M. A. Cousin, Dynamin I phosphorylation by GSK3 controls activity-dependent bulk endocytosis of synaptic vesicles. Nat. Neurosci. 13, 845–851 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.C. R. Reis, P. H. Chen, S. Srinivasan, F. Aguet, M. Mettlen, S. L. Schmid, Crosstalk between Akt/GSK3β signaling and dynamin-1 regulates clathrin-mediated endocytosis. EMBO J. 34, 2132–2146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.S. Gkountela, F. Castro-Giner, B. M. Szczerba, M. Vetter, J. Landin, R. Scherrer, I. Krol, M. C. Scheidmann, C. Beisel, C. U. Stirnimann, C. Kurzeder, V. Heinzelmann-Schwarz, C. Rochlitz, W. P. Weber, N. Aceto, Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell 176, 98–112.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.E. A. Platz, S. Yegnasubramanian, J. O. Liu, C. R. Chong, J. S. Shim, S. A. Kenfield, M. J. Stampfer, W. C. Willett, E. Giovannucci, W. G. Nelson, A novel two-stage, transdisciplinary study identifies digoxin as a possible drug for prostate cancer treatment. Cancer Discov. 1, 68–77 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Z. Meng, C. R. Garrett, Y. Shen, L. Liu, P. Yang, Y. Huo, Q. Zhao, A. R. Spelman, C. S. Ng, D. Z. Chang, L. Cohen, Prospective randomised evaluation of traditional Chinese medicine combined with chemotherapy: A randomised phase II study of wild toad extract plus gemcitabine in patients with advanced pancreatic adenocarcinomas. Br. J. Cancer 107, 411–416 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Q. Li, R. L. Liang, Q. R. Yu, D. Q. Tian, L. N. Zhao, W. W. Wang, H. Xiao, X. J. Yong, X. D. Peng, Efficacy and safety of cinobufacini injection combined with vinorelbine and cisplatin regimen chemotherapy for stage III/IV non-small cell lung cancer: A protocol for systematic review and meta-analysis of randomized controlled trials. Medicine (Baltimore). 99, e21539, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.M. Mounir, M. Lucchetta, T. C. Silva, C. Olsen, G. Bontempi, X. Chen, H. Noushmehr, A. Colaprico, E. Papaleo, New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. PLoS Comput. Biol. 15, e1006701 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.M. E. Ritchie, B. Phipson, D. Wu, Y. Hu, C. W. Law, W. Shi, G. K. Smyth, limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.M. C. Lin, J. J. Lin, C. L. Hsu, H. F. Juan, P. J. Lou, M. C. Huang, Erratum: GATA3 interacts with and stabilizes HIF-1α to enhance cancer cell invasiveness. Oncogene 36, 4380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.A. Durrans, D. Gao, R. Gupta, K. R. Fischer, H. Choi, T. El Rayes, S. Ryu, A. Nasar, C. F. Spinelli, W. Andrews, O. Elemento, D. Nolan, B. Stiles, S. Rafii, N. Narula, R. Davuluri, N. K. Altorki, V. Mittal, Identification of reprogrammed myeloid cell transcriptomes in NSCLC. PLoS One. 10, e0129123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.K. Yang, J. Zhou, Y. Chen, Y. Chen, L. Chen, P. Zhang, L. Ma, Z. Jiang, J. Bian, W. Yin, Angiotensin II contributes to intratumoral immunosuppression via induction of PD-L1 expression in non-small cell lung carcinoma. Int. Immunopharmacol. 84, 106507 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Figs. S1 to S14
Tables S1 to S6
References
Data files S1 to S3