Abstract
MicroRNA (miRNA) homeostasis is crucial for the posttranscriptional regulation of their target genes during development and in disease states. miRNAs are derived from primary transcripts and are processed from a hairpin precursor intermediary to a mature 22-nucleotide duplex RNA. Loading of the duplex into the Argonaute (AGO) protein family is pivotal to miRNA abundance and its posttranscriptional function. The Integrator complex plays a key role in protein coding and noncoding RNA maturation, RNA polymerase II pause-release, and premature transcriptional termination. Here, we report that loss of Integrator results in global destabilization of mature miRNAs. Enhanced ultraviolet cross-linking and immunoprecipitation of Integrator uncovered an association with duplex miRNAs before their loading onto AGOs. Tracing miRNA fate from biogenesis to stabilization by incorporating 4-thiouridine in nascent transcripts pinpointed a critical role for Integrator in miRNA assembly into AGOs.
Integrator complex loss resulted in a global miRNA decrease due to defective Integrator-dependent Argonaute protein loading.
INTRODUCTION
MicroRNAs (miRNAs) are a class of small ~22-nucleotide RNAs that are generated through a complex biogenesis pathway (1–4). The canonical miRNAs consist of multiple classes depending on their precise pathways by which they are generated. The primary transcripts of miRNAs are derived from a protein coding transcript or a noncoding RNA, which is then processed into a precursor hairpin small RNA (smRNA) of approximately 70 nucleotides through the action of ribonuclease III (RNase III) enzyme DROSHA in a complex with a double-stranded RNA binding protein DiGeorge Syndrome Critical Region 8 (DGCR8), known as the Microprocessor complex (2, 3, 5). While most abundant miRNAs are initially produced by Microprocessor, there are two classes of miRNAs that are independent of DROSHA/DGCR8. These include miRNAs known as mirtrons whose precursor processing is funneled through the RNA splicing machinery and miRNAs whose primary transcripts are 5′-end capped noncoding RNAs (6–9). Regardless of how precursor miRNAs are generated, with the single exception of miR-451 (10, 11), canonical miRNAs are processed to a ~22-nucleotide duplex smRNA by a second RNase III enzyme DICER in a complex with a double-stranded RNA binding protein transactivation response element RNA-binding protein (TRBP) or protein activator of protein kinase R (PACT)(1, 4, 12).
While DROSHA is predominantly a nuclear protein, DICER primarily resides in the cytoplasm of mammalian cells (1, 13–15). Once the 22-nucleotide mature miRNA duplex is processed by the DICER/TRBP complex, the precise mode of its loading onto effector RNA silencing Argonaute (AGO) proteins in vivo has been under investigation. Numerous studies have suggested a role for heat shock protein 90 (HSP90) and HSP70 chaperones in AGO loading of the duplex RNA (16–19). Once the duplex RNA is associated with AGO, the identity of the nucleotide on the 5′-end and the thermodynamic stability of the two ends of the duplex are thought to be important for AGO-mediated strand separation (20–22). While one strand known as the “guide strand” associates with and is stabilized by AGO proteins, the other strand—the so-called “passenger strand”—is degraded (23). Nascent RNA sequencing (RNA-seq) in a time course experiment could track the faith of two strands, highlighting the stabilization of the guide strand by virtue of its association with AGOs (24). miRNA stabilization into RNA-induced silencing complex (RISC) is critical for modulation of miRNA levels and consequently the regulation of miRNA target mRNA stability and translation. Many studies showed that perturbing the levels of specific miRNAs, or the activity of the miRNA machinery, has marked effects on the level of hundreds of miRNA targets (25–27). In addition, decreasing or increasing the availability of endogenous targets reduces or elevates mature miRNA levels, respectively (23).
The Integrator complex is a conserved metazoan multisubunit protein complex that associates with the C-terminal domain of the largest subunit of RNA polymerase II (RNAPII). Integrator has a function in the regulation of gene expression and RNA processing and was first shown in the 3′-end processing of the U-rich small nuclear RNAs (UsnRNAs) involved in spliceosome formation (28). We further showed a critical role for Integrator in RNAPII pause-release and the biogenesis of enhancer RNAs (29, 30). Xie et al. (31) identified a miRNA-specific 3′-end processing signal (miR 3′ box) in Herpesvirus saimiri. They confirmed direct interaction between primary miRNA (pri-miRNA) substrates and Integrator and showed that Integrator has a function in transcription-independent maturation of herpesvirus miRNA 3′ ends. We recently reported that Integrator is required for the cleavage of stalled RNAPII transcripts, generating small promoter-associated RNAs (32). The intriguing resemblance of a ~20-nucleotide subpopulation of this smRNA class and miRNAs (32) led us to investigate the effect of Integrator depletion on miRNAs.
In this study, we observed that depletion of multiple Integrator subunits led to a major loss of canonical miRNAs in HeLa and human embryonic kidney (HEK) 293T cells, without affecting pri-miRNA transcription or precursor miRNA processing. By tracing miRNA fate from biogenesis to stabilization through metabolic sequencing, we determined Integrator to be essential for miRNA stability after maturation. We confirmed direct association of Integrator with duplex miRNAs before AGO loading through enhanced ultraviolet (UV) cross-linking and immunoprecipitation (eCLIP). In vitro reconstitution of AGO2-mediated miRNA-targeted cleavage confirmed the potentiation of miRNA-induced cleavage of a complementary target by increasing concentrations of Integrator complex. The absence of Integrator resulted in decreased association of miRNAs with AGOs and a consequent global loss of canonical miRNAs. Together, our studies point to a fundamental role for Integrator complex in maintaining miRNA homeostasis.
RESULTS
Integrator depletion leads to the loss of mature miRNAs
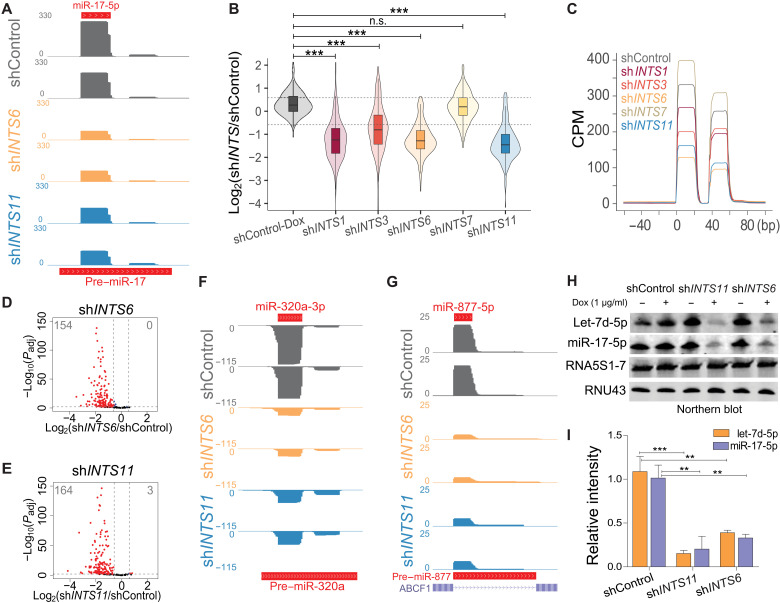
In the course of experiments analyzing the consequence of INTS11 depletion on the steady-state levels of smRNAs in HeLa cells using genome-wide smRNA sequencing (smRNA-seq), we noticed a profound diminution in canonical miRNA levels (exemplified by miR-17; Fig. 1A). We used a smRNA-seq platform that does not require primer ligation, minimizing miRNA distortions due to bias in primer ligation efficiencies [see Materials and Methods, SMARTer smRNA-seq kit (Takara, #635030)]. Guided by the abundance of miRNAs detected in smRNA-seq of cells treated with control short hairpin RNA (shControl), we selected the 200 most expressed mature miRNAs in HeLa cells. These encompassed miRNAs whose primary transcripts are processed by the Microprocessor complex. We extended this list for five Microprocessor-independent miRNAs, including mirtrons (miR-877-5p and miR-1226-3p) (33) and miRNAs with capped 5′-ends (miR-320a-3p/5p and miR-484) (9). Integrator is a modular multiprotein complex containing an endonucleolytic catalytic core composed of INTS11/4/9 (34, 35). To gain insight into the Integrator role in miRNA biogenesis and stability, we performed smRNA-seq following depletion of multiple subunits of Integrator including, INTS11 (a component of catalytic core), INTS1 and INTS7 (components of the backbone module), INTS6 (a key subunit bridging the association of backbone with protein phosphatase 2A and the INTS5/8 shoulder module), and INTS3 (a putative DNA repair module known to associate with INTS6) (fig. S1, A and B) (36–40). Notably, we observed a global loss of canonical miRNA following INTS1, INTS3, INTS6, and INTS11 depletion, while INTS7 knockdown did not affect miRNA steady state (Fig. 1, B and C, and fig. S1, A and C to F). Because of the modular nature of Integrator and multiple interactions among different subunits (32, 41), it is difficult to fully assess the impact of depletion of any individual subunit on the complex composition (fig. S1B). Overall, depletion of INTS6 or INTS11 displayed the greatest loss in miRNA steady state (Fig. 1, A to E, and fig. S1, C to G), with DROSHA-independent miRNAs also being reduced [e.g., 5′m7G-capped miR-320a-3p (9) or mirtrons miR-877-5p and miR1226-3p (33); Fig. 1, F and G, and fig. S1, H and I). We confirmed these findings by TaqMan-qPCR (quantitative polymerase chain reaction) miRNA detection in induced shINTS6 or shINTS11 HeLa cells and siINTS6-, siINTS11-, or siDROSHA-transfected HEK293T cells (fig. S1, J and K,). Last, Northern blot analyses of miRNAs let-7d-5p and miR-17-5p confirmed the specific loss of miRNA steady state following depletion of INTS11 or INTS6 (Fig. 1, H and I).
Fig. 1. Integrator absence leads to global miRNA loss.
(A) smRNA-seq profile at the canonical miRNA locus miR-17. The y-axis represents counts per million (CPM). (B) Box and violin plot depicting the log2 fold change of 205 expressed miRNAs determined by smRNA-seq in the indicated knockdown or uninduced shControl HeLa cells, calculated against induced shControl cells. ***P < 0.001, one-way analysis of variance (ANOVA), followed by Tukey’s post hoc test. n.s., not significant. Gray dotted lines indicate 1.5× threshold. (C) Global average smRNA-seq profiles around 112 5p-miRNAs aligned at their start site. (D and E) Volcano plot comparing statistical significance and miRNA log2 fold change between control and knockdown cells. Padj refers to adjusted P value as calculated by DESeq2. (D) ShINTS6. (E) ShINTS11. (F and G) smRNA-seq profiles at DROSHA-independent miRNA loci. The y-axis represents CPM. (F) 5′-Capped miR-320a locus. (G) Mirtron miR-877 locus. (H) Northern blot miRNA detection from uninduced and induced [+1 μg/ml doxycycline (Dox)] shControl, shINTS6, and shINTS11 HeLa cells. Twenty micrograms of RNA was loaded on a 15% tris-borate EDTA (TBE)–urea gel and probed for let-7d-5p, miR-17-5p, 5S ribosomal RNA, and RNU43. (I) Quantification of Northern blot signal intensity relative to shControl + Dox (n = 3). **P < 0.01 and ***P < 0.001, unpaired t test.
Integrator loss does not affect primary or precursor miRNA levels
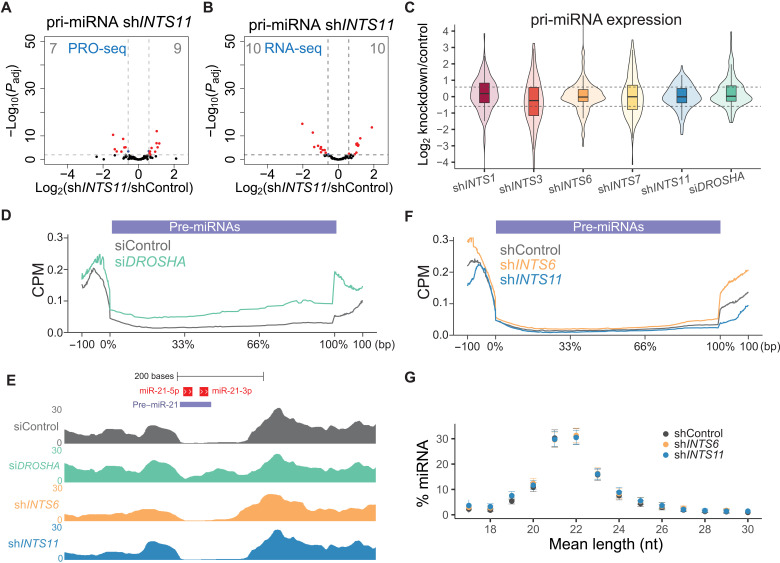
Because Integrator has been shown to widely affect transcription levels of protein coding genes, RNAPII pause-release, and processing of noncoding RNAs (28, 29, 32, 42), we assessed whether our observed miRNA loss was related to alterations in pri-miRNA transcript levels. Because of efficient DROSHA processing, primary transcripts are often inaccurately annotated (43). To reliably capture primary transcripts corresponding to the studied 205 mature miRNAs, we performed de novo transcript assembly from a HeLa siDROSHA RNA-seq dataset, stipulating that the loss of DROSHA cleavage pattern allows for a more precise detection of primary transcripts. We combined the resulting transcript annotation with Ensembl-annotated primary transcripts focusing on genomic loci containing our miRNAs of interest. Careful comparisons and manual curation resulted in a list of 112 expressed pri-miRNA transcripts that correspond to our mature miRNAs. Differential analyses of the expression levels of curated pri-miRNA transcripts revealed that depletion of INTS11 did not significantly influence nascent transcription, as measured by precision run-on sequencing (PRO-seq; Fig. 2A), or the steady state of pri-miRNA detected by RNA-seq (Fig. 2B). Depletion of INTS1, INTS3, INTS6, INTS7, or DROSHA did not change the average steady-state levels of pri-miRNAs (Fig. 2C).
Fig. 2. miRNA loss does not depend on Integrator’s endonucleolytic cleavage activity.
(A and B) Volcano plot of statistical significance against log2 fold change between shControl and shINTS11 cells quantifying 112 pri-miRNAs in (A) PRO-seq (transcriptional elongation) or (B) total RNA-seq. Significantly regulated pri-miRNAs are depicted in red. Padj refers to adjusted P value as calculated by DESeq2. (C) Box and violin plot depicting the log2 fold change of pri-miRNA expression obtained by total RNA-seq in the indicated knockdown cells, calculated against pri-miRNA levels in shControl or siControl cells. One hundred twelve pri-miRNAs were extracted from Ensembl or newly annotated transcripts based on siDROSHA RNA-seq (see Materials and Methods for details). (D) Cumulative total RNA-seq read densities across annotated pre-miRNAs ± 100 bp for siControl and siDROSHA samples. (E) Example total RNA-seq profiles of indicated knockdowns depicting pre-miRNA excision at the miR-21 locus. Mature miR-21 are indicated in red, and the annotated precursor is indicated in light blue. The y-axis represents CPM. (F) Cumulative total RNA-seq read densities across annotated pre-miRNAs ± 100 bp for shControl, shINTS6, and shINTS11 samples. (G) Mean length nucleotide (nt) percentage per miRNA ranging from 18 to 30 nucleotides detected in smRNA-seq of shControl, shINTS6, and shINTS11 cells. Means ± SEM.
Next, we assessed whether depletion of Integrator subunits disrupted the processing of pri-miRNAs to precursor miRNA hairpin governed by Microprocessor (DROSHA/DGCR8) (2, 3, 5). As expected, while depletion of DROSHA impaired the processing of primary to precursor miRNA (Fig. 2, D and E), INTS6 or INTS11 depletion had no effect on the biogenesis of precursor miRNAs (Fig. 2, E and F). In addition, INTS6 or INTS11 depletion did not affect the average length of mature miRNAs, suggesting that the processing of precursor to mature miRNAs by DICER/TRBP was not impaired (Fig. 2G). Together, we did not find evidence of Integrator function in modulating either the transcription or processing of primary or precursor miRNAs to their mature species.
We next inquired whether genes belonging to the miRNA machinery, as defined by Gene Ontology (GO) terms, were differentially expressed following Integrator depletion. We detected some fluctuations in the expression of components of the miRNA machinery (fig. S2A) and in the abundance of the major miRNA-related proteins (fig. S2B). Notably, we detected decreased AGO2 protein levels following INTS11 depletion (fig. S2B). Conversely, AGO2 levels after INTS6 depletion were slightly up-regulated, despite the shared loss of miRNAs following INTS6 and INTS11 depletion, suggesting that fluctuation in AGO levels could not explain the global miRNA reduction following Integrator depletion. Furthermore, we could not identify any consistent alterations that explained the profound loss of mature miRNAs following Integrator depletion. Last, we determined whether INTS11 catalytic activity played a role in modulation of mature miRNA levels. We found that both ectopic expression of wild-type or catalytic inactive E203Q mutant INTS11 rescued the loss of miRNAs following depletion of endogenous INTS11 (fig. S2, C to E), indicating that Integrator’s miRNA function was independent of its endonucleolytic activity. Together, these results point to a role for Integrator in regulation of mature miRNA levels at a step following processing by DICER/TRBP complex.
Integrator depletion impairs miRNA stabilization
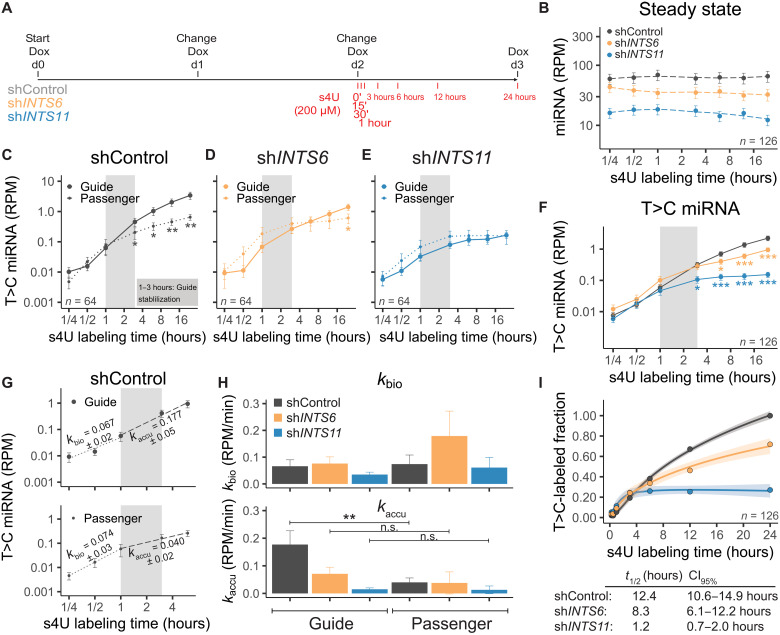
To further confirm that Integrator deficiency did not affect miRNA transcription and processing and to precisely pinpoint Integrator’s role in miRNA fate, we used thiol (SH)–linked alkylation for metabolic sequencing of RNA (SLAM-seq) to determine smRNA dynamics (24) following depletion of INTS6 or INTS11 (Fig. 3A). Briefly, after 2 days of short hairpin RNA (shRNA) induction to assure sufficient INTS knockdown, 4-thiouridine (s4U) was incorporated for increasing time periods (15 min to 24 hours) during transcription (Fig. 3A). Subsequent carboxyamidomethylation [+iodoacetamide (IAA)] allowed us to trace newly labeled miRNAs via the appearance of T>C conversions after sequencing (T>C-labeled miRNAs) from a pool of labeled and unlabeled miRNAs (steady state). While 24 hours of s4U treatment alone did not affect miRNA levels (fig. S3, A and B), global miRNA loss was still observed upon INTS6 or INTS11 depletion at steady state (Fig. 3B and fig. S3, C and D). T>C-labeled miRNAs were only detected in samples treated with s4U and IAA (fig. S3, E to G). A total number of 126 from our list of 205 expressed miRNAs were captured containing at least one T>C conversion after 24 hours of s4U treatment in shControl cells (fig. S3, H and I). smRNA SLAM-seq allows determination of miRNA fate from transcription, miRNA maturation, to miRNA stabilization by tracing the separating duplex miRNAs in guide and passenger miRNA strands (24). Because AGOs asymmetrically load miRNAs based on the identity of the 5′-end nucleotide and the thermodynamic stability of duplex ends (44, 45), the guide miRNA is more likely to be loaded into AGOs than the passenger miRNA, which will be reflected in their relative abundance. Plotting the average RPM (reads per million) of T>C-labeled miRNAs separated for guide (n = 32) and passenger (n = 32) strand from shControl samples allowed us to distinguish miRNA biogenesis (15 min to 1 hour), initiation of RISC loading (1 to 3 hours), and miRNA stability (3 to 24 hours; Fig. 3C), as previously shown in Drosophila melanogaster (24). In shControl-treated cells, the guide and passenger strands initially followed a similar trajectory indicative of their biogenesis from their hairpin precursor (15 min to 1 hour; Fig. 3C). However, the two strands displayed a distinct trajectory within 1 to 3 hours, which is further enhanced from 3 to 24 hours (Fig. 3C). Because the guide strand accumulated, there was a significant decrease in the levels of passenger strand over time, such that the two strands displayed a significant difference in abundance (Fig. 3C). In contrast to control knockdown, depletion of INTS6 (Fig. 3D) or INTS11 (Fig. 3E) impaired the separation of guide and passenger miRNA strands starting at 1- to 3-hour time points and extending throughout the 24 hours.
Fig. 3. Depletion of Integrator reduces miRNA stabilization.
(A) Scheme of INTS knockdown and s4U labeling. D0, day 0. (B) Steady state (unlabeled and T>C labeled) of miRNA expression over time. n = 126. (C to E) T>C-labeled miRNA abundance over time, separated for 32 guide or 32 passenger miRNAs. Means ± SEM. *P < 0.05 and **P < 0.01, Mann-Whitney-Wilcoxon test. (C) ShControl. (D) ShINTS6. (E) ShINTS11. (F) Combined T>C-labeled miRNA abundance. (G) Example of linear regression on shControl guide or passenger miRNAs. miRNA biogenesis rates (kbio) determined from 15 min to 1 hour or accumulation rates (kaccu) from 1 to 6 hours. Slope ± SE is indicated. (H) Histogram of kbio and kaccu. Means ± SEM. **P < 0.01, one-way ANOVA, followed by Tukey’s post hoc test on single miRNAs with kbio or kaccu > 0. (I) Single exponential saturation kinetics to calculate median half-life t1/2 (hours) as depicted in the table below including 95% confidence interval (CI). Shades indicate SEM.
This observation was further confirmed when depicting the average of all detected miRNAs (n = 126; Fig. 3F and fig. S3J), attesting to a highly compromised miRNA stabilization in the absence of INTS6 or INTS11. To quantify the changes in guide and passenger concentrations over time, we determined biogenesis (kbio) and accumulation rates (kaccu) by linear regression of T>C-labeled miRNAs on early (15 min to 1 hour) or intermediate (1 to 6 hours) time points for either guide or passenger miRNAs (Fig. 3G), as described previously (24). While knockdown of INTS6 appeared to result in an increase of passenger miRNA kbio, we did not detect any statistically significant changes for either kbio of guide or passenger miRNAs following INTS11 or INTS6 depletion (Fig. 3H, top). Conversely, as expected, kaccu was significantly higher for guide miRNAs as compared to that of passenger miRNAs in the control knockdown condition (Fig. 3H, bottom). Depletion of INTS6 or INTS11 abrogated the difference between guide and passenger strand kaccu (Fig. 3H, bottom). Concomitantly, median half-life estimations by single exponential saturation kinetics revealed reduced miRNA half-lives following shINTS6 treatment (t1/2 = 8.3 hours) compared to that of shControl (t1/2 = 12.4 hours), with greatest decrease observed following shINTS11 treatment (t1/2 = 1.2 hours) (Fig. 3I). Together, these results revealed that, in the absence of INTS6 or INTS11, miRNAs are severely destabilized, resulting in a profound deceased half-life.
Integrator loss reduces RISC loading
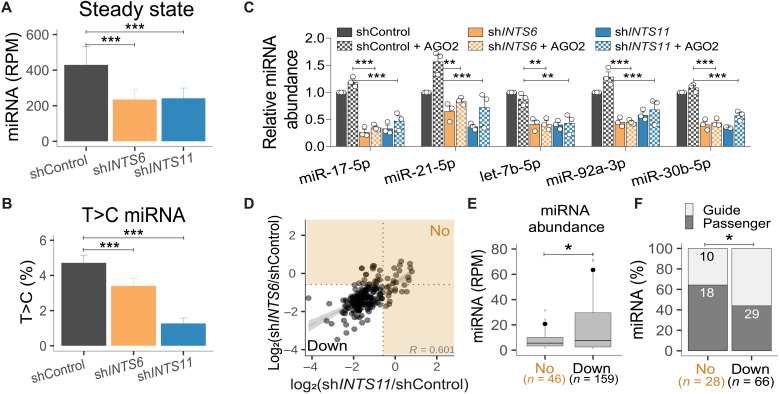
miRNAs display a long half-life, which is thought to reflect their binding and stabilization by AGOs (46). Our miRNA kinetics experiments indicated that the absence of Integrator leads to miRNA destabilization, resulting in similar abundances of guide and passenger miRNAs (Fig. 3, D and E) and decreasing miRNA half-lives (Fig. 3I). We determined whether Integrator may have a role in association of duplex miRNAs with AGOs during the loading process. Analyses of miRNAs associated with AGO2 using TaqMan-qPCR following RNA IP (RIP) revealed that AGO2-loaded miRNAs were markedly reduced after INTS11 or INTS6 depletion (fig. S4, A and B). To confirm that the absence of the Integrator complex affects the newly generated miRNAs from loading into AGO2, we performed s4U labeling for 24 hours, followed by AGO2 RIP to detect RISC-loaded miRNAs in shControl, shINTS6-, and shINTS11-treated cells. We observed a significant decrease in the steady-state levels of miRNAs following INTS6 or INTS11 depletion (Fig. 4A), confirming our RIP-qPCR results. In addition, AGO2 association of T>C-labeled, newly generated miRNAs were significantly reduced upon INTS6 or INTS11 knockdown (Fig. 4B and fig. S4C). Collectively, these results reflect an impairment of AGO2 loading following loss of Integrator subunits INTS6 or INTS11.
Fig. 4. Integrator depletion decreases AGO2 loading.
(A) Steady-state miRNA abundance from AGO2 RIP after 24 hours + s4U. n = 122. (B) T>C miRNA percentage. Means ± SEM. (C) miRNA TaqMan qPCR before and after AGO2 overexpression (see fig. S4D). miRNA levels relative to ath-miR-159a spike-in and shControl. Means ± SEM, n = 3. **P < 0.01 and ***P < 0.001, one-way ANOVA, followed by Dunnett’s multiple comparisons test. (D) Scatterplot of the miRNA log2 fold change in shINTS6 and shINTS11 from Fig. 1 (D and E). Integrator-unregulated miRNAs are indicated by orange shaded area. (R, Spearman correlation coefficient). (E) ShControl miRNA abundance for unregulated (No) and down-regulated (Down) miRNAs. *P < 0.05, Welch two-sample t test. Black circles indicate mean. (F) Percentage of guide and passenger miRNAs. Absolute miRNA numbers are indicated. *P < 0.05, Fisher’s exact test.
In previous experiments, we had observed diminished AGO2 levels following INTS11 but not INTS6 knockdown (fig. S2B). We consequently determined whether increased AGO2 concentrations could alter the steady state of miRNAs in shControl conditions or following INTS6 or INTS11 depletion (Fig. 4C and fig. S4D). We measured the steady-state concentrations of five selected miRNAs (Fig. 4C). While we noticed increased levels of miR-21-5p upon AGO2 ectopic expression in shControl, enhanced expression of AGO2 did not prevent the loss of five miRNAs following depletion of INTS6 or INTS11 (Fig. 4C).
Previous experiments highlighted the cytoplasmic compartment as a main cellular venue for miRNA loading and therefore its stabilization in RISC (47–49). While nuclear miRNA functions have been described (50), they are thought to rely on active RISC transport to the nucleus (51, 52). Integrator has been described to predominantly function in the nucleus with direct links to active transcription (28, 30, 32, 53–55). However, Integrator subunits have been shown to reside in the cytoplasmic fraction depending on the cell type and cell cycle stage (40, 56). While we found a predominant nuclear localization for Integrator subunits, we also found varying concentrations of individual INTS in the cytoplasmic fractions of HEK293T or HeLa extracts (fig. S4E). We therefore determined whether Integrator acts upon a specific population of miRNAs, depending on their subcellular localization. We analyzed smRNA-seq from nuclear and cytoplasmic fractions following INTS6 or INTS11 depletion and detected miRNA loss in both compartments, albeit stronger in the nucleus (fig. S4F). We consequently conclude that Integrator acts on miRNA abundance in both cellular compartments.
To further determine characteristics of miRNAs that may predict their behavior upon Integrator depletion, we compared classes of “unregulated” and “down-regulated” miRNAs based on the degree to which they are depleted after INTS6 and INTS11 knockdown. We detected a total of 46 miRNAs that were not significantly down-regulated following either INTS6 or INTS11 depletion (Fig. 4D, “No” versus “Down”). Seventeen miRNAs remained unregulated in the absence of both INTS6 and INTS11. Collectively, 159 miRNAs were reduced more that 1.5-fold following INTS6 or INTS11 loss (Fig. 4D, Down). miRNAs that were down-regulated after Integrator depletion were significantly more abundant than miRNAs that were less affected by Integrator perturbations (Fig. 4E) and contained a higher proportion of guide strand miRNAs (Fig. 4F). These results further confirm a more pronounced effect of Integrator on AGO-loaded guide miRNAs, which also display a higher abundance.
INTS11 binds duplex miRNAs
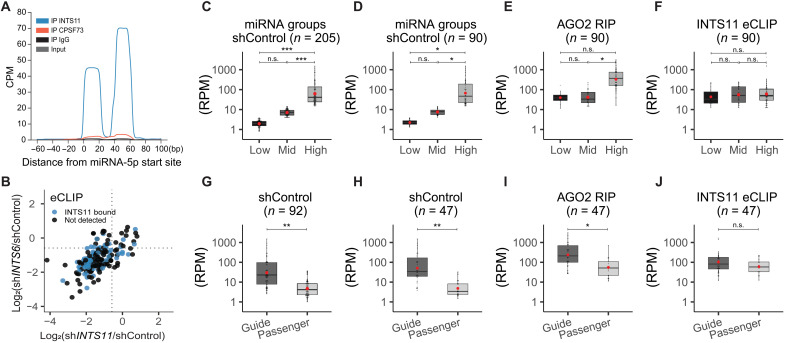
Analyses of miRNA fate following the biogenesis of newly synthesized miRNAs led to the hypothesis that Integrator functions at a step between DICER cleavage of the precursor hairpin and miRNA loading into AGOs. Therefore, we assessed whether Integrator may interact with miRNAs following their processing by DICER/TRBP complex. To examine whether Integrator associates with miRNAs and to pinpoint the species of Integrator-associated miRNAs, we performed eCLIP (57). We optimized eCLIP conditions for the detection of miRNAs by increasing initial RNase concentrations and isolated INTS11 or its homolog CPSF73, a member of the cleavage and polyadenylation specificity factor (CPSF) complex, which served as a negative control (fig. S5A). We identified miRNAs by quantifying the number of reads that overlap with our 205 human mature miRNAs expressed in HeLa in “reads per million mapped reads” (RPM). To maximize detectable read numbers, we pooled our two eCLIP replicates for each sample [INTS11, CPSF73, immunoglobulin G (IgG), and input] (Fig. 5A). Individual eCLIP repeats are shown in fig. S5B. In addition, we used AGO2-bound miRNAs from our AGO2 RIP data from untreated shControl cells as a positive control for the characteristics of AGO2-loaded miRNAs. Among the 205 expressed canonical miRNAs in HeLa cells, we captured 146 miRNAs in the AGO2 RIP. Analyses of eCLIP revealed the presence of 90 miRNAs in the INTS11 samples (Fig. 5A). It is likely that differences in the number of miRNAs detected in AGO2 RIP and INTS11 eCLIP reflected the technical differences among the two experimental methodologies. Nevertheless, in comparison to 90 miRNAs detected following INTS11 eCLIP, control eCLIP performed using antibodies against CPSF73, IgG, or size-matched input represented 11, 4, and 8 miRNAs, respectively. INTS11-associated miRNAs measured by eCLIP included not only the down-regulated miRNAs following INTS11 or INTS6 depletion but also the miRNAs that were not significantly changed following Integrator depletion (Fig. 5B). This observation indicates that the physical interaction of miRNAs with INTS11 may not be predictive of their functional loss upon Integrator depletion.
Fig. 5. Integrator-bound miRNAs are independent of miRNA abundance.
(A) Global average of INTS11, CPSF73, IgG, and size-matched input eCLIP profiles around 112 5p-miRNAs aligned at their start site. (B) Scatterplot of the log2 fold change of 205 miRNAs in after INTS6 (y-axis) or INTS11 (x-axis) depletion (see also Fig. 4D) with concomitant labeling of INTS11-bound miRNAs (detected by eCLIP, n = 90). (C to F) Classification according miRNA abundance. (C) Two hundred five detected miRNAs from shControl smRNA-seq were separated by tertiles to define “low-,” “mid-,” and “high-”expression miRNAs. (D) Same as in (C), calculated with the subset of 90 miRNAs also detected from INTS11 eCLIP. (E) Abundance of the 90 miRNAs subset detected from AGO2 RIP, according to previously defined expression classes. (F) Abundance of miRNAs detected in eCLIP. (G to J) Classification according miRNA type (guide or passenger miRNA). (G) Abundance of 92 miRNA couples detected in shControl smRNA-seq and separated by type. (H) Same as (G), calculated with the subset of 47 miRNAs and also detected from INTS11 eCLIP. (I) Abundance separated by type of the 47 miRNA subset detected from AGO2 RIP. (J) INTS11 eCLIP miRNA abundance separated by type. Red circles indicate mean. Statistics were calculated using either one-way ANOVA followed by Tukey’s post hoc test (C to F) or Welch two-sample t test (G to J). *P < 0.05, **P < 0.01, and ***P < 0.001.
To further characterize the 90 INTS11-bound miRNAs detected by eCLIP, we determined whether they correlated with global miRNA expression levels (Fig. 5, C to F). We also examined whether they displayed a preference for guide or passenger miRNA strand (Fig. 5, G to J). We divided the 205 miRNAs detected in HeLa cells by tertiles into low-expressing (low, n = 69), mid-expressing (mid, n = 68), and high-expressing miRNAs (high, n = 68) (Fig. 5C). The miRNA expression levels were significantly different between the groups low-high and mid-high (Fig. 5C). The 90 miRNAs found associated with INTS11 using eCLIP showed a similar distribution following analyses of their steady-state levels using smRNA-seq (Fig. 5D). The differences in miRNAs concentrations in their steady state was similarly reflected in the concentration of miRNAs found in AGO2 RIP attesting to AGO association as a key driver of miRNA abundance (Fig. 5E). In contrast, the INTS11-associated miRNAs did not represent the steady-state levels of miRNAs and were found to be present at similar levels among all tertiles (Fig. 5F).
Next, we assessed the differences among the two strands of miRNAs, the “guide” and “passenger” strands. As described previously, we identified 92 miRNA couples and defined the more abundant partner as guide and the less abundant one as the passenger miRNA. As expected, when separated according to abundance in smRNA-seq, we observed a significant difference between guide and passenger strands with the guide strands showing higher abundance (Fig. 5G). Among these 92 miRNAs, 47 miRNAs were detected as INTS11-bound miRNAs in eCLIP samples. We analyzed the abundance of these 47 guide or passenger miRNAs in control steady state, AGO2 RIP, and INTS11 eCLIP (Fig 5, H to J). We found a greater representation of guide strand in miRNA steady state measured through smRNA-seq and in the miRNAs detected in AGO2 RIP, consistent with expected increased abundance of AGO2-bound guide strand versus the passenger strand (Fig. 5, H and I). In contrast to AGO2-associated miRNAs, the abundance of guide and passenger strands in INTS11-bound miRNAs was not significantly different (Fig. 5J). In addition, we did not detect any sequences corresponding to the hairpin miRNAs beyond the duplex 22-nucleotide mature miRNA (Fig. 5A and fig. S5B), indicating that INTS11 did not associate with miRNA precursor hairpin. Together, these results strongly support the hypothesis that INTS11 associates with the miRNA duplex following its processing by DICER and before separation of the duplex and stabilization of the guide strand through AGO association.
Integrator interacts with AGOs
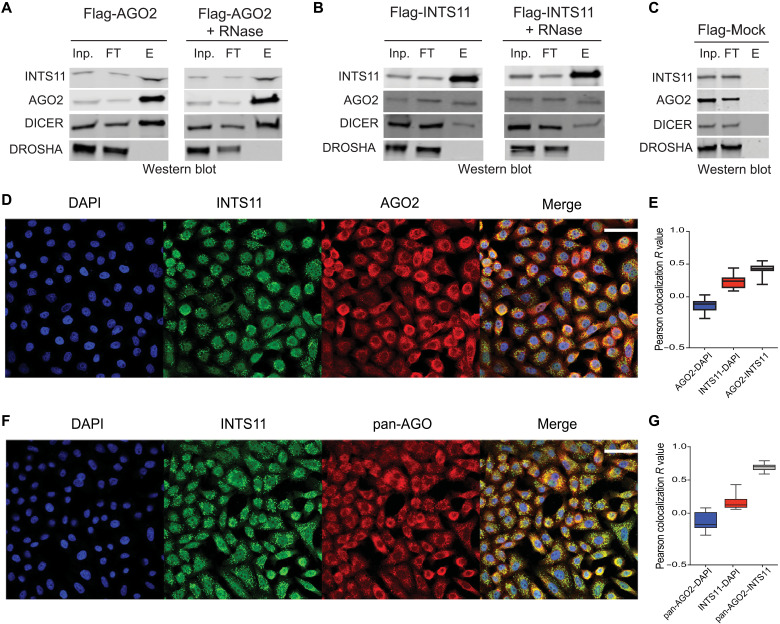
We had previously developed stable cell lines expressing Flag-INTS11 and Flag-AGO2 (1, 28, 29) in HEK-293T cells. To assess whether AGOs physically associate with the Integrator complex, we isolated Flag-AGO2 from the cytoplasmic fraction of HEK293T cells (Fig. 6A). While we found an association of AGO2 with DICER and INTS11, DROSHA was absent (Fig. 6A). In addition, treatment of the affinity-purified fractions with RNase A/T1 did not affect the association of AGO2 with either DICER or INTS11 (Fig. 6A, right). We next isolated Flag-INTS11 from the cytoplasmic fraction of HEK-293T cells stably expressing Flag-INTS11 and found an RNase-independent association of AGO2 and DICER with INTS11, while DROSHA was absent (Fig. 6B). Control Flag-mock purification from untagged HEK-293T cells did not yield any aforementioned components (Fig. 6C). These results highlight a physical association of a fraction of DICER, AGO2, and INTS11 in the cytoplasm of HEK293T cells. To further characterize the association of INTS11 and AGOs, we used immunofluorescence analysis to visualize their cellular localization. While as expected, we found a predominant localization of in the cytoplasm AGOs (using either an antibody against AGO2 or an antibody against pan-AGO) and INTS11 in the nucleus of HeLa cells, and there was a substantial colocalization of AGOs and INTS11, mainly in the cytoplasm (Fig. 6, D to G). Together, these results point to a physical proximity of Integrator and AGOs in the cytoplasmic fraction of human cells.
Fig. 6. Characterization of the interaction between Integrator and AGO2.
(A) Flag-AGO2 affinity purification, either without (left) or with (right) RNase treatment. (B) Flag-INTS11 affinity purification, either without (left) or with (right) RNase treatment. Inp., input; FT, flowthrough; E, elution. (C) Flag-Mock affinity purification. (D) Representative immunofluorescence of INTS11 (green; Abcam, #ab75276) and AGO2 (red; Abcam, #ab57113) and merged images including DAPI (4′,6-diamidino-2-phenylindole; blue) in induced shControl HeLa cells. Scale bar, 50 μm. (E) Quantified Pearson colocalization values of AGO2-INTS11 compared to AGO2-DAPI and INTS11-DAPI using ImageJ 1.53o plug-in Coloc 2. (F) Representative immunofluorescence of INTS11 (green; Abcam, #ab75276) and pan-AGO (red; Abcam, #MABE56) and merged images including DAPI (blue) in induced shControl HeLa cells. Scale bar, 50 μm. (G) Quantified colocalization of pan-AGO2–INTS11 compared to pan-AGO2–DAPI and INTS11-DAPI.
Integrator potentiates RISC activity
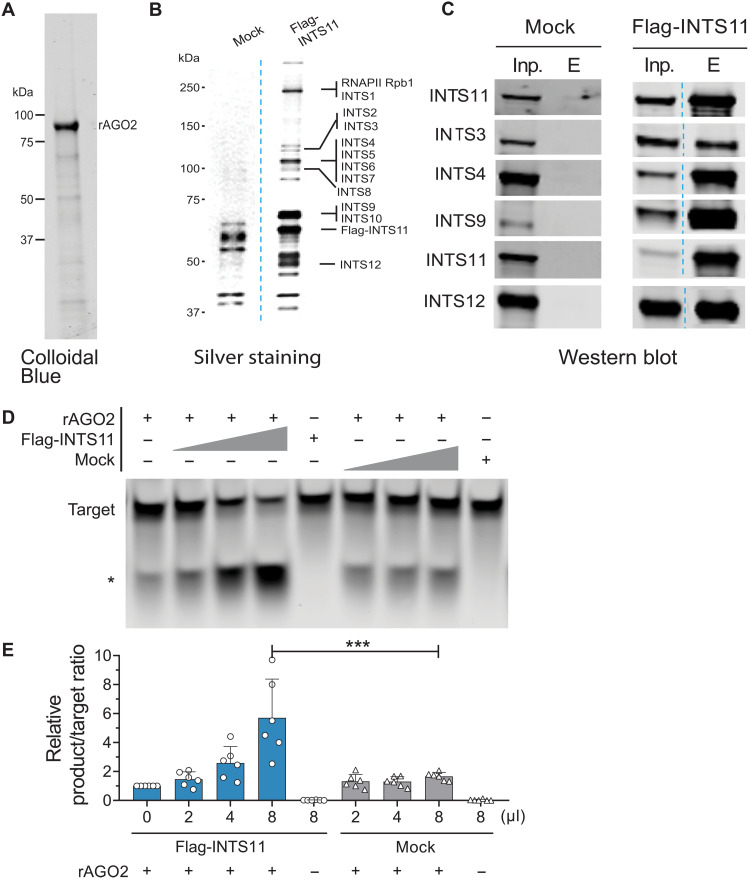
To gain mechanistic insight into the role of Integrator in AGO2 functions, we developed an AGO2-dependent miRNA target cleavage assay in vitro, using a dme-let-7a miRNA duplex, a complementary target sequence, and recombinant AGO2 (rAGO2) (Fig. 7A). We affinity-purified the Integrator complex from the nuclear fraction of Flag-INTS11 HEK293T cells or performed a control nuclear mock Flag purification from untagged HEK293T cells (Fig. 7, B and C). While the highest concentration of the affinity-purified Integrator did not exhibit any cleavage activity on its own, addition of increasing concentrations of affinity-purified Integrator specifically enhanced the cleavage activity of rAGO2 (Fig. 7, D and E). The control mock-purified fraction was devoid of any stimulatory effect (Fig. 7, D and E). These results indicated that, while in vitro, rAGO2 displayed a cleavage activity towards the target sequence, its activity was significantly enhanced in the presence of the Integrator complex. Together with our previous findings of direct INTS11-miRNA duplex binding and INTS11-AGO interaction, we propose a role for Integrator in assisting the loading of miRNAs into AGO, which is reflected in enhanced AGO2 activity in vitro.
Fig. 7. Purified Integrator complex enhances AGO2 target cleavage efficiency.
(A) rAGO2 purchased from Active Motif (#31486) by Colloidal Blue staining. (B) Silver stain of affinity-purified Integrator complex. Integrator complex was purified from nuclear lysate of HEK293Tcells stably overexpressing Flag-INTS11. Mock indicates the same Flag-IP purification steps from parental HEK293T cells. The indicated Integrator subunits were assigned as identified by Baillat et al. (28). (C) Western blot detection of indicated INTS. (D) AGO2 cleavage assay in the presence of dme-let-7a miRNA-duplex, 5′ IRDye-700 labeled guide complementary target RNA, rAGO2 and increasing concentrations of affinity purified (Flag-INTS11) Integrator complex or Mock. *Cleaved product. (E) Quantification of the product/target ratio relative to rAGO2 alone (lane 1). Means ± SEM, n = 6. One-way ANOVA, Dunnett’s multiple comparisons against rAGO2 + 8 μl of Mock condition. ***P < 0.001.
DISCUSSION
We serendipitously found that depletion of several Integrator subunits led to the global loss of miRNAs. Our analyses of nascent RNA-seq (PRO-seq) and steady-state levels of miRNA primary transcripts indicated that the observed loss of mature miRNAs was not the result of changes in the primary transcripts. In addition, because all classes of miRNAs, including mirtrons and 5′-capped miRNAs (DROSHA-independent miRNAs), were diminished, the defect could not have resulted from the processing of primary transcripts to precursor miRNAs by Microprocessor complex. Furthermore, while we could readily observe the defect in pri-miRNA processing following DROSHA depletion, knockdown of Integrator subunits had no effect in precursor formation (Fig. 2, D to F). Time course experiments following the fate of nascent miRNA synthesis pinpointed a defect in AGO stabilization of miRNAs as a key culprit for miRNA loss (Fig. 3).
Analyses of miRNA steady state or miRNAs that are loaded onto AGO proteins by RIP can distinguish the differences among the two strands of the miRNAs with a guide strand displaying a higher concentration. This is thought to arise from stabilization of the guide strand upon AGO association and the degradation of the passenger strand through the action of RNA exonucleases (23, 58). Analyses of miRNAs associated with INTS11 in eCLIP experiments indicated an equivalent abundance for guide and passenger strands, highlighting the interaction of duplex miRNA with INTS11 before strand separation and stabilization by AGO (Fig. 5, G to J). These results were also consistent with the metabolic sequencing of s4U-labeled miRNAs pointing to the destabilization of the mature miRNAs upon Integrator loss. Collectively, these results indicate a role for Integrator associating with duplex miRNAs and stabilization of the AGO-associated strand of the duplex miRNA.
We have previously shown that Integrator plays a role in pause-release of RNAPII during early rounds of transcriptional initiation (29). We demonstrated that Integrator promotes premature termination of transcription through cleavage of nonproductive stalled RNAPII, allowing for renewed transcriptional initiation and productive RNAPII elongation (32). In addition, Integrator played a key role in growth factor signaling orchestrating the responsiveness to mitogen-activated protein kinase signaling (59). While we have uncovered numerous functions for this multiprotein complex in transcriptional control and RNA processing in the nucleus, the current results point to a role in posttranscriptional regulation for Integrator in both nuclear and cytoplasmic compartments through promotion of miRNA stability, expanding the repertoire of Integrator functions.
Integrator complex was previously implicated in the processing of viral miRNAs (60) displaying similarities to UsnRNA in their 3′ end, a process dependent on INTS11’s endonucleolytic activity (31). However, a global role for Integrator in the canonical miRNA pathway was not anticipated, since miRNA biogenesis was studied in detail biochemically and structurally (4, 13, 49). In this work, we found a key role for Integrator in binding the duplex miRNA and through its association with AGO proteins, facilitating the handing over of the processed mature miRNA duplex to the effector silencing complexes. Our eCLIP results are consistent with Integrator association with miRNA duplex following its production by DICER/TRBP and before its loading into the AGO2. Because DICER/TRBP are enriched in the cytoplasm, it is likely that Integrator before its nuclear localization interacts with the duplex miRNA in the cytoplasmic fraction, where AGO proteins are also enriched. Our studies suggest that, while approximately 40% of INTS11 display cytoplasmic localization, around 10 to 20% of other Integrator subunits reside in the cytoplasm of HeLa or HEK293T cells (fig. S4E). At present, it is not clear whether Integrator assembles as a holo-complex in the nuclear fraction or that, similar to other multiprotein complexes such as small nuclear ribonucleoprotein particles, it is assembled in the cytoplasm and then transported to the nucleus (61, 62). Because INTS11 displayed the greatest cytoplasmic localization, associated with duplex miRNAs, and its depletion exhibited the largest decrease in miRNA levels, we surmise that it fulfills a central role as a component of the Integrator complex in controlling miRNA stability. Other subunits such as INTS1, INTS3, and INTS6 may play auxiliary roles or are required for complex integrity and/or INTS11 stability. While multiple lines of experimental evidence in vivo suggest that Integrator regulation of miRNA stability is a direct function, it is formally possible that depletion of Integrator subunits affects an unknown component of miRNA machinery, resulting in global decreased miRNA levels.
Addition of affinity-purified Integrator complex to rAGO2 could potentiate its cleavage activity of a complementary target RNA in a reconstituted in vitro assay. While such an observation is consistent with a direct effect of Integrator on AGO2 loading and activity in vitro, the formal proof has to wait the analysis of fully assembled recombinant Integrator complex. Future biochemical and structural characterization of Integrator and AGO complexes should shed further light into the precise interplay of the Integrator complex in posttranscriptional gene silencing through regulation of AGO proteins.
MATERIALS AND METHODS
Cell lines
HeLa [the American Type Culture Collection (ATCC), #CCL-2] and HEK293T (ATCC, #CRL-3216) cells were maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Gibco, #11965-084), supplemented with 10% fetal bovine serum (FBS; Atlas Biologicals, #F-0500-D) and the respective antibiotics, if required. Cells were regularly tested for mycoplasma. HeLa-inducible shControl (containing an shRNA targeting green fluorescent protein), shINTS1, and shINTS11 cells have been previously described by Gardini et al. (29), and shRNA-resistant N-terminal Flag-tagged wild-type and E203Q mutant INTS11 have been previously described by Beckedorff et al. (32). HeLa-inducible shINTS3, shINTS6, and shINTS7 single clones were established by lentiviral infection with Tet-pLKO-puro vector (Addgene, #21915) containing the respective shRNA sequences (shINTS3: GCTGTGACCTCATTCGCTACA, shINTS6: ACCACTAATGATTCGATAATA, and shINTS7: GCAGTAAAGAGACTTGCTATT) and puromycin (2.5 μg/ml; InvivoGen, #ant-pr) selection. shINTS-stable cells were maintained in puromycin (2.5 μg/ml), wild type, and E203Q in the presence of puromycin and G418 (200 μg/ml; InvivoGen, #ant-gn-5). shRNA expression was induced by adding doxycycline (Dox; Selleckchem, #S4163) at 1 μg/ml for 3 days with medium replaced every 24 hours.
Cell transfections
HeLa cells were transfected with 20 nmol of siControl (Ambion, #4390847) or siDROSHA (Ambion, #s26491) using Lipofectamine RNAiMAX (Invitrogen, #13778030) according to the manufacturer’s instructions. HEK293T cells were transfected with 30 nmol of siControl, equimolar amounts of combined siINTS6 (#s25483 and #s25484), siINTS11 (#s29893, #s29894, and #s29895), or siDROSHA (#s26490 and #s26491). For transient AGO2 overexpression in HEK293T cells, 3.2 μg of pFlag-CMV-AGO2 plasmid was transfected using Lipofectamine 2000 (Invitrogen, #11668019). Cells were harvested 3 days after transfection.
Silver staining
Four to 12% gel (Invitrogen) was fixed in 50% methanol and 10% acetic acid for 1 hour. Fixation was continued in 10% methanol and 7% acetic acid for 1 hour. The gel then was washed in 10% glutaraldehyde for 15 min, followed by three times washing in Milli-Q water for 15 min. Silver staining was performed for 15 min in 100 ml of staining solution (185 μl of NaOH, 2.8 ml of NH4OH, and 1 g of AgNO3, brought it up to 100 ml with Milli-Q water). The gel was washed three times for 2 min and then developed in 100 ml of developing solution (0.5 ml of 1% citric acid and 52 μl of 37% formaldehyde in 100 ml Milli-Q water). Gel staining was stopped with 50% methanol and 5% acetic acid.
Colloidal blue staining
Colloidal blue staining was done on 4 to 12% gel (Invitrogen) and then stained according to the manufacturer’s protocol by the Colloidal Blue Staining Kit (Invitrogen, #LC6025).
Immunoblot detection
For whole-cell radioimmunoprecipitation assay (RIPA) lysates, cell pellets were lysed in 1 volume of RIPA buffer [150 mM NaCl, 5 mM EDTA, 50 mM tris (pH 8.0), 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS] in the presence of Halt protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, #1861282) for 15 min on ice and centrifuged at maximum speed for 15 min at 4°C. The protein concentration of the lysate was measured using the Pierce PCA Protein Assay Kit (Thermo Fisher Scientific, #23225), and equal amount of proteins (10 to 30 μg) was separated by 4 to 15% Criterion TGX Stain-Free precast polyacrylamide gels (Bio-Rad, #5678084). After transfer on nitrocellulose membranes, we detected our protein of interest using the following antibodies: INTS1 (Bethyl Laboratories, #A300-361A), INTS3 (Sigma Prestige, #HPA074391), INTS6 (Novus Biologicals, #NB10086990), INTS7 (Bethyl Laboratories, #A300-271A), INTS11 (Sigma Prestige, #HPA029025), glyceraldehyde-3-phosphate dehydrogenase (Abcam, #ab8245), DROSHA (Abcam, #ab12286), DICER (Abcam, #ab14601), DGCR8 (Abcam, #ab90579), AGO1 (Cell Signaling, #D84G10), AGO2 (Abcam, #ab57113), AGO3 (Sigma-Aldrich, #SAB4200112), AGO4 (Cell Signaling, #D10F10), and lamin B1 (Proteintech, #66095-1-1g). Antibodies were used in 1:1000 dilutions.
Subcellular fractionation
Nuclear and cytoplasmic fractions of HEK293T or HeLa cells were prepared as described by Bhatt et al. (63). Briefly, cells were washed with phosphate-buffered saline, and 1 × 107 cells were lysed in 400 μl of cytoplasmic lysis buffer (10 mM tris-HCl, 15 mM NaCl, and 0.15% NP-40), layered on a 500-μl sucrose cushion (10 mM tris-HCl, 15 mM NaCl, and 24% sucrose, w/v), and then centrifuged at 3.500g for 10 min at 4°C. A nuclear protein was extracted from the pellet using 1 volume of RIPA buffer including protease inhibitor cocktail (Thermo Fisher Scientific, #1861282) as described above, and 20 μg of nuclear and cytoplasmic protein was inserted for immunoblot as described above. Immunoblot detection of lamin B1 served as nuclear control for efficient fractionation.
RNA isolation
Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific, #15596026) according to the manufacturer’s instructions. Nuclear and cytoplasmic fractions were prepared as described above, and TRIzol LS reagent (Thermo Fisher Scientific, #10296010) was used for extraction of cytoplasmic RNA. Genomic DNA was removed by Turbo deoxyribonuclease (DNase) treatment (Invitrogen, #AM1907).
miRNA detection by TaqMan qPCR
Ten nanograms of total RNA containing 5 pM ath-miR-159a spike-in was reverse-transcribed using a TaqMan Advanced miRNA cDNA synthesis kit (Applied Biosystems, #A28007), and miRNAs were detected using specific probes for ath-miR159a (478411_mir), miR-17-5p (478447_mir), miR-21-5p (477975_mir), let-7b-5p (478576_mir), miR-320a-3p (478594_mir), miR-877-5p (478206_mir), miR-1226-3p (478640_mir), miR-92a-3p (477827_mir), miR-30b-5p, (478007_mir), miR-19a-5p (478750_mir), miR-182-5p (477935_mir), and RNU43_FAMMGB (Thermo Fisher Scientific, custom order) and SsoAdvanced Universal Probes Supermix (Bio-Rad, #1725281). TaqMan Advanced miRNA assay probe efficiencies were verified at 100 ± 9.5% (Table 1). Relative miRNA expression was calculated against RNU43 or ath-miR-159a spike-in and shControl using the ΔΔct method.
Table 1. Primer efficiency estimation of TaqMan probes used in our analysis.
| TaqMan Advanced miRNA qPCR primer | Primer efficiency E (%) |
|---|---|
| ath-miR-159a | 99.9 |
| let-7b-5p | 96.8 |
| miR-17-5p | 99.7 |
| miR-21-5p | 94.8 |
| miR-30b-5p | 99.0 |
| miR-92a-3p | 91.9 |
| miR-182-5p | 97.2 |
| miR-320a-3p | 98.9 |
| miR-877-5p | 97.8 |
| miR-1226-3p | 90.5 |
Northern blot
Northern blot analysis was performed on the basis of the method that has been previously described, with some modifications (64). Twenty micrograms of total RNA was separated on 15% tris-borate EDTA (TBE)–urea gels (Bio-Rad, #4566055) and then transferred to a positively charged nylon membrane (Roche, #11209299001). The membrane was cross-linked twice with 254 nm of UV light at 120 mJ/cm2 using a CL-1000 UV cross-linker. After prehybridization with 10 ml of ExpressHyb hybridization solution (Clontech) for 40 min at 37°C, the membrane was hybridized overnight with 15 pmol of IRDye-labeled oligonucleotide probes [hsa-miR-17-5p (/5IRD700/ctacctgcactgtaagcactttg), hsa-let-7d-5p (/5IRD700/aactatgcaacctactacctct), RNU43 (/5IRD800/CAGCACACAGTTTCTGTCCGCCCGTC), and RNA5S1 (/5IRD800/cccaggcggtctcccatccaagtactaaccaggcccgaccc)] in 10 ml of ExpressHyb solution at 37°C. The membrane was washed twice with 2× SSC and 0.1% SDS and once with 1× SSC and 0.1% SDS (10 min at room temperature for each) and visualized using the Odyssey CLx Imaging System followed by quantification using Image Studio software.
smRNA library preparation and genome mapping
smRNA libraries were prepared using the SMARTer smRNA-seq kit (Takara, #635030) with 700 ng of total RNA. We omitted the size selection of input RNA to make sure to capture different smRNA species [e.g., small nucleolar RNAs (snoRNAs) are ranging from 60 to 170 nucleotides (65) and promoter RNAs from 18 to 70 base pairs (bp), with a mean read length of 40 bp (32)] but performed bead-based library size selection, as recommended by the manufacturer’s protocol. smRNA-seq from nuclear and cytoplasmic RNA was performed from 700 ng containing 35 ng (5%) of D. melanogaster RNA as spike-in. Experiments were performed in two independent biological replicates that were sequenced in the same sequencing run to avoid for sequencing bias. This resulted in five different datasets, each including its own controls (Table 2).
Table 2. Overview of the smRNA-seq datasets.
Set affiliation, sample name, and file name (as submitted in GSE178127) are indicated.
| Set | Sample name | File name |
|---|---|---|
| Set 1 | shControl_r1 | smrna_hela_set1_shcontrol_r1.fastq.gz |
| Set 1 | shControl_r2 | smrna_hela_set1_shcontrol_r2.fastq.gz |
| Set 1 | shINTS1_r1 | smrna_hela_set1_shINTS1_r1.fastq.gz |
| Set 1 | shINTS1_r2 | smrna_hela_set1_shINTS1_r2.fastq.gz |
| Set 1 | shINTS11_r1 | smrna_hela_set1_shINTS11_r1.fastq.gz |
| Set 1 | shINTS11_r2 | smrna_hela_set1_shINTS11_r1.fastq.gz |
| Set 2 | shControl_r1 | smrna_hela_set2_shcontrol_r1.fastq.gz |
| Set 2 | shControl_r2 | smrna_hela_set2_shcontrol_r2.fastq.gz |
| Set 2 | shINTS3_r1 | smrna_hela_set2_shINTS3_r1.fastq.gz |
| Set 2 | shINTS3_r2 | smrna_hela_set2_shINTS3_r2.fastq.gz |
| Set 2 | shINTS7_r1 | smrna_hela_set2_shINTS7_r1.fastq.gz |
| Set 2 | shINTS7_r2 | smrna_hela_set2_shINTS7_r2.fastq.gz |
| Set 3 | shControl_r1 | smrna_hela_set3_shcontrol_r1.fastq.gz |
| Set 3 | shControl_r2 | smrna_hela_set3_shcontrol_r2.fastq.gz |
| Set 3 | shINTS6_r1 | smrna_hela_set3_shINTS6_r1.fastq.gz |
| Set 3 | shINTS6_r2 | smrna_hela_set3_shINTS6_r2.fastq.gz |
| Set 4 | shControl_r1 | smrna_hela_set4_shcontrol_r1.fastq.gz |
| Set 4 | shControl_r2 | smrna_hela_set4_shcontrol_r2.fastq.gz |
| Set 4 | shControl_noDox_r1 | smrna_hela_set4_shcontrol_nodox_r1.fastq.gz |
| Set 4 | shControl_noDox_r2 | smrna_hela_set4_shcontrol_nodox_r2.fastq.gz |
| Set 4 | shINTS11_r1 | smrna_hela_set4_shINTS11_r1.fastq.gz |
| Set 4 | shINTS11_r2 | smrna_hela_set4_shINTS11_r2.fastq.gz |
| Set 4 | WT_r1 | smrna_hela_set4_wt_r1.fastq.gz |
| Set 4 | WT_r2 | smrna_hela_set4_wt_r2.fastq.gz |
| Set 4 | E203Q_r1 | smrna_hela_set4_eq_r1.fastq.gz |
| Set 4 | E203Q_r2 | smrna_hela_set4_eq_r2.fastq.gz |
| Set 5 | siControl_r1 | smrna_hela_set5_sicontrol_r1.fastq.gz |
| Set 5 | siControl_r2 | smrna_hela_set5_sicontrol_r2.fastq.gz |
| Set 5 | siDrosha_r1 | smrna_hela_set5_sidrosha_r1.fastq.gz |
| Set 5 | siDrosha_r2 | smrna_hela_set5_sidrosha_r2.fastq.gz |
We carefully validated the result of each independent dataset (fig. S6A) and performed principal components analysis, a method to emphasize variations in datasets, using DESeq2 on all samples and replicates (fig. S6B). Because of the similarity of our sample replicates and to assure better comparability across samples, we merged our fastq files from different sets that carried the same name for subsequent analyses. To account for varying read numbers and simplify data normalization in the following, we randomly subsampled all samples to 30 million reads. This approach results in two independent files per sample and projected to downstream data processing.
Sequencing reads were trimmed for adapter (AAAAAAAAAA) as recommend by a SMARTer smRNA-seq kit (Takara, #635030) protocol using Cutadapt (66) (v1.18), and reads shorter than 17 bp were omitted. Reads were aligned against human elements in RepBase (v23.08) with STAR (67) (v2.5.3a), and the unmapped output was then mapped against the human genome (hg19), allowing three mismatches and keeping all uniquely aligned reads. For UCSC Genome Browser visualization (https://genome.ucsc.edu/) (68), all tracks were normalized by CPM (counts per million) using deepTools2 (69) (v3.2.1).
miRNA detection and data analysis
miRNAs were quantified using miRDeep2 (70) (v2.0.0.7), and the top 200 expressed known canonical miRNAs in shControl samples were selected for further analysis. We extended this list by microprocessor-independent miRNAs (mirtrons: miR-877-5p and miR-1226-3p) (33) and miRNAs with capped 5′-ends (miR-320a-3p/5p and miR-484) (9). The final list of 205 miRNAs was analyzed in all datasets. Differential expression was calculated using DESeq2 (71) and R (version 3.6.1). Differentially expressed miRNAs were determined by a cutoff of 1.5-fold and q value of 0.01 (table S1). Significances were either calculated by DESeq2 or using one-way analysis of variance (ANOVA) testing, followed by Tukey multiple pairwise comparisons in R. Graphics were generated using ggplot2 (72). Boxplots were represented with the median, the lower and upper hinges correspond to the first and third quartiles, and the whiskers represent 1.5× the interquartile range to both sides. Global average smRNA-seq profiles were based on CPM-normalized bigwig files. miRNA lengths were determined by mapping reads after removal of repetitive regions to miRNA precursors as described below (the “smRNA SLAM-seq: Bioinformatic processing” section), and the mapped sequence lengths were retrieved.
Total RNA library preparation and genome mapping
Total RNA-seq libraries were generated using a TruSeq Stranded Total RNA library preparation kit (Illumina, #20020596) with 500 ng of DNase-treated RNA, including ribosomal RNA depletion. Sequencing was performed using NovaSeq to at least 50 million reads. Resulting fastq files were processed with Trimmomatic (73) v0.32 and aligned to human genome (hg19) using spliced transcripts alignment to a reference (STAR) (67) aligner v2.5.3a with default parameters. For UCSC Genome Browser (68) visualization (https://genome.ucsc.edu/), all tracks were normalized by CPM using deepTools2 (69) (v3.2.1). RNA-seq by expectation-maximization (RSEM) (74) v1.2.31 was used to obtain expected gene counts against the human Ensembl reference (release 87). Differential expression of shINTS compared to shControl was determined using DESeq2 (71). miRNA-related genes were determined by selection-relevant GO terms (75) containing “miRNA” (GO annotations: 0070883, 0070878, 0031054, 0035280, 1990428, 0035196, 2000634, 0031053, 0035281, and 2000631) and extracting a list of 42 unique gene names.
Pre-miRNA determination
The miRNA precursor file was obtained from miRBase (76) v22, extracting entries annotated as “primary transcript” (corresponds to pre-miRNA) and lifting over to hg19 using CrossMap (77). The resulting file was crossed with our list of 205 expressed miRNAs [BEDTools (78) intersect, v2.29.0] to keep only relevant entries (n = 176; GSE178127: 176_precursor_cleaned_hg19.bed).
Pri-miRNA determination and quantification
To assess expression of pri-miRNAs, we used RNA-seq from siDROSHA-transfected HeLa cells as basis for de novo transcriptome assembly using StringTie (79) (v2.0). We retrieved transcripts overlapping with our 176 annotated miRNA precursors of interest (see above) from the newly annotated transcript file, Ensembl annotation GRCh37.87 and GRCh38.99 (lifted to hg19 using CrossMap) (77). After curating the resulting annotation file manually to remove unrelated transcripts and annotated precursor miRNAs, we obtained a final reference of 112 expressed primary transcripts in HeLa cells (GSE178127: primir_final_annotation_v87_v99_StringTie_hg19_manualClean.gtf). We mapped shControl and shINTS RNA-seq data to the new primary transcript reference using STAR v2.5.3a and performed RSEM and DESeq2 as described above to assess differential pri-miRNA expression.
smRNA SLAM-seq: s4U treatment and carboxyamidomethylation
smRNA SLAM-seq and data analysis were described by Reichholf et al. (24). ShControl, shINTS6, and shINTS11 cells were seeded at a density of 1 × 106 cells in 10-cm dishes at day −1 in DMEM (Gibco, #11965-084) and supplemented with 10% FBS (Atlas Biologicals, #F-0500-D). shRNA induction was started at day 0 by adding Dox-containing medium (1 μg/ml; Selleckchem, #S4163); the medium was changed every 24 hours. Two days after shRNA induction (corresponds to time point 0 min), cells were additionally treated with 200 μM s4U (Cayman chemical, #16373). Parallel controls without s4U treatment were performed. Medium was exchanged every 3 hours during metabolic labeling to ensure homogeneous incorporation, and cells were kept from light exposure. At the respective time points (0, 15, 30 min, 1, 3, 6, 12, and 24 hours), cells were lysed directly on plate using a TRIzol reagent (Thermo Fisher Scientific, #15596026), and samples were stored at −80°C until further processing. While protected from light, RNA was extracted according to the manufacturer’s instructions in the presence of 0.1 mM dithiothreitol (DTT). Carboxyamidomethylation was performed as in described by Herzog et al. (80). Forty nanograms of RNA was treated with 10 mM IAA (Sigma-Aldrich, #I1149-5g) dissolved in 100% ethanol in the presence of 50 mM NaPO4 and 50% dimethyl sulfoxide at 50°C for 50 min. After quenching the reaction with 1 M DTT, RNA was ethanol-precipitated, followed by DNase treatment (Invitrogen, #AM1907). smRNA libraries were prepared as described above using 800 ng of RNA containing 40 ng of D. melanogaster RNA as spike-in and sequenced with the NovaSeq 6000 system (Illumina) to 40 to 90 million reads per sample.
smRNA SLAM-seq: Bioinformatic processing
smRNA-seq reads were treated as described above and mapped to repetitive regions. The resulting unmapped reads were mapped to a fasta file of 176 expressed miRNA precursors (as determined above) extended for 20 bp at their 3′ end (GSE178127: 176_precursor_cleaned_hg19_ext20bp.fasta) while allowing for six mismatches using STAR v2.5.3a (67). Minus strand miRNA precursors were annotated as reverse complement to allow for all miRNAs to be treated as plus strand. After mapping smRNA-seq reads to the precursor file, the numbers of detected miRNAs were normalized by RPM (relative to the number of input reads, i.e., the number of reads after removal of reads mapping to repetitive regions and Drosophila spike-in), and a minimum threshold of 1 miRNA per million reads in all samples was applied (resulting in 126 miRNAs). Reads containing s4U-induced T>C conversions with a minimum base quality score of 27 were detected and analyzed as described by Reichholf et al. (24) (https://github.com/breichholf/smRNAseq; last accessed 9 September 2022). Background (0-min time point) was subtracted from the final RPM-normalized reads. Median half-life was calculated based the T>C-labeled fraction per time point, relative to shControl 24 hours, by nonlinear regression one-phase decay analyses performed in GraphPad Prism 8.0.
Flag affinity purification
HEK293T cells (1.2 × 109) stably overexpressing Flag-INTS11 (28) and Flag-AGO2 (1) were cultured in DMEM media (Gibco, #11965-084) containing puromycin and supplemented with 10% FBS (Atlas Biologicals, #F-0500-D). For Flag-INTS11 purification, nuclear lysate was extracted using 10 ml of buffer containing 20 mM tris-HCl (pH 7.9), 1.5 mM MgCl2, 0.42 M NaCl, 25% glycerol, 0.5 mM DTT, 0.2 mM EDTA, and 0.2 mM phenylmethylsulfonyl fluoride (PMSF). For Flag-AGO2 and Flag-INTS11, cytoplasmic lysate was extracted using 30 ml of buffer containing 10 mM tris-HCl (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, and 0.2 mM PMSF. Both complexes were purified from extract using 1 ml of anti-FLAG M2 affinity gel (Sigma-Aldrich). After washing twice with 12 ml of buffer BC500 [20 mM tris (pH 7.6), 0.2 mM ETDA, 10 mM 2-mercaptoethanol, 10% glycerol, 0.2 mM PMSF, and 0.5 M KCl] and three times with 12 ml of buffer BC150 [20 mM tris (pH 7.6), 0.2 mM ETDA, 10 mM 2-mercaptoethanol, 10% glycerol, 0.2 mM PMSF, and 150 mM KCl], the affinity columns were eluted with 500 μl of FLAG peptide (0.5 μg/μl; Sigma-Aldrich, #F3290).
In case of affinity purification of Flag-INTS11 and Flag-AGO2 following RNase A/T1 treatment, the protocol was the same with the mentioned affinity purification except treating the fractions with RNase A/T1 (Thermo Fisher Scientific, #MAN0012005) during their incubation with anti-FLAG M2 affinity gel for 1 hour at room temperature and then continued overnight at 4°C.
RNA immunoprecipitation
RIP was performed as described by Peritz et al. (81). Protein A/G magnetic beads (Thermo Fisher Scientific, #26126) were rotated 16 hours with 10 μg of AGO2 antibody (Abcam, #ab57113). Cells were lysed in fresh polysome lysis buffer [100 mM KCl, 5 mM MgCl2, 10 mM Hepes (pH 7.0), 0.5% NP-40, 1 mM DTT, SUPERase-in RNase inhibitor (0.04 U/ml; Ambion, #AM2694), Halt protease, and phosphatase inhibitor cocktail (Thermo Fisher Scientific, #1861282)], and lysate was cleared by centrifugation. Protein concentration was determined using BCA protein assay, and 350 μg of protein lysate was inserted in the IP reaction. After 16 hours, beads were directly lysed in 400 μl of TRIzol reagent (Thermo Fisher Scientific, #15596026), and RNA isolation was performed following the manufacturer’s instructions. For s4U-containing samples, IPs were performed in the absence of light, and 2 pmol of synthetic ath-miR159a RNA was spiked-in after IP to correct for isolation bias. RNA was isolated in the presence of 0.1 mM DTT and resuspended in H2O and 1 mM DTT, and the entire IP was inserted in carboxyamidomethylation reaction as described above. After DNase treatment and final phenol/chloroform purification, equal volumes of IP RNA (400 to 600 ng of RNA per IP) were subjected to smRNA library preparation as described previously. Sequencing data were treated as described above with the adjustment of mapping to ath-miR159a precursor before hsa-miRNA precursor mapping as described above, with the relative number of ath-miR-159a mapped reads serving as correction factor.
In case of RIP-qPCR, adjusted amount of IP RNAs (normalized based on AGO2 IP band intensity that were detected by Western blot) were applied for cDNA synthesis using a TaqMan Advanced miRNA cDNA synthesis kit (Applied Biosystems, #A28007). Five picomolar ath-miR-159a spike-in was used as control. miRNA expressions were detected by TaqMan-qPCR using specific mentioned probes and SsoAdvanced Universal Probes Supermix (Bio-Rad, #1725281). miRNA levels were normalized to shControl and ath-miR-159a spike-in using the ΔΔct method (means ± SEM, n = 3).
Enhanced UV cross-linking and immunoprecipitation
eCLIP was performed in duplicates as previously described by Van Nostrand et al. (57) and optimized for the detection of mature miRNAs by increasing RNase I (Ambion, #AM2294) concentration from 40 to 200 U/ml. Briefly, 2 × 107 HeLa cells were cross-linked by UV-C irradiation (254 nm, 400 mJ/cm2) and lysed on ice followed by sonication. The lysate was subjected to RNase I (Ambion, #AM2294) digest (200 U/ml) in the presence of murine RNase inhibitor [New England Biolabs (NEB), #M0314L] and Turbo DNase (4 U/ml; Ambion, #AM2238). Four micrograms of antibody [INTS11 (Sigma Prestige, #HPA029025), CPSF73 (Bethyl Laboratories, #A301-091A), and rabbit IgG isotype control (Invitrogen, #02-6102)] was preincubated with Dynabeads M-280 Sheep Anti-Rabbit IgG (Invitrogen, #11204D) for 1 hour and added to the lysates for IP at 4°C for 16 hours. Two percent of the lysate was removed and stored as size-matched input controls. Co-IP RNA was dephosphorylated, followed by on-bead 3′ RNA adapter ligation using high-concentration T4 RNA ligase I (NEB, #M0204L). IP efficiency was verified by immunoblot of 20% of the IP samples. Input controls and 80% of the immunoprecipitated protein-RNA complexes were run on a NuPAGE 4 to 12% Bis-Tris Plus Gels (Invitrogen, #NP0321BOX) and transferred to nitrocellulose membrane (Invitrogen, #IB23001), and the desired size range (protein size: +75 kDa) was cut from the membrane for IP and size-matched input samples. To extract RNA, nitrocellulose membranes were finely fragmented and treated with urea/proteinase K, followed by acid phenol-chloroform extraction and purification using RNA Clean & Concentrator column cleanup (Zymo Research, #R1014). Input samples were also dephosphorylated and ligated to 3′ RNA adapter. After reverse transcription (AffinityScript reverse transcriptase, Agilent, #600107), excess oligonucleotides were removed with exonuclease (ExoSAP-IT, Affymetrix, #78201), and the remaining RNA was hydrolyzed by NaOH. A 3′ DNA linker was ligated to the cDNA, and the resulting library was PCR amplified using Q5 Ultra II Master Mix (NEB, #M0544S). The library was size selected by agarose gel electrophoresis and column-purified (QIAGEN, MinElute, #28606). Single-end sequencing was performed to an average of 30 million reads per sample using Illumina NovaSeq 6000. Data were processed according to Van Nostrand et al. (57) and https://github.com/YeoLab/eclip (last accessed 28 January 2021). After double adapter trimming [Cutadapt (66) v1.14)] resulting reads were first mapped against the repetitive genome using STAR (67) (v2.7.6a), the unmapped output was aligned against the human genome (hg19). PCR duplicates were removed by UMI-tools (82) (v1.0.0), and the samples were visualized in UCSC (68).
AGO2 rescue experiment
HeLa cells (1.5 × 105) were seeded in a six-well plate. The following day, Dox (1 μg/ml) treatment was started and continued for 3 days. A Sigma-Aldrich pFLAG-CMV-2 (N-terminal FLAG) plasmid overexpressing AGO2 was transiently transfected into the cells 1 day after starting Dox treatment. The cells were harvested, and total RNA was extracted after 3 days. Ten nanograms of total RNA was applied for cDNA synthesis using a TaqMan Advanced miRNA cDNA synthesis kit (Applied Biosystems, #A28007). Five picomolar ath-miR-159a spike-in was added as control. miRNA expression was detected by TaqMan-qPCR using specific mentioned probes and SsoAdvanced Universal Probes Supermix (Bio-Rad, #1725281). miRNA TaqMan-qPCR was performed, and miRNA levels were relatively normalized to ath-miR-159a spike-in and shControl.
AGO2 cleavage assay
AGO2 cleavage assay was performed as previously described by Gregory et al. (83) and the following modifications. Oligo RNAs were purchased from Integrated DNA Technologies (IDT): dme-let-7a-5p_guide (/5Phos/UGAGGUAGUAGGUUGUAUAGU), dme-let-7a-3p_passenger
(/5Phos/UAUACAAUGUGCUAGCUUUCU), and dme-let-7a_target (/5IRD700/UAUACAACCUACUACCUCAUU). miRNA duplex was prepared by mixing equal volumes of both dme-let-7a-5p_guide and dme-let-7a-3p_passenger oligos in annealing buffer [10 mM tris (pH 8), 50 mM NaCl, and 1 mM EDTA], incubating at 95°C for 3 min and cooling gradually to room temperature for 1 hour. Either 0.025 μg of rAGO2 (Active Motif, #31486) or rAGO2 in combination with increasing concentrations of affinity-purified Flag-INTS11 was preincubated with the 5 nM miRNA duplex in buffer containing 3.2 mM MgCl2, 1 mM adenosine triphosphate, 20 mM creatine phosphate, RNasin (0.2 U/μl), 20 mM tris-HCl (pH 8), 0.1 M KCl, and 10% glycerol for 30 min at 37°C. Then, 10 nM dme-let-7a_target was added, and the cleavage reaction was incubated for 90 min at 37°C and stopped by adding proteinase K for 30 min at room temperature. Samples were loaded onto a 15% TBE-urea gels (Bio-Rad, #4566055), visualized using the Odyssey CLx Imaging System, and quantified by Image Studio Lite (v5.2).
Immunofluorescence and image processing
Cells were fixed at 10 min in ice-cold methanol/ethanol, blocked for 3 hours in 10% bovine serum, and followed by 5% BSA with primary antibody 1:200 dilution for AGO antibodies (AGO ab57113 and pan-AGO MABE56) and 1:100 dilution for INTS11 (ab75276). Secondary Antibodies were Alexa Fluor 568 goat anti-mouse A11031 (1:500) and Alexa Fluor 488 goat anti-rabbit A32731 (1:1000). DAPI (4′,6-diamidino-2-phenylindole) was diluted 1 μl/2 ml for 5 min. Images were captured sequentially using a Nikon A1R confocal microscope using a 60× 1.4 numerical aperture plan APO oil objective (0.1 μm per pixel resolution). Cells were manually segmented (n = 50), and Pearson colocalization values were calculated using ImageJ 1.53o plug-in Coloc 2 (https://github.com/fiji/Colocalisation_Analysis; last accessed 1 November 2022). Coloc 2 is a plugin for colocalization analysis, which implements and performs the pixel intensity correlation. The scale bar is 50 μm.
Acknowledgments
We thank all members of the Shiekhattar Lab for discussions. We thank B. Reichholf for comments on s4U small RNA SLAM-seq analysis. We thank the Sylvester Comprehensive Cancer Center Onco-Genomics Core Facility for high-throughput sequencing.
Funding: This work was supported by the University of Miami Miller School of Medicine, Sylvester Comprehensive Cancer Center and grants R01 GM078455 and DP1 CA228041 from the National Institute of Health to R.S. Research reported in this publication was funded by the National Cancer Institute of the National Institutes of Health under award number P30CA240139.
Author contributions: N.K. performed small RNA-seq, small-RNA SLAM-seq, RIP-seq, eCLIP experiments, and bioinformatic analyses. S.D. performed smRNA-seq, RNA-seq, RIP-qPCR, AGO2-rescue, Flag affinity purifications, immunoblots, in vitro AGO2 cleavage, Northern blotting, silver staining, and colloidal blue staining experiments. P.R.C. performed PRO-seq and Northern blotting experiments. M.G.V. established the shINTS6 cell line and performed TaqMan probe efficiency measurements. F.B. and H.G.D.S. processed RNA-seq. F.B. and G.S.G. performed immunofluorescence experiments and image processing. M.M.T. performed one independent biological repeat of eCLIP experiment. E.B. set up smRNA-seq. N.K., S.D., and R.S. designed the experiments and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All sequencing data generated in this study are made available at the Gene Expression Omnibus (GEO). The accession number for the raw and processed data reported here is GSE178127. Our previously reported PRO-seq dataset is available under the accession GSE125535. Code generated during this study is available at https://doi.org/10.5281/zenodo.7517172 or https://github.com/Shiekhattarlab/miRNApaper.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Legend for table S1
Other Supplementary Material for this : manuscript includes the following:
Table S1
REFERENCES AND NOTES
- 1.T. P. Chendrimada, R. I. Gregory, E. Kumaraswamy, J. Norman, N. Cooch, K. Nishikura, R. Shiekhattar, TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436, 740–744 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.R. I. Gregory, K.-p. Yan, G. Amuthan, T. Chendrimada, B. Doratotaj, N. Cooch, R. Shiekhattar, The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240 (2004). [DOI] [PubMed] [Google Scholar]
- 3.J. Han, Y. Lee, K.-H. Yeom, Y.-K. Kim, H. Jin, V. N. Kim, The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 18, 3016–3027 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.R. C. Wilson, A. Tambe, M. A. Kidwell, C. L. Noland, C. P. Schneider, J. A. Doudna, Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol. Cell 57, 397–407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.A. M. Denli, B. B. Tops, R. H. Plasterk, R. F. Ketting, G. J. Hannon, Processing of primary microRNAs by the Microprocessor complex. Nature 432, 231–235 (2004). [DOI] [PubMed] [Google Scholar]
- 6.K. Okamura, J. W. Hagen, H. Duan, D. M. Tyler, E. C. Lai, The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130, 89–100 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.J. G. Ruby, C. H. Jan, D. P. Bartel, Intronic microRNA precursors that bypass Drosha processing. Nature 448, 83–86 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.P. Sheng, C. Fields, K. Aadland, T. Wei, O. Kolaczkowski, T. Gu, B. Kolaczkowski, M. Xie, Dicer cleaves 5′-extended microRNA precursors originating from RNA polymerase II transcription start sites. Nucleic Acids Res. 46, 5737–5752 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.M. Xie, M. Li, A. Vilborg, N. Lee, M.-D. Shu, V. Yartseva, N. Šestan, J. A. Steitz, Mammalian 5′-capped microRNA precursors that generate a single microRNA. Cell 155, 1568–1580 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.S. Cheloufi, C. O. Dos Santos, M. M. Chong, G. J. Hannon, A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465, 584–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D. Cifuentes, H. Xue, D. W. Taylor, H. Patnode, Y. Mishima, S. Cheloufi, E. Ma, S. Mane, G. J. Hannon, N. D. Lawson, A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 328, 1694–1698 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.A. Grishok, A. E. Pasquinelli, D. Conte, N. Li, S. Parrish, I. Ha, D. L. Baillie, A. Fire, G. Ruvkun, C. C. Mello, Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34 (2001). [DOI] [PubMed] [Google Scholar]
- 13.V. N. Kim, MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 6, 376–385 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Y. Lee, C. Ahn, J. Han, H. Choi, J. Kim, J. Yim, J. Lee, P. Provost, O. Rådmark, S. Kim, The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 (2003). [DOI] [PubMed] [Google Scholar]
- 15.P. Provost, D. Dishart, J. Doucet, D. Frendewey, B. Samuelsson, O. Rådmark, Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 21, 5864–5874 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.S. Iwasaki, T. Kawamata, Y. Tomari, Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol. Cell 34, 58–67 (2009). [DOI] [PubMed] [Google Scholar]
- 17.K. Nakanishi, Anatomy of RISC: How do small RNAs and chaperones activate Argonaute proteins? Wiley Interdiscip. Rev. RNA 7, 637–660 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.J. M. Pare, N. Tahbaz, J. López-Orozco, P. LaPointe, P. Lasko, T. C. Hobman, Hsp90 regulates the function of argonaute 2 and its recruitment to stress granules and P-bodies. Mol. Biol. Cell 20, 3273–3284 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.N. Tahbaz, F. A. Kolb, H. Zhang, K. Jaronczyk, W. Filipowicz, T. C. Hobman, Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep. 5, 189–194 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.F. Frank, N. Sonenberg, B. Nagar, Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465, 818–822 (2010). [DOI] [PubMed] [Google Scholar]
- 21.M. Ghildiyal, J. Xu, H. Seitz, Z. Weng, P. D. Zamore, Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA 16, 43–56 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.M. B. Warf, W. E. Johnson, B. L. Bass, Improved annotation of C. elegans microRNAs by deep sequencing reveals structures associated with processing by Drosha and Dicer. RNA 17, 563–577 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.S. Chatterjee, M. Fasler, I. Büssing, H. Großhans, Target-mediated protection of endogenous microRNAs in C. elegans. Dev. Cell 20, 388–396 (2011). [DOI] [PubMed] [Google Scholar]
- 24.B. Reichholf, V. A. Herzog, N. Fasching, R. A. Manzenreither, I. Sowemimo, S. L. Ameres, Time-resolved small RNA sequencing unravels the molecular principles of microRNA homeostasis. Mol. Cell 75, 756–768. e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.K. K.-H. Farh, A. Grimson, C. Jan, B. P. Lewis, W. K. Johnston, L. P. Lim, C. B. Burge, D. P. Bartel, The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 310, 1817–1821 (2005). [DOI] [PubMed] [Google Scholar]
- 26.A. J. Giraldez, Y. Mishima, J. Rihel, R. J. Grocock, S. Van Dongen, K. Inoue, A. J. Enright, A. F. Schier, Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79 (2006). [DOI] [PubMed] [Google Scholar]
- 27.L. P. Lim, N. C. Lau, P. Garrett-Engele, A. Grimson, J. M. Schelter, J. Castle, D. P. Bartel, P. S. Linsley, J. M. Johnson, Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433, 769–773 (2005). [DOI] [PubMed] [Google Scholar]
- 28.D. Baillat, M.-A. Hakimi, A. M. Näär, A. Shilatifard, N. Cooch, R. Shiekhattar, Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123, 265–276 (2005). [DOI] [PubMed] [Google Scholar]
- 29.A. Gardini, D. Baillat, M. Cesaroni, D. Hu, J. M. Marinis, E. J. Wagner, M. A. Lazar, A. Shilatifard, R. Shiekhattar, Integrator regulates transcriptional initiation and pause release following activation. Mol. Cell 56, 128–139 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.F. Lai, A. Gardini, A. Zhang, R. Shiekhattar, Integrator mediates the biogenesis of enhancer RNAs. Nature 525, 399–403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.M. Xie, W. Zhang, M.-D. Shu, A. Xu, D. A. Lenis, D. DiMaio, J. A. Steitz, The host Integrator complex acts in transcription-independent maturation of herpesvirus microRNA 3′ ends. Genes Dev. 29, 1552–1564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.F. Beckedorff, E. Blumenthal, L. Ferreira daSilva, Y. Aoi, P. R. Cingaram, J. Yue, A. Zhang, S. Dokaneheifard, M. G. Valencia, G. Gaidosh, The human integrator complex facilitates transcriptional elongation by endonucleolytic cleavage of nascent transcripts. Cell Rep. 32, 107917 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.C. R. Sibley, Y. Seow, S. Saayman, K. K. Dijkstra, S. El Andaloussi, M. S. Weinberg, M. J. Wood, The biogenesis and characterization of mammalian microRNAs of mirtron origin. Nucleic Acids Res. 40, 438–448 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.T. R. Albrecht, S. P. Shevtsov, Y. Wu, L. G. Mascibroda, N. J. Peart, K.-L. Huang, I. A. Sawyer, L. Tong, M. Dundr, E. J. Wagner, Integrator subunit 4 is a “Symplekin-like” scaffold that associates with INTS9/11 to form the integrator cleavage module. Nucleic Acids Res. 46, 4241–4255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Y. Wu, T. R. Albrecht, D. Baillat, E. J. Wagner, L. Tong, Molecular basis for the interaction between Integrator subunits IntS9 and IntS11 and its functional importance. Proc. Natl. Acad. Sci. U.S.A. 114, 4394–4399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.I. Fianu, Y. Chen, C. Dienemann, O. Dybkov, A. Linden, H. Urlaub, P. Cramer, Structural basis of Integrator-mediated transcription regulation. Science 374, 883–887 (2021). [DOI] [PubMed] [Google Scholar]
- 37.K.-L. Huang, D. Jee, C. B. Stein, N. D. Elrod, T. Henriques, L. G. Mascibroda, D. Baillat, W. K. Russell, K. Adelman, E. J. Wagner, Integrator recruits protein phosphatase 2A to prevent pause release and facilitate transcription termination. Mol. Cell 80, 345–358.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.S. J. Vervoort, S. A. Welsh, J. R. Devlin, E. Barbieri, D. A. Knight, S. Offley, S. Bjelosevic, M. Costacurta, I. Todorovski, C. J. Kearney, The PP2A-Integrator-CDK9 axis fine-tunes transcription and can be targeted therapeutically in cancer. Cell 184, 3143–3162.e32 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.F. Zhang, T. Ma, X. Yu, A core hSSB1-INTS complex participates in the DNA damage response. J. Cell Sci. 126, 4850–4855 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.H. Zheng, Y. Qi, S. Hu, X. Cao, C. Xu, Z. Yin, X. Chen, Y. Li, W. Liu, J. Li, Identification of Integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase. Science 370, eabb5872 (2020). [DOI] [PubMed] [Google Scholar]
- 41.L. F. Dasilva, E. Blumenthal, F. Beckedorff, P. R. Cingaram, H. Gomes Dos Santos, R. R. Edupuganti, A. Zhang, S. Dokaneheifard, Y. Aoi, J. Yue, Integrator enforces the fidelity of transcriptional termination at protein-coding genes. Sci. Adv. 7, eabe3393 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.J. Barra, G. S. Gaidosh, E. Blumenthal, F. Beckedorff, M. M. Tayari, N. Kirstein, T. K. Karakach, T. H. Jensen, F. Impens, K. Gevaert, E. Leucci, R. Shiekhattar, J.-C. Marine, Integrator restrains paraspeckles assembly by promoting isoform switching of the lncRNA NEAT1. Sci. Adv. 6, eaaz9072 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.T.-C. Chang, M. Pertea, S. Lee, S. L. Salzberg, J. T. Mendell, Genome-wide annotation of microRNA primary transcript structures reveals novel regulatory mechanisms. Genome Res. 25, 1401–1409 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.A. Khvorova, A. Reynolds, S. D. Jayasena, Functional siRNAs and miRNAs exhibit strand bias. Cell 115, 209–216 (2003). [DOI] [PubMed] [Google Scholar]
- 45.D. S. Schwarz, G. Hutvágner, T. Du, Z. Xu, N. Aronin, P. D. Zamore, Asymmetry in the assembly of the RNAi enzyme complex. Cell 115, 199–208 (2003). [DOI] [PubMed] [Google Scholar]
- 46.J. H. Park, S.-Y. Shin, C. Shin, Non-canonical targets destabilize microRNAs in human Argonautes. Nucleic Acids Res. 45, 1569–1583 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.G. Hutvagner, P. D. Zamore, A microRNA in a multiple-turnover RNAi enzyme complex. Science 297, 2056–2060 (2002). [DOI] [PubMed] [Google Scholar]
- 48.H. Kobayashi, Y. Tomari, RISC assembly: Coordination between small RNAs and Argonaute proteins. Biochim. Biophys. Acta 1859, 71–81 (2016). [DOI] [PubMed] [Google Scholar]
- 49.T. Treiber, N. Treiber, G. Meister, Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 20, 5–20 (2019). [DOI] [PubMed] [Google Scholar]
- 50.A. A. Sarshad, A. H. Juan, A. I. C. Muler, D. G. Anastasakis, X. Wang, P. Genzor, X. Feng, P.-F. Tsai, H.-W. Sun, A. D. Haase, Argonaute-miRNA complexes silence target mRNAs in the nucleus of mammalian stem cells. Mol. Cell 71, 1040–1050.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.R. Kalantari, J. A. Hicks, L. Li, K. T. Gagnon, V. Sridhara, A. Lemoff, H. Mirzaei, D. R. Corey, Stable association of RNAi machinery is conserved between the cytoplasm and nucleus of human cells. RNA 22, 1085–1098 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.T. Ohrt, J. Mütze, W. Staroske, L. Weinmann, J. Höck, K. Crell, G. Meister, P. Schwille, Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res. 36, 6439–6449 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.N. D. Elrod, T. Henriques, K.-L. Huang, D. C. Tatomer, J. E. Wilusz, E. J. Wagner, K. Adelman, The integrator complex attenuates promoter-proximal transcription at protein-coding genes. Mol. Cell 76, 738–752.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.J. R. Skaar, A. L. Ferris, X. Wu, A. Saraf, K. K. Khanna, L. Florens, M. P. Washburn, S. H. Hughes, M. Pagano, The Integrator complex controls the termination of transcription at diverse classes of gene targets. Cell Res. 25, 288–305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.D. C. Tatomer, N. D. Elrod, D. Liang, M.-S. Xiao, J. Z. Jiang, M. Jonathan, K.-L. Huang, E. J. Wagner, S. Cherry, J. E. Wilusz, The Integrator complex cleaves nascent mRNAs to attenuate transcription. Genes Dev. 33, 1525–1538 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.J. N. Jodoin, P. Sitaram, T. R. Albrecht, S. B. May, M. Shboul, E. Lee, B. Reversade, E. J. Wagner, L. A. Lee, Nuclear-localized Asunder regulates cytoplasmic dynein localization via its role in the integrator complex. Mol. Biol. Cell 24, 2954–2965 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.E. L. Van Nostrand, T. B. Nguyen, C. Gelboin-Burkhart, R. Wang, S. M. Blue, G. A. Pratt, A. L. Louie, G. W. Yeo, Robust, cost-effective profiling of RNA binding protein targets with single-end enhanced crosslinking and immunoprecipitation (seCLIP), in mRNA Processing (Springer, 2017), pp. 177–200. [DOI] [PMC free article] [PubMed]
- 58.K.-W. Tsai, C.-M. Leung, Y.-H. Lo, T.-W. Chen, W.-C. Chan, S.-Y. Yu, Y.-T. Tu, H.-C. Lam, S.-C. Li, L.-P. Ger, Arm selection preference of microRNA-193a varies in breast cancer. Sci. Rep. 6, 28176 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.J. Yue, F. Lai, F. Beckedorff, A. Zhang, C. Pastori, R. Shiekhattar, Integrator orchestrates RAS/ERK1/2 signaling transcriptional programs. Genes Dev. 31, 1809–1820 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.D. Cazalla, M. Xie, J. A. Steitz, A primate herpesvirus uses the integrator complex to generate viral microRNAs. Mol. Cell 43, 982–992 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.D. J. Shaw, P. Eggleton, P. J. Young, Joining the dots: Production, processing and targeting of U snRNP to nuclear bodies. Biochim. Biophys. Acta 1783, 2137–2144 (2008). [DOI] [PubMed] [Google Scholar]
- 62.C. L. Will, R. Lührmann, Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13, 290–301 (2001). [DOI] [PubMed] [Google Scholar]
- 63.D. M. Bhatt, A. Pandya-Jones, A.-J. Tong, I. Barozzi, M. M. Lissner, G. Natoli, D. L. Black, S. T. Smale, Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 150, 279–290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.B. R. Miller, T. Wei, C. J. Fields, P. Sheng, M. Xie, Near-infrared fluorescent northern blot. RNA 24, 1871–1877 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.H. Jorjani, S. Kehr, D. J. Jedlinski, R. Gumienny, J. Hertel, P. F. Stadler, M. Zavolan, A. R. Gruber, An updated human snoRNAome. Nucleic Acids Res. 44, 5068–5082 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.M. Martin, Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011). [Google Scholar]
- 67.A. Dobin, C. A. Davis, F. Schlesinger, J. Drenkow, C. Zaleski, S. Jha, P. Batut, M. Chaisson, T. R. Gingeras, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.W. J. Kent, C. W. Sugnet, T. S. Furey, K. M. Roskin, T. H. Pringle, A. M. Zahler, D. Haussler, The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.F. Ramírez, D. P. Ryan, B. Grüning, V. Bhardwaj, F. Kilpert, A. S. Richter, S. Heyne, F. Dündar, T. Manke, deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.M. R. Friedländer, S. D. Mackowiak, N. Li, W. Chen, N. Rajewsky, miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40, 37–52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.M. I. Love, W. Huber, S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.H. Wickham, ggplot2: Elegant Graphics for Data Analysis (Springer, 2016).
- 73.A. M. Bolger, M. Lohse, B. Usadel, Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.B. Li, C. N. Dewey, RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.R. P. Huntley, T. Sawford, P. Mutowo-Meullenet, A. Shypitsyna, C. Bonilla, M. J. Martin, C. O’Donovan, The GOA database: Gene ontology annotation updates for 2015. Nucleic Acids Res. 43, D1057–D1063 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.A. Kozomara, M. Birgaoanu, S. Griffiths-Jones, miRBase: From microRNA sequences to function. Nucleic Acids Res. 47, D155–D162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.H. Zhao, Z. Sun, J. Wang, H. Huang, J.-P. Kocher, L. Wang, CrossMap: A versatile tool for coordinate conversion between genome assemblies. Bioinformatics 30, 1006–1007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.A. R. Quinlan, I. M. Hall, BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.M. Pertea, G. M. Pertea, C. M. Antonescu, T.-C. Chang, J. T. Mendell, S. L. Salzberg, StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.V. A. Herzog, B. Reichholf, T. Neumann, P. Rescheneder, P. Bhat, T. R. Burkard, W. Wlotzka, A. von Haeseler, J. Zuber, S. L. Ameres, Thiol-linked alkylation of RNA to assess expression dynamics. Nat. Methods 14, 1198–1204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.T. Peritz, F. Zeng, T. J. Kannanayakal, K. Kilk, E. Eiríksdóttir, U. Langel, J. Eberwine, Immunoprecipitation of mRNA-protein complexes. Nat. Protoc. 1, 577–580 (2006). [DOI] [PubMed] [Google Scholar]
- 82.T. Smith, A. Heger, I. Sudbery, UMI-tools: Modeling sequencing errors in unique molecular Identifiers to improve quantification accuracy. Genome Res. 27, 491–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.R. I. Gregory, T. P. Chendrimada, N. Cooch, R. Shiekhattar, Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123, 631–640 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Legend for table S1
Table S1