Abstract
Probiotics interfere with pathogenic microorganisms or reinstate the natural microbiome. Streptococcus mutans and Candida albicans are well-known emerging pathogenic bacteria/fungi for dental caries. In this study, three probiotic Lactobacilli strains (Lactobacillus plantarum 8014, L. plantarum 14917, and Lactobacillus salivarius 11741) were tested on S. mutans and C. albicans clinical isolates using a multispecies biofilm model simulating clinical cariogenic conditions. The ten pairs of clinical isolates of S. mutans and C. albicans were obtained from children with severe early childhood caries. Our study findings show a remarkable inhibitory effect of L. plantarum 14917 on S. mutans and C. albicans clinical isolates, resulting in significantly reduced growth of S. mutans and C. albicans, a compromised biofilm structure with a significantly smaller microbial and extracellular matrix and a less virulent microcolony structure. FurTre, plantaricin, an antimicrobial peptide produced by L. plantarum, inhibited the growth of S. mutans and C. albicans. The mechanistic assessment indicated that L. plantarum 14917 had a positive inhibitory impact on the expression of S. mutans and C. albicans virulence genes and virulent structure, such as C. albicans hypha formation. Future utilization of L. plantarum 14917 and/or its antimicrobial peptide plantaricin could lead to a new paradigm shift in dental caries prevention.
Keywords: Streptococcus mutans, Candida albicans, Lactobacillus plantarum, probiotics, multispecies biofilm
1. Introduction
Probiotics are living bacteria found in dairy products, chewing gum, and gummies and confer health benefits on the host when delivered in adequate quantities [1,2,3]. Probiotics potentially interfere with pathogenic microorganisms or reinstate the natural microbiome [4]. Probiotics, when ingested, compete for adhesion sites of pathogens and nutrients that can co-aggregate and produce antimicrobial compounds, thus inhibiting the adhesion and the growth of pathogens on biofilm [5,6]. Probiotic Lactobacilli produce antimicrobial agents such as organic acids, hydrogen peroxide, and antifungal compounds such as fatty acids and bacteriocins, which have bactericidal and bacteriostatic characteristics [7].
Oral microorganisms are the most common culprits of dental caries after host and environmental factors [8]. Streptococcus mutans is the most widely known pathogenic bacteria causing dental caries [9], due to its acidogenicity, aciduric property, and capability of forming an extracellular matrix that comprises the main component of dental plaques/biofilms. Furthermore, studies have demonstrated the potential cariogenic role of fungi, highlighting the presence of high levels of Candida species in children with early childhood caries (ECC) [10,11,12,13], and C. albicans yields an organic acid that is believed to play a role in caries formation [14].
Studies have indicated a potential role of probiotics in managing oral health diseases, such as caries disease, periodontal disease, halitosis [15,16], and candidiasis [17,18]. Few studies have demonstrated the inhibitory effect of probiotic Lactobacillus on S. mutans and C. albicans in vitro [14,19,20]. Intriguingly, our previous work examined the impact of four probiotic Lactobacillus strains on S. mutans and C. albicans wild-type strains using a thorough multispecies biofilm model that mimicked a high-caries-risk clinical condition [21]. These tested Lactobacillus strains were Lactobacillus rhamnosus ATCC 2836, Lactobacillus plantarum ATCC 8014, Lactobacillus plantarum ATCC 14917, and Lactobacillus salivarius ATCC 11741 [21]. Among the examined probiotic species, L. plantarum and L. salivarius showed better growth suppression of C. albicans and S. mutans wild-type strains, disrupting the formation of virulent biofilms with fewer bacteria and exopolysaccharides (EPSs) and the development of virulent microcolonies structures [21]. Our results provide data and a rationale for further assessment of the effect of the probiotic L. plantarum on clinically isolated S. mutans and C. albicans, since various studies have shown that clinically isolated bacteria and yeast have a higher tolerance to antimicrobial reagents.
Hence, the objective of this study was to examine the effect of three probiotic Lactobacillus species (L. salivarius and L. plantarum) on the growth of clinically isolated S. mutans and C. albicans from children with severe early childhood caries (S-ECC) in multispecies planktonic and biofilm conditions and further assessed mechanistic interactions between probiotic Lactobacillus species and cariogenic S. mutans and C. albicans. Promising study results will lead to a new paradigm of dental caries prevention using probiotics.
2. Results
2.1. Characteristics of S-ECC Children whose S. mutans and C. albicans Were Isolated
The demographic–socioeconomic–oral health conditions of the S-ECC children the C. albicans and S. mutans isolated from are shown in Table 1. The S-ECC children had an average age of 3.5 ± 1.0 years of age, with an equal number of males and females. The majority of the children brushed their teeth daily and did not attend daycare. The average plaque index was 1.8 ± 0.6. The average decayed teeth number and decayed surface number were 11.7 ± 5.1 and 27.2 ± 17.4, respectively.
Table 1.
Demographics data and characteristics of study participants (n = 10).
| Items | Mean (SD) or % |
|---|---|
| Age (Year) | 3.5 ± 1.0 |
| Gender (Male) | 50% |
| Race: Caucasian | 60% |
| African American | 30% |
| Asian | 10% |
| Ethnicity: Hispanic | 10% |
| Brushing Frequency (Daily) | 90% |
| Attending Daycare (Yes) | 20% |
| Plaque Index | 1.8 ± 0.6 |
| dt | 11.7 ± 5.1 |
| mt | 0.2 ± 0.6 |
| ft | 0.1 ± 0.3 |
| dmft | 12 ± 4.9 |
| ds | 27.2 ± 17.4 |
| ms | 1.0 ± 3.2 |
| fs | 0.1 ± 0.3 |
| dmfs | 28.3 ± 16.5 |
Note: clinical isolates were obtained from the children with ECC. dt: decayed teeth. mt: missing teeth. ft: filled teeth. dmft: decayed/missing/filled teeth. ds: decayed surfaces. ms: missing surfaces. fs: filled surfaces. dmfs: decayed/missing/filled surfaces.
2.2. L. plantarum 14917 Inhibited the Growth of S. mutans and C. albicans Clinical Isolates in Planktonic Condition
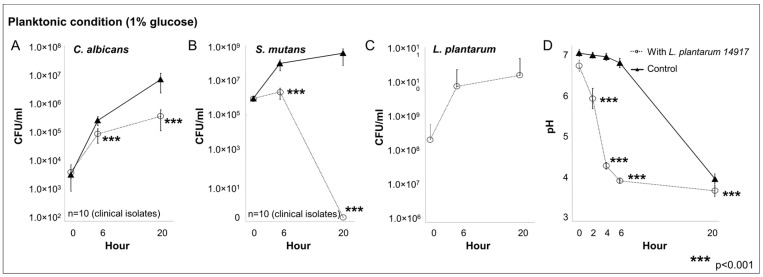
The growth of C. albicans, S. mutans, and L. plantarum 14917 in multispecies planktonic conditions is plotted in Figure 1. L. plantarum inhibited the growth of C. albicans (Figure 1A) by <1-log after 6 h and approximately 2-log at 20 h (p < 0.05 at 20 h). L. plantarum significantly inhibited the growth of S. mutans (Figure 1B) at 6 h by 1-log, and completely inhibited the growth of S. mutans after 20 h (p < 0.05 at 20 h). L. plantarum (Figure 1C) maintained a stable growth during the 20 h of interaction with S. mutans and C. albicans. The culture medium pH (Figure 1D) dropped rapidly with the addition of L. plantarum in the planktonic condition, and both groups reached the same acidic level at 20 h.
Figure 1.
Inhibitory effect of L. plantarum on clinically isolated C. albicans and S. mutans from children with early childhood caries in multispecies planktonic condition. C. albicans and S. mutans clinical strains were isolated from 10 children with early childhood caries (ECC). Experiments were repeated in triplicate. Each planktonic multispecies condition included the C. albicans and S. mutans isolated from the same ECC child, with/without added L. plantarum 14917. The control group only consisted of C. albicans and S. mutans. (A) L. plantarum inhibited the growth of C. albicans. (B) L. plantarum significantly inhibited the growth of S. mutans. (C) L. plantarum maintained a stable growth during the 20 h. (D) The culture medium pH.
Of note is that among the three Lactobacillus spp. tested in the screening step against C. albicans and S. mutans isolated from two S-ECC children, the inhibitory effects of L. plantarum 14917 and L. salivarius were similar, but superior to that of L. plantarum 8014 (See Supplemental Figure S2).
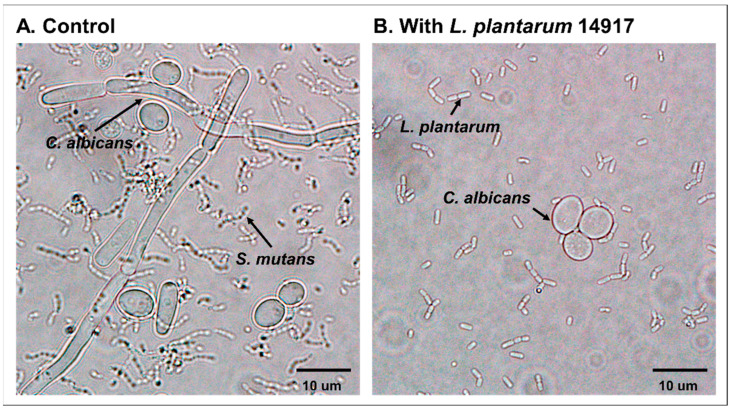
2.3. Inhibition of C. albicans Hypha Formation by L. plantarum
The inhibition of C. albicans’ switching from yeast to the hyphal form was observed in the planktonic condition when treated with L. plantarum 14917 (Figure 2). The addition of L. plantarum 14917 reduced the growth of C. albicans and inhibited the switching from yeast to the hypha and pseudohypha form.
Figure 2.
Inhibition of C. albicans hypha formation by L. plantarum 14917. (A) S. mutans and C. albicans grown in 1% glucose at 24 h. (B) S. mutans and C. albicans grown in 1% glucose with added L. plantarum 14917 at 24 h.
2.4. L. plantarum 14917 Inhibited Biofilm Formation by S. mutans and C. albicans Clinical Isolates
During the screening step, L. plantarum 14917 and L. salivarius 11741 were added to the biofilms formed by S. mutans and C. albicans isolated from two S-ECC children, respectively. Both L. plantarum 14917 and L. salivarius 11741 inhibited the growth of C. albicans and S. mutans (Figures S3 and S4). Although both L. plantarum 14917 and L. salivarius 11741 became the dominant species after 48 h of incubation, L. plantarum 14917 had a higher composition at 24 h (40%, Figure S3E) compared to L. salivarius 11741 (20%, Figure S3E). Furthermore, the growth of L. plantarum 14917 within the multispecies biofilms remained at a higher level than L. salivarius 11741 by 2-log at 72 h (Figure S3C). For the reasons stated above, L. plantarum 14917 advanced to the assessment of impact on biofilm structure and mechanistic interaction assessment.
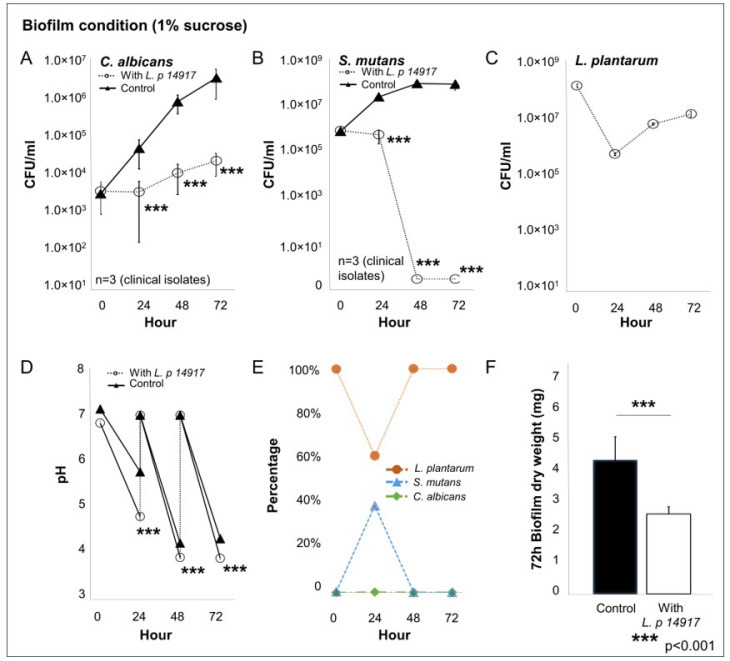
The growth of C. albicans and S. mutans was significantly inhibited by L. plantarum 14917 and is plotted in Figure 3. In the group treated with L. plantarum 14917, C. albicans was reduced by 3-log compared to the control group (Figure 3A). At 48 h, after two administrations of L. plantarum 14917, S. mutans was inhibited entirely in biofilms of the treatment group (Figure 3B), and the number of L. plantarum dropped during the first 24 h due to the low-sucrose (0.1%) condition and remained growing steady after 24 h in the high-sucrose (1%) condition (Figure 3C). The pH of the culture medium was significantly decreased when L. plantarum 14917 was added at 24, 48, and 72 h, compared to the control group (p < 0.05) (Figure 3D). L. plantarum 14917 became the dominant species after 48 h of incubation (Figure 3E). Biofilm formation was significantly reduced when L. plantarum 14917 was added, with a significant reduction in dry weight at 72 h (Figure 3F). In addition, the total biofilm proteins formed by the clinically isolated S. mutans and C. albicans with/without L. plantarum 14917 treatment were quantified. The biofilm total protein of the control group (S. mutans and C. albicans) was 338.4 ± 19.6 µg/disc versus 199.4 ± 52.7 µg/disc for the L. plantarum 14917-treated group. The addition of L. plantarum 14917 reduced the total protein in multispecies biofilms formed by S. mutans and C. albicans clinical isolates (from 3 independent experiments, n = 12, p < 0.001).
Figure 3.
Interaction of L. plantarum 14917 and clinically isolated C. albicans and S. mutans in multispecies biofilms. Multispecies biofilms were formed by L. plantarum 14917 and clinically isolated C. albicans and S. mutans from three children with ECC. The treated group was grown with added L. plantarum 14917. (A–C) The growth of C. albicans, S. mutans, and L. plantarum 14917 in multispecies biofilms is plotted. (D) The pH of the culture medium. (E) The composition of each microorganism. (F) The dry weight of biofilms at 72 h.
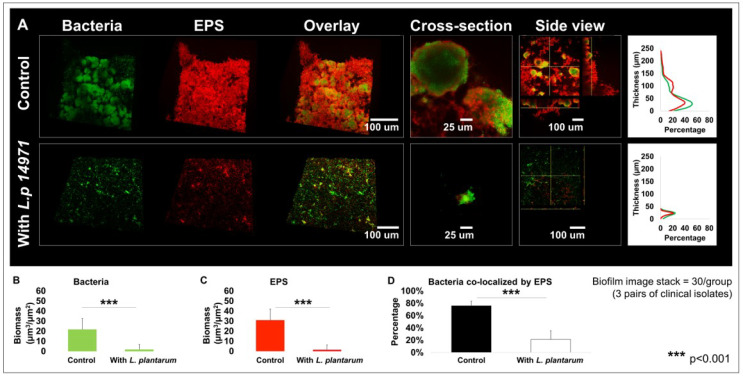
2.5. L. plantarum 14917 Altered 3D Structure of Biofilms Formed by S. mutans and C. albicans Clinical Isolates
Since L. plantarum 14917 demonstrated better inhibition of C. albicans and S. mutans isolates in planktonic and biofilm conditions, it was assessed for its impact on biofilm structure and mechanistic interaction. L. plantarum 14917 significantly reduced cariogenic biofilm formation, as measured by bacteria and EPS biomass, compared to the control group (C. albicans–S. mutans duo-species biofilm). The 72 h multispecies biofilms are shown in Figure 4A. In contrast to the complex and thick biofilms formed in the control group, the L. plantarum 14917-treated biofilms significantly reduced the biofilm thickness and biomass of both bacteria and EPSs (Figure 4B,C, p < 0.05). Furthermore, the horizontal coverage (Figure 4A) of the control group was also much broader than that of the treatment group, with nearly 60% bacterial coverage and 40 % EPS coverage in the most abundant layer (~30–40 µm above the substrate), while the treated group only had 20% coverage at the most abundant layer (~10 µm above the substrate). In addition, bacteria co-localized by EPSs were significantly fewer in number in the treatment group (Figure 4D, p < 0.05).
Figure 4.
Changes in multispecies biofilm 3D structure caused by L. plantarum 14917. Biofilms were formed by C. albicans and S. mutans only (control) and treated with L. plantarum 14917 in 1% sucrose condition and visualized via two-photon laser confocal microscope at 72 h. Amira software was used to reconstruct the images as 3D structures, to visualize bacterial channels (green), EPS channels (red), and the bacteria/EPS overlay. (A) Biofilm structure. The layer distribution of the biofilms indicating that the biofilms grown in the control group were much thicker than those of the treatment group. (B) Biomass formed by bacteria. (C) Biomass formed by EPSs. (D) Bacteria co-localized by EPSs. Biofilm parameters were calculated using data from three biofilms formed by C. albicans and S. mutans isolated from three ECC children. For each biofilm, 10 randomly selected points of biofilms were visualized.
Furthermore, the added L. plantarum 14917 also significantly impacted the microcolony formation in the 72 h old multispecies biofilms. In contrast to the well-formed mushroom-shaped microcolonies in the control group, L. plantarum 14917-treated biofilms had significantly compromised microcolony structure (Figure 4A). Microcolonies are considered virulent and functional structures of biofilms. Surface-attached and free-floating microcolonies were quantified, and their numbers and sizes were compared between the control and L. plantarum 14917-treated biofilms. Numerous and large microcolonies were detected in the control group, while the intervention of L. plantarum 14917 resulted in fewer and smaller microcolonies (Table 2).
Table 2.
Quantitative assessment of microcolonies in multispecies biofilms.
| Microcolony Parameters | Control (n = 30) | With L. plantarum 14917 (n = 30) |
|---|---|---|
| Number of attached microcolonies | 15.3 ± 4.1 | 9.2 ± 6.7 ** |
| Area of attached microcolonies (um2) | 972.2 ± 924.4 | 283.6 ± 84.3 ** |
| Volume of attached microcolonies (um3) × 103 | 3471.5 ± 2334.8 | 25.6 ± 27.5 *** |
| Number of free microcolonies | 287.4 ± 71.1 | 213.3 ± 77.5 *** |
| Diameter of free microcolonies (um) | 38.0 ± 4.3 | 28.4 ± 3.0 *** |
| Volume of free microcolonies (um3) × 103 | 925.0 ± 255.0 | 145.8 ± 111.8 *** |
Note: ** p < 0.01, *** p < 0.001 when comparing the control and L. plantarum 14917-treated biofilms. n = 30 indicates that 30 biofilm image stacks were used for each group (control and treatment).
2.6. Plantaricin Inhibited the Growth of S. mutans and C. albicans Clinical Isolates
S. mutans (103 CFU/mL) and C. albicans (101 CFU/mL) were treated with plantaricin at a concentration ranging from 0 to 400 µg/mL and grown for 24 h in 1% glucose conditions. The MIC assay was carried out on three children’s S. mutans and C. albicans clinical isolates. Among the three pairs of S. mutans and C. albicans isolates, the MIC of plantaricin was 400 µg/mL for all three S. mutans, 200 µg/mL for two C. albicans, and 25 µg/mL for the other C. albicans strain. The clear culture was plated and incubated for an additional 48 h. Results reveal that the inhibitory effect on all these S. mutans and C. albicans isolates was bacteriostatic and fungistatic.
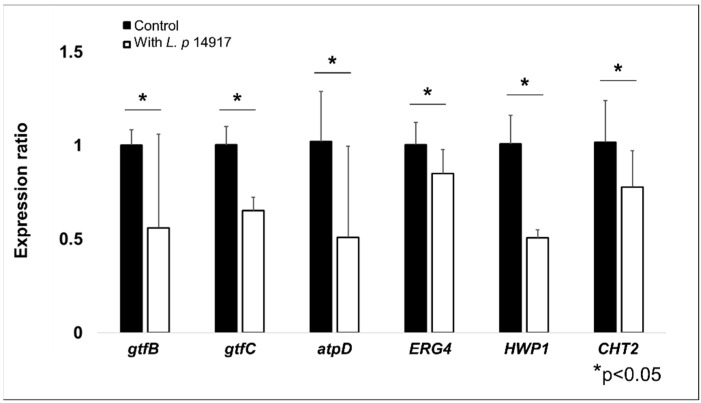
2.7. L. plantarum 14917 Downregulated C. albicans and S. mutans Virulence Genes in Biofilms
To assess the transcriptional regulations of S. mutans and C. albicans virulence genes in biofilms treated with L. plantarum 14917, qRT-PCR was performed for biofilms at 50 h, 2 h after the fresh culture medium was changed (and the sucrose concentration was changed from 0.1% to 1%). S. mutans genes related to EPS formation (gtfB and gtfC) and the atpD gene related to the assembled ATPase complex were significantly downregulated at 50 h. Meanwhile, genes related to C. albicans resistance fungal cell wall chitin remodeling (CHT2), resistance to antifungal medication (ERG4), and yeast hypha formation (HWP1) were also significantly downregulated following the culture medium change. The expressions of S. mutans (gtfB, gtfC, and atpD) and C. albicans (ERG4, HWP1, and CHT2) genes related to cariogenicity were downregulated by approximately 50–80% in the biofilms treated with L. plantarum 14917 for 50 h, compared to the control group (see Figure 5).
Figure 5.
Regulation of S. mutans and C. albicans virulence genes by L. plantarum 14917 in multispecies biofilms. The expression of S. mutans (gtfB, gtfC, and atpD) and C. albicans (ERG4, HWP1, and CHT2) genes related to carcinogenicity were reduced in the biofilms treated with L. plantarum 14917 for 50 h, compared to the control group (S. mutans and C. albicans). * p < 0.05 when comparing the control and L. plantarum 14917-treated biofilms.
3. Discussion
Dysbiosis of microflora at various body sites has reported associations with disease conditions, including gastrointestinal diseases, dermatitis, and oral diseases, such as dental caries. Probiotics were initially introduced to the healthcare field to maintain or improve a healthy gut by modulating or restoring gut flora [22,23,24,25]. The usage of probiotics later expanded to many other medical fields [26], such as treating and preventing vaginal and urinary tract infections in women, controlling the development of food allergies or eczema in children, and stimulating and strengthening the human immune system [27].
Several studies have reported the inhibitory effect of probiotics on S. mutans and C. albicans in planktonic or single-species biofilm conditions [28,29,30,31]. The most commonly used probiotics include Lactobacilli and Bifidobacterium, both of which produce lactic acid and other bioactive substances, including hydrogen peroxide, carbon peroxide, diacetyl, low-molecular-weight antimicrobial substances, bacteriocins, and adhesion inhibitors that could potentially affect the growth of oral microorganisms [29]. For example, Lactobacillus rhamnosus GG, Lactobacillus reuteri, Lactobacillus casei, Lactobacillus plantarum, and Lactobacillus salivarius inhibit the growth of S. mutans in vitro and in vivo [32,33,34]. Moreover, L. reuteri, L. rhamnosus, L. casei, L. paracasei, L. fermentum, and L. acidophilus inhibit the growth of C. albicans in vitro and in vivo [35,36,37]. Since the co-existence of S. mutans and C. albicans in the oral cavity leads to more pathogenic microbial eco-community and potentially elevates the caries risk of individuals, an ideal probiotic regimen is one that controls S. mutans and C. albicans simultaneously. Limited studies, however, have assessed the effect of probiotics on S. mutans and C. albicans in a multispecies setting [14,19,20].
3.1. Susceptibility of Clinically Isolated S. mutans and C. albicans to Common Antimicrobial Agents
Studies from various researchers have shown that clinically isolated bacteria and yeast have a higher tolerance to antimicrobial reagents, such as antibiotics and antifungal medications; this developed resistance poses a challenge in bacteria and yeast control in clinical settings and requires a better antimicrobial regimen. Similar phenomena have been seen for S. mutans; for example, the study reported by AL-Shami et al. [38] showed significant levels of penicillin, erythromycin, amoxicillin, clindamycin, and lincomycin resistance in S. mutans clinical isolates in dental patients. Similarly, Pasquantonio et al. [39] reported that 14% of 50 clinically isolated S. mutans had resistance to penicillin, although quinolones and rifampicin were confirmed to have an excellent activity against oral streptococci.
Clinically isolated C. albicans have also shown less susceptibility to antifungal medication. Jahanshiri et al. reported that the levels of resistance of 32 C. albicans isolates from head and neck cancer patients to fluconazole, itraconazole, ketoconazole, and amphotericin B were 62.5%, 81.25%, 93.75%, and 87.50%, respectively [40]. Aitken-Saavedra J et al. tested the susceptibility of C. albicans clinical strains of type II diabetic patients to fluconazole therapy and nystatin [41]. At higher pH, C. albicans yeast is less susceptible to nystatin, whereas at lower pH, C. albicans is more susceptible to treatment, suggesting that it might have greater effectiveness in patients with higher salivary acidification. Fluconazole had no significant results in their study. Biofilms are considered to provide a protective environment that enhances C. albicans’ resistance to antifungal medications. CHANDRA et al. reported that C. albicans cells that underwent 72 h of biofilm treatment were highly resistant against amphotericin B, fluconazole, nystatin, and chlorhexidine in an in vitro model, and this increased virulence is similar to what occurs in vivo [42]. Furthermore, topical antifungal agents such as fluconazole have negligible effects on C. albicans in mixed oral biofilms, due to the protective microbial exopolysaccharide matrix. However, antifungal susceptibility was enhanced by introducing povidone iodine, which inhibited exopolysaccharide matrix formation, resulting in a reduction in C. albicans [43].
Since clinically isolated bacteria and yeast were more resistant to antimicrobial reagents, it is possible that our experimental isolates were in the same situation. We want to emphasize that, while clinically isolated S. mutans and C. albicans may be more resistant to antimicrobial reagents, the tested probiotics significantly inhibited the growth of S. mutans and C. albicans isolates in both planktonic and biofilm conditions.
3.2. Equal Effectiveness of L. plantarum 14917 on Clinical Isoalted C. albicans and S. mutans Compared to Wild-Type Strains
Our previous work [21] assessed the inhibition of probiotic Lactobacillus spp. on the growth and biofilm formation of wild-type C. albicans and S. mutans. The results of the current study indicate that the inhibitory effect of L. plantarum on clinically isolated C. albicans and S. mutans is similar to its effect on the wild-type strains. Of note is that the inoculation concentration of S. mutans we used in the current study (106 CFU/mL) was even 1-log higher than that used in our previous wild-type study (105 CFU/mL), which demonstrated a higher challenge for L. plantarum during the inhibitory interactions. In terms of the change in the pH level in the planktonic and the culture medium of the biofilm model over time, no significant differences were noted between the current study, which used clinical isolates, and the previous study, which used C. albicans and S. mutans laboratory strains.
Regarding the effectiveness of growth inhibition in the planktonic model, the findings of this study are also similar to those of the study of Zeng et al., in which L. plantarum 14917 demonstrated significant inhibition of C. albicans by 1-log for at six hours, and 2-log at 20 h. S. mutans laboratory and clinical strains were significantly inhibited by 2-log at 6 h, and completely inhibited at 20 h for the clinical isolate in the planktonic model. Both studies showed comparable and significant inhibition of the cariogenic biofilm at 72 h (1% sucrose condition) in terms of the morphological structure of the biofilm, dry weight, biomass of bacteria and EPSs, bacteria co-localization by EPS, and the number, size, and surface area of microcolonies. The mechanistic assessment indicated that L. plantarum 14917 inhibited the expression of S. mutans (gtfB, gtfC, and atpD) and C. albicans (ERG4, HWP1, and CHT2) virulence genes and virulent structures, such as C. albicans hypha formation, which is consistent with our previous findings on wild-type C. albicans and S. mutans [21].
L. plantarum 14917 demonstrated equal effectiveness in inhibiting clinically isolated C. albicans and S. mutans from S-ECC children compared to wild-type strains, indicating L. plantarum 14917’s strong potential to be incorporated into a future clinical caries prevention and control regimen to target cariogenic pathogens.
Despite the promising finding that L. plantarum inhibited the growth of C. albicans and S. mutans clinical isolates and their biofilm formation, we recognize the study’s limitations. Although clinical isolates of C. albicans and S. mutans were used, the study was conducted using an in vitro model; further animal and clinical studies are required to verify their safety and efficacy in an oral biofilm complex, as well as to observe long-term oral microbial virulence.
4. Materials and Methods
4.1. Study Design
This study was designed in six steps to screen the best-performing probiotic Lactobacillus spp. on S. mutans and C. albicans clinical isolates. The study scheme is shown in Figure S1. In Step 1, the inhibitory effect of Lactobacilli was assessed in the planktonic condition against C. albicans and S. mutans isolates from two S-ECC children. The best-performing Lactobacillus advanced to Step 2 to verify its inhibitory effect against C. albicans and S. mutans isolates from an additional eight S-ECC children. In Step 3, two of the three Lactobacilli with a higher inhibition of S. mutans and C. albicans in the planktonic condition were further tested with C. albicans and S. mutans isolated from two S-ECC children in a multispecies biofilm condition. In Step 4, the better-performing Lactobacillus advanced to Step 5 to assess its effect on cariogenic biofilm structure. Moreover, molecular assays were used to assess the mechanistic interactions between Lactobacillus, S. mutans, and C. albicans in biofilms in Step 5. Lastly, the effect of plantaricin, an antimicrobial peptide produced by L. plantarum, on the growth of S. mutans and C. albicans was examined in Step 6.
4.2. Bacterial Strains and Starter Preparation
The microorganisms used in this study were L. plantarum ATCC 8014, L. plantarum ATCC 14917, L. salivarius ATCC 11741, the pairs of S. mutans and C. albicans isolated from ten preschool children with S-ECC. The clinical isolates were archived from a previous study [44]. The current study did not involve human subject interaction.
Lactobacillus, S. mutans, and C. albicans were recovered from frozen stock using MRS agar (BD Difco™, 288210), blood agar (TSA with Sheep Blood, Thermo Scientific™ R01202, Wilmington, DE, USA), and YPD agar (BD Difco™, 242720) for 48 h, respectively. Altogether, 3–5 colonies of each species were inoculated in 10 mL of broth for overnight incubation (5% CO2, 37 °C). Lactobacillus spp. were grown in MRS broth (BD Difco™, 288130); S. mutans was grown in TSBYE broth (3% Tryptic Soy, 0.5% Yeast Extract Broth, BD Bacto™ 286220 and Gibco™ 212750) with 1% (w/v) glucose; and C. albicans was grown in YPD broth (BD Difco™, 242820). On the following day, 0.5 mL of the overnight starters was added to individual glass tubes with fresh broth and incubated for 3–4 h to reach the mid-exponential phase with desirable optical density (OD). The morning starters were then ready for the preparation of planktonic and biofilm models in this study.
4.3. Planktonic Model
Interactions between S. mutans, C. albicans, and Lactobacillus species were first evaluated in planktonic conditions using the method established in our previous study [21]. Briefly, the inoculation quantity of S. mutans (105 CFU/mL) and C. albicans (103 CFU/mL) was chosen for simulating high-caries-risk conditions in the clinical setting; the inoculation quantity of Lactobacillus (108 CFU/mL) was used for inhibiting the growth of S. mutans and C. albicans. A pair of S. mutans and C. albicans obtained from the same child and one of the Lactobacilli were inoculated in 10 mL of TSBYE broth with 1% (w/v) glucose and incubated for 20 h (5% CO2, 37 °C). The growth of each microorganism was assessed using blood agar at 0, 6, and 20 h. Figure S5 indicates distinct morphological differences between S. mutans, C. albicans, and Lactobacillus spp. The pH values were measured at 0, 2, 4, 6, and 20 h.
4.4. Mixed-Species Biofilm Model
A mixed-species biofilm model was used to assess the effect of Lactobacilli on the biofilm formation of S. mutans and C. albicans. The biofilm method involved using saliva-coated hydroxyapatite discs (0.50″ diameter × 0.05″ thickness, Clarkson Chromatography Products, Inc., South Williamsport, PA) following the concept of “ecological plaque-biofilm” [45]. The discs were placed in a vertical position using a custom-made disc holder to mimic the caries-prone smooth tooth surfaces in the oral cavity [9,21,46,47,48].
The mixture of S. mutans, C. albicans, and Lactobacilli was inoculated in 2.8 mL of TSBYE broth with 0.1% (w/v) sucrose, and incubated at 37 °C and 5% CO2. The organisms were grown for the first 24 h to allow the initial biofilm to form. At 24 h of biofilm growth, the discs with biofilms were transferred to a fresh culture medium containing 1% (w/v) sucrose to induce cariogenic challenges. The culture medium was replaced once every 24 h until the end of the experimental period (at 72-h), and Lactobacilli (108 CFU/mL) were added to the fresh culture medium daily during the replacement for treatment groups. The pH of the culture medium was measured at selected time points using a standard pH electrode. The biofilms were processed for microbiological properties, dry weight, confocal imaging assays, and qRT-PCR at specific time points, according to the study design, to assess the regulation of Lactobacilli on the expression of C. albicans and S. mutans virulence genes.
4.5. Microbiological Analysis of the Mixed-Species Bacterial Population
The biofilms were homogenized by sonication, as detailed previously [21,48,49,50]. The homogenized suspension was used to determine the number of viable cells by plating on blood agar using an automated EddyJet Spiral Plater (IUL, SA, Barcelona, Spain). Three species were differentiated by colony morphology in conjunction with microscopic examination of cells from selected colonies [51]. The homogenized suspension was also used for the measurement of dry weight, as detailed previously [21,48,50]. Duplicated discs were used in each run. Independent assays were repeated three times.
4.6. Laser Scanning Confocal Fluorescence Microscopy (LCSFM) Imaging of Biofilm Matrix
We assessed two essential components of the biofilm matrix, bacteria and exopolysaccharides (EPSs), using LCSFM, as detailed previously [21,48,50]. Briefly, 1 μM Alexa Fluor® 647-labeled dextran conjugate (Molecular Probes, Invitrogen Corp., Carlsbad, CA, USA) was added to the culture medium at the beginning and during the development of the biofilms for exopolysaccharide visualization. The bacterial species and fungal species were labeled with SYTO® 9 green fluorescent nucleic acid stain (485/498 nm; Molecular Probes). The images were obtained using an Olympus FV 1000 two-photon laser scanning microscope (Olympus, Tokyo, Japan) equipped with a 10× (0.45 numerical aperture) water immersion objective lens. Each biofilm formed on the HA disc was scanned at 5 positions randomly [52]. Twenty image stacks were collected for each experimental condition.
4.7. Computational Analyses of the Confocal Biofilm Images
To visualize the morphology and 3D architecture of the biofilms, each structural component (EPSs and bacteria) was rendered in 3D using Amira 5.0.2 (Mercury Computer Systems Inc., Chelmsford, MS, USA) [52,53]. For biofilm quantitative analysis, COMSTAT and DUOSTAT (http://www.imageanalysis.dk, accessed on 1 January 2020.) were employed. We calculated the biomass, quantity, and dimensions (volume, diameter, and height) of microcolonies using the software COMSTAT, and we utilized DUOSTAT to determine the co-localization of two biofilm components (EPSs and bacterial cells, for example) across the 3D biofilm architecture [48].
4.8. Assessment of Total Protein in Multispecies Biofilms
The biofilms were homogenized via sonication, as detailed previously [21,48,49,50]. The 48 h old biofilms were harvested from the 1 HA disc in 1 mL of 0.89% w/w saline. A total of 150 µL of homogenized suspension was used for total protein assessment using a Pierce™ BCA Protein Assay Kit (Thermo Scientific™, Wilmington, DE, USA) with a microplate reader (Tecan Infinite® 200 PRO, Männedorf, Switzerland). Three independent experiments were conducted for each condition.
4.9. Inhibition of S. mutans and C. albicans by Plantaricin
Bacteriocins, antimicrobial molecules produced by L. plantarum, are also known as plantaricins [54]. Peptide plantaricin-149 (acetate) powers (Creative Peptides, Shirley, NY, USA) were dissolved in ddH2O to prepare plantaricin solutions. S. mutans (103 CFU/mL) and C. albicans (101 CFU/mL) from 3 S-ECC children were selected and treated with the plantaricin at a concentration of (0–400 µg/mL). The mixtures of plantaricin with S. mutans or C. albicans were grown for 24 h in TSBYE with 1% glucose in 96-well plates. Clear culture after 24 hours’ incubation indicated no growth of microorganisms. Therefore, the minimal inhibition concentration (MIC) of plantaricin-149 was defined as the lowest concentration that inhibited the growth of S. mutans and C. albicans.
4.10. Real-Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
The mass of biofilms was harvested from four discs at 50 h for each condition. The discs were immersed in RNALater (Applied Biosystems/Ambion, Austin, TX, USA) for 1 h, followed by biomass removal with a spatula. RNAs were extracted and purified with a MasterPure complete DNA and RNA purification kit (epicenter, Lucigen, WI, USA). Raw RNA product was quantified using a NanoDrop One Microvolume UV-Vis Spectrophotometer (Thermo Scientific™, Wilmington, DE, USA). A 2nd purification was performed with DNase I (Thermo Fisher Scientific, Invitrogen™ 18068015, Wilmington, DE, USA).
Then, cDNAs were synthesized using 0.4 μg of purified RNA and the BioRad iScript cDNA synthesis kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The resulting cDNAs and negative controls were amplified by quantitative amplification using Applied Biosystems™ PowerTrack™ SYBR Green Master Mix and a QuantStudio™ 3 Real-Time PCR System (Thermo Fisher Scientific, Wilmington, DE, USA). Each 20 µL reaction mixture included template cDNA, 10 µM of each primer, and a 2× SYBR-Green mix (containing SYBR-Green and Taq DNA Polymerase). Unique core genes of S. mutans and C. albicans were used as an internal reference for comparative expression calculation: gyrA for S. mutans genes [55] and ACT1 for C. albicans. Expression of virulence genes of S. mutans (gtfB, gtfC, and atpD) and C. albicans (CHT2, HWP1, and ERG4) was assessed using the methods detailed previously [21].
4.11. Assessment of Morphology of C. albicans
The 24 h old planktonic culture medium was observed under a light microscope (Olympus BX43, 214, Tokyo, Japan) with 100X oil objective (Olympus UPlanFL N 100X, Tokyo, Japan) to assess the morphological changes in C. albicans. A total of 20 µL of culture medium was placed on the glass slide and visualized without staining.
4.12. Statistical Analysis
To compare the abundance of S. mutans, C. albicans, and Lactobacillus spp. in planktonic and biofilms, the CFU values were first converted to natural log values. Zero values of the CFU remained at zero. The natural log values were compared between groups using the t-test for two groups or ANOVA for more than two groups, after assessing the normality of data. For other parameters, such as biomass (bacteria and EPS), the number and size of microcolonies, and the pH value of the biofilms at specific time points, normality tests were performed first. For normally distributed data, the comparisons between groups were tested using the t-test for two groups and one-way ANOVA for more than two groups followed by the post hoc test. For non-normally distributed data, the Kruskal–Wallis test was used to compare the outcomes of more than two groups, and Mann–Whitney tests were used for the comparison of two groups. Statistical tests were two-sided with a significant level of 5%. IBM SPSS was used for statistical analyses.
4.13. Ethical Approval
Ethical approval of the study and the written consent/permission forms were obtained from the Research Subject Review Board at the University of Rochester (RSRB00056870, date of approval: 17 May 2015).
5. Conclusions
Our study findings show a remarkable inhibitory effect of L. plantarum 14917 on S. mutans and C. albicans clinical isolates, resulting in a reduced biofilm structure with a significantly smaller microbial and extracellular matrix and less virulent microcolonies structure. The mechanistic assessment indicated that L. plantarum 14917 had a positive inhibitory impact on the expression of S. mutans and C. albicans virulence genes and virulent structures, such as C. albicans hypha formation. Future utilization of L. plantarum 14917 and/or its antimicrobial peptide plantaricin could lead to a new paradigm shift in dental caries prevention.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24032991/s1.
Author Contributions
Conceptualization, Y.Z., A.F. and J.X.; Data curation, Y.Z., A.F., Y.W. and S.Q.; Formal analysis, Y.Z., A.F., N.A., T.T.W. and S.Q.; Funding acquisition, J.X.; Investigation, Y.Z., A.F., N.A. and Y.W.; Methodology, Y.Z., N.A., T.T.W. and J.X.; Project administration, J.X.; Resources, J.X.; Supervision, J.X.; Validation, Y.Z., A.F. and T.T.W.; Visualization, Y.Z.; Writing—original draft, Y.Z. and A.F.; Writing—review and editing, Y.Z., A.F. and J.X. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Ethical approval of the study and the written consent/permission forms were obtained from the Research Subject Review Board at the University of Rochester (RSRB00056870, date of approval: 17 May 2015).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All data generated or analyzed during this study are included in this article. Further enquiries can be directed to the corresponding author.
Conflicts of Interest
All authors declare no conflict of interest.
Funding Statement
This research was funded by NIDCR K23DE027412 and R01DE031025. The funding agencies had no role in the study design, data collection, analyses, decision to publish, or preparation of the manuscript.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Heimer M., Teschler M., Schmitz B., Mooren F.C. Health Benefits of Probiotics in Sport and Exercise—Non-existent or a Matter of Heterogeneity? A Systematic Review. Front. Nutr. 2022;9:804046. doi: 10.3389/fnut.2022.804046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korada S.K., Yarla N.S., Mishra V., Daim M.A., Sharma B., Gm A., Reggi R., Palmery M., Peluso I., Kamal M.A. Single Probiotic versus Multiple Probiotics—A Debate On Current Scenario for Alleviating Health Benefits. Curr. Pharm. Des. 2018;24:4150–4153. doi: 10.2174/1381612824666181012124136. [DOI] [PubMed] [Google Scholar]
- 3.Kechagia M., Basoulis D., Konstantopoulou S., Dimitriadi D., Gyftopoulou K., Skarmoutsou N., Fakiri E.M. Health benefits of probiotics: A review. ISRN Nutr. 2013;2013:481651. doi: 10.5402/2013/481651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwendicke F., Dorfer C., Kneist S., Meyer-Lueckel H., Paris S. Cariogenic effects of probiotic Lactobacillus rhamnosus GG in a dental biofilm model. Caries Res. 2014;48:186–192. doi: 10.1159/000355907. [DOI] [PubMed] [Google Scholar]
- 5.Haukioja A. Probiotics and oral health. Eur. J. Dent. 2010;4:348–355. doi: 10.1055/s-0039-1697851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rastogi P., Saini H., Dixit J., Singhal R. Probiotics and oral health. Natl. J. Maxillofac. Surg. 2011;2:6–9. doi: 10.4103/0975-5950.85845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Twetman L., Larsen U., Fiehn N.-E., Stecksén-Blicks C., Twetman S. Coaggregation between probiotic bacteria and caries-associated strains: An in vitro study. Acta Odontol. Scand. 2009;67:284–288. doi: 10.1080/00016350902984237. [DOI] [PubMed] [Google Scholar]
- 8.Nishikawara F., Nomura Y., Imai S., Senda A., Hanada N. Evaluation of cariogenic bacteria. Eur. J. Dent. 2007;1:31–39. doi: 10.1055/s-0039-1698309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng Y., Nikitkova A., Abdelsalam H., Li J., Xiao J. Activity of quercetin and kaemferol against Streptococcus mutans biofilm. Arch. Oral Biol. 2019;98:9–16. doi: 10.1016/j.archoralbio.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Carvalho F.G., Silva D.S., Hebling J., Spolidorio L.C., Spolidorio D.M. Presence of mutans streptococci and Candida spp. in dental plaque/dentine of carious teeth and early childhood caries. Arch. Oral Biol. 2006;51:1024–1028. doi: 10.1016/j.archoralbio.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Signoretto C., Burlacchini G., Faccioni F., Zanderigo M., Bozzola N., Canepari P. Support for the role of Candida spp. in extensive caries lesions of children. New Microbiol. 2009;32:101–107. [PubMed] [Google Scholar]
- 12.Raja M., Hannan A., Ali K. Association of oral candidal carriage with dental caries in children. Caries Res. 2010;44:272–276. doi: 10.1159/000314675. [DOI] [PubMed] [Google Scholar]
- 13.Yang X.Q., Zhang Q., Lu L.Y., Yang R., Liu Y., Zou J. Genotypic distribution of Candida albicans in dental biofilm of Chinese children associated with severe early childhood caries. Arch. Oral Biol. 2012;57:1048–1053. doi: 10.1016/j.archoralbio.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Hasslof P., Hedberg M., Twetman S., Stecksen-Blicks C. Growth inhibition of oral mutans streptococci and candida by commercial probiotic lactobacilli—An in vitro study. BMC Oral Health. 2010;10:18. doi: 10.1186/1472-6831-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meurman J.H., Stamatova I. Probiotics: Contributions to oral health. Oral Dis. 2007;13:443–451. doi: 10.1111/j.1601-0825.2007.01386.x. [DOI] [PubMed] [Google Scholar]
- 16.Comelli E.M., Guggenheim B., Stingele F., Neeser J.R. Selection of dairy bacterial strains as probiotics for oral health. Eur. J. Oral Sci. 2002;110:218–224. doi: 10.1034/j.1600-0447.2002.21216.x. [DOI] [PubMed] [Google Scholar]
- 17.Ahola A.J., Yli-Knuuttila H., Suomalainen T., Poussa T., Ahlstrom A., Meurman J.H., Korpela R. Short-term consumption of probiotic-containing cheese and its effect on dental caries risk factors. Arch. Oral Biol. 2002;47:799–804. doi: 10.1016/S0003-9969(02)00112-7. [DOI] [PubMed] [Google Scholar]
- 18.Hatakka K., Ahola A.J., Yli-Knuuttila H., Richardson M., Poussa T., Meurman J.H., Korpela R. Probiotics reduce the prevalence of oral candida in the elderly—A randomized controlled trial. J. Dent. Res. 2007;86:125–130. doi: 10.1177/154405910708600204. [DOI] [PubMed] [Google Scholar]
- 19.Krzysciak W., Koscielniak D., Papiez M., Vyhouskaya P., Zagorska-Swiezy K., Kolodziej I., Bystrowska B., Jurczak A. Effect of a Lactobacillus salivarius Probiotic on a Double-Species Streptococcus mutans and Candida albicans Caries Biofilm. Nutrients. 2017;9:1242. doi: 10.3390/nu9111242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Q.R., Stamatova I., Kainulainen V., Korpela R., Meurman J.H. Interactions between Lactobacillus rhamnosus GG and oral micro-organisms in an in vitro biofilm model. BMC Microbiol. 2016;16:149. doi: 10.1186/s12866-016-0759-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng Y., Fadaak A., Alomeir N., Wu T.T., Rustchenko E., Qing S., Bao J., Gilbert C., Xiao J. Lactobacillus plantarum Disrupts S. mutans–C. albicans Cross-Kingdom Biofilms. Front. Cell Infect. Microbiol. 2022;12:872012. doi: 10.3389/fcimb.2022.872012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arvola T., Laiho K., Torkkeli S., Mykkanen H., Salminen S., Maunula L., Isolauri E. Prophylactic Lactobacillus GG reduces antibiotic-associated diarrhea in children with respiratory infections: A randomized study. Pediatrics. 1999;104:e64. doi: 10.1542/peds.104.5.e64. [DOI] [PubMed] [Google Scholar]
- 23.Kelesidis T., Pothoulakis C. Efficacy and safety of the probiotic Saccharomyces boulardii for the prevention and therapy of gastrointestinal disorders. Ther. Adv. Gastroenterol. 2012;5:111–125. doi: 10.1177/1756283X11428502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D’Haens G.R., Jobin C. Fecal Microbial Transplantation for Diseases Beyond Recurrent Clostridium Difficile Infection. Gastroenterology. 2019;157:624–636. doi: 10.1053/j.gastro.2019.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iannitti T., Palmieri B. Therapeutical use of probiotic formulations in clinical practice. Clin. Nutr. 2010;29:701–725. doi: 10.1016/j.clnu.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Gonzalez N., Battista N., Prete R., Corsetti A. Health-Promoting Role of Lactiplantibacillus plantarum Isolated from Fermented Foods. Microorganisms. 2021;9:349. doi: 10.3390/microorganisms9020349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sales-Campos H., Soares S.C., Oliveira C.J.F. An introduction of the role of probiotics in human infections and autoimmune diseases. Crit. Rev. Microbiol. 2019;45:413–432. doi: 10.1080/1040841X.2019.1621261. [DOI] [PubMed] [Google Scholar]
- 28.Caglar E., Kargul B., Tanboga I. Bacteriotherapy and probiotics’ role on oral health. Oral Dis. 2005;11:131–137. doi: 10.1111/j.1601-0825.2005.01109.x. [DOI] [PubMed] [Google Scholar]
- 29.Meurman J.H. Probiotics: Do they have a role in oral medicine and dentistry? Eur. J. Oral Sci. 2005;113:188–196. doi: 10.1111/j.1600-0722.2005.00191.x. [DOI] [PubMed] [Google Scholar]
- 30.Cagetti M.G., Mastroberardino S., Milia E., Cocco F., Lingstrom P., Campus G. The Use of Probiotic Strains in Caries Prevention: A Systematic Review. Nutrients. 2013;5:2530–2550. doi: 10.3390/nu5072530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin T.H., Lin C.H., Pan T.M. The implication of probiotics in the prevention of dental caries. Appl. Microbiol. Biotechnol. 2018;102:577–586. doi: 10.1007/s00253-017-8664-z. [DOI] [PubMed] [Google Scholar]
- 32.Nase L., Hatakka K., Savilahti E., Saxelin M., Ponka A., Poussa T., Korpela R., Meurman J.H. Effect of long-term consumption of a probiotic bacterium, Lactobacillus rhamnosus GG, in milk on dental caries and caries risk in children. Caries Res. 2001;35:412–420. doi: 10.1159/000047484. [DOI] [PubMed] [Google Scholar]
- 33.Laleman I., Detailleur V., Slot D.E., Slomka V., Quirynen M., Teughels W. Probiotics reduce mutans streptococci counts in humans: A systematic review and meta-analysis. Clin. Oral Investig. 2014;18:1539–1552. doi: 10.1007/s00784-014-1228-z. [DOI] [PubMed] [Google Scholar]
- 34.Wasfi R., Abd El-Rahman O.A., Zafer M.M., Ashour H.M. Probiotic Lactobacillus sp. inhibit growth, biofilm formation and gene expression of caries-inducing Streptococcus mutans. J. Cell Mol. Med. 2018;22:1972–1983. doi: 10.1111/jcmm.13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraft-Bodi E., Jorgensen M.R., Keller M.K., Kragelund C., Twetman S. Effect of Probiotic Bacteria on Oral Candida in Frail Elderly. J. Dent. Res. 2015;94:181S–186S. doi: 10.1177/0022034515595950. [DOI] [PubMed] [Google Scholar]
- 36.Matsubara V.H., Wang Y., Bandara H., Mayer M.P.A., Samaranayake L.P. Probiotic lactobacilli inhibit early stages of Candida albicans biofilm development by reducing their growth, cell adhesion, and filamentation. Appl. Microbiol. Biotechnol. 2016;100:6415–6426. doi: 10.1007/s00253-016-7527-3. [DOI] [PubMed] [Google Scholar]
- 37.Rossoni R.D., de Barros P.P., de Alvarenga J.A., Ribeiro F.C., Velloso M.D.S., Fuchs B.B., Mylonakis E., Jorge A.O.C., Junqueira J.C. Antifungal activity of clinical Lactobacillus strains against Candida albicans biofilms: Identification of potential probiotic candidates to prevent oral candidiasis. Biofouling. 2018;34:212–225. doi: 10.1080/08927014.2018.1425402. [DOI] [PubMed] [Google Scholar]
- 38.Al-Shami I.Z., Al-Hamzi M.A., Al-Shamahy H.A., Majeed A.L.A.A. Efficacy of some Antibiotics against Streptococcus Mutans Associated with Tooth decay in Children and their Mothers. Online J. Dent. Oral Health. 2019;2 [Google Scholar]
- 39.Pasquantonio G., Condò S., Cerroni L., Bikiqu L., Nicoletti M., Prenna M., Ripa S. Antibacterial activity of various antibiotics against oral streptococci isolated in the oral cavity. Int. J. Immunopathol. Pharmacol. 2012;25:805–809. doi: 10.1177/039463201202500331. [DOI] [PubMed] [Google Scholar]
- 40.Jahanshiri Z., Manifar S., Moosa H., Asghari-Paskiabi F., Mahmoodzadeh H., Shams-Ghahfarokhi M., Razzaghi-Abyaneh M. Oropharyngeal candidiasis in head and neck cancer patients in Iran: Species identification, antifungal susceptibility and pathogenic characterization. J. Mycol. Med. 2018;28:361–366. doi: 10.1016/j.mycmed.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 41.Aitken-Saavedra J., Lund R.G., González J., Huenchunao R., Perez-Vallespir I., Morales-Bozo I., Urzúa B., Tarquinio S.C., Maturana-Ramírez A., Martos J. Diversity, frequency and antifungal resistance of Candida species in patients with type 2 diabetes mellitus. Acta Odontol. Scand. 2018;76:580–586. doi: 10.1080/00016357.2018.1484154. [DOI] [PubMed] [Google Scholar]
- 42.Chandra J., Kuhn D.M., Mukherjee P.K., Hoyer L.L., McCormick T., Ghannoum M.A. Biofilm formation by the fungal pathogen Candida albicans: Development, architecture, and drug resistance. J. Bacteriol. 2001;183:5385–5394. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D., Liu Y., Benhamou R.I., Sanchez H., Simon-Soro A., Li Y., Hwang G., Fridman M., Andes D.R., Koo H. Bacterial-derived exopolysaccharides enhance antifungal drug tolerance in a cross-kingdom oral biofilm. ISME J. 2018;12:1427–1442. doi: 10.1038/s41396-018-0113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao J., Moon Y., Li L., Rustchenko E., Wakabayashi H., Zhao X., Feng C., Gill S.R., McLaren S., Malmstrom H., et al. Candida albicans Carriage in Children with Severe Early Childhood Caries (S-ECC) and Maternal Relatedness. PLoS ONE. 2016;11:e0164242. doi: 10.1371/journal.pone.0164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marsh P.D. Are dental diseases examples of ecological catastrophes? Microbiology. 2003;149:279–294. doi: 10.1099/mic.0.26082-0. [DOI] [PubMed] [Google Scholar]
- 46.Koo H., Xiao J., Klein M.I., Jeon J.G. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J. Bacteriol. 2010;192:3024–3032. doi: 10.1128/JB.01649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao J., Hara A.T., Kim D., Zero D.T., Koo H., Hwang G. Biofilm three-dimensional architecture influences in situ pH distribution pattern on the human enamel surface. Int. J. Oral Sci. 2017;9:74–79. doi: 10.1038/ijos.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao J., Klein M.I., Falsetta M.L., Lu B., Delahunty C.M., Yates J.R., 3rd, Heydorn A., Koo H. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 2012;8:e1002623. doi: 10.1371/journal.ppat.1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao J., Zeng Y., Rustchenko E., Huang X., Wu T.T., Falsetta M.L. Dual transcriptome of Streptococcus mutans and Candida albicans interplay in biofilms. J. Oral Microbiol. 2023;15:2144047. doi: 10.1080/20002297.2022.2144047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alomeir N., Zeng Y., Fadaak A., Wu T.T., Malmstrom H., Xiao J. Effect of Nystatin on Candida albicans–Streptococcus mutans duo-species biofilms. Arch. Oral Biol. 2023;145:105582. doi: 10.1016/j.archoralbio.2022.105582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guggenheim B., Giertsen E., Schupbach P., Shapiro S. Validation of an in vitro biofilm model of supragingival plaque. J. Dent. Res. 2001;80:363–370. doi: 10.1177/00220345010800011201. [DOI] [PubMed] [Google Scholar]
- 52.Xiao J., Koo H. Structural organization and dynamics of exopolysaccharide matrix and microcolonies formation by Streptococcus mutans in biofilms. J. Appl. Microbiol. 2010;108:2103–2113. doi: 10.1111/j.1365-2672.2009.04616.x. [DOI] [PubMed] [Google Scholar]
- 53.Klein M.I., Xiao J., Heydorn A., Koo H. An analytical tool-box for comprehensive biochemical, structural and transcriptome evaluation of oral biofilms mediated by mutans streptococci. J. Vis. Exp. 2011:2512. doi: 10.3791/2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sabo S.D., Vitolo M., Gonzalez J.M.D., Oliveira R.P.D. Overview of Lactobacillus plantarum as a promising bacteriocin producer among lactic acid bacteria. Food Res. Int. 2014;64:527–536. doi: 10.1016/j.foodres.2014.07.041. [DOI] [PubMed] [Google Scholar]
- 55.Zeng L., Burne R.A. Comprehensive mutational analysis of sucrose-metabolizing pathways in Streptococcus mutans reveals novel roles for the sucrose phosphotransferase system permease. J. Bacteriol. 2013;195:833–843. doi: 10.1128/JB.02042-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article. Further enquiries can be directed to the corresponding author.